Abstract
The induction of neutralizing antibodies is a promising way to prevent retrovirus infections. Neutralizing antibodies are mainly directed against the envelope proteins, which consist of two molecules, the surface envelope (SU) protein and the transmembrane envelope (TM) protein. Antibodies broadly neutralizing the human immunodeficiencvy virus-1 (HIV-1) and binding to the TM protein gp41 of the virus have been isolated from infected individuals. Their epitopes are located in the membrane proximal external region (MPER). Since there are difficulties to induce such neutralizing antibodies as basis for an effective AIDS vaccine, we performed a comparative analysis immunising with the TM proteins of different viruses from the family Retroviridae. Both subfamilies, the Orthoretrovirinae and the Spumaretrovirinae were included. In this study, the TM proteins of three gammaretroviruses including (1) the porcine endogenous retrovirus (PERV), (2) the Koala retrovirus (KoRV), (3) the feline leukemia virus (FeLV), of two lentiviruses, HIV-1, HIV-2, and of two spumaviruses, the feline foamy virus (FFV) and the primate foamy virus (PFV) were used for immunisation. Whereas in all immunisation studies binding antibodies were induced, neutralizing antibodies were only found in the case of the gammaretroviruses. The induced antibodies were directed against the MPER and the fusion peptide proximal region (FPPR) of their TM proteins; however only the antibodies against the MPER were neutralizing. Most importantly, the epitopes in the MPER were localized in the same position as the epitopes of the antibodies broadly neutralizing HIV-1 in the TM protein gp41 of HIV-1, indicating that the MPER is an effective target for the neutralization of retroviruses.
Introduction
In most cases retroviruses induce tumors and/or immunodeficiencies in the infected host. Exceptional retroviruses are non-pathogenic, for example the foamy viruses. To fight the AIDS pandemic with the high number of infected individuals worldwide, an effective vaccine is required. Vaccines may be also useful in the case of other retrovirus infection, including infection of animals. The existence of different vaccines protecting from leukemia caused by the feline leukemia virus (FeLV)Citation1-Citation5 is an example giving hope for vaccines against other retroviral infections including HIV-1. FeLV causes leukemia, lymphoma and opportunistic infections on the basis of a severe immunodeficiency induced by the virus. Unfortunately all FeLV based vaccines do not induce sterilizing immunity, but prevent from antigenemia and prevent the outbreak of the disease.Citation4,Citation6 Immunisation with a combination of the surface envelope (SU) protein and the transmembrane envelope ™ protein of FeLV increased the titer of neutralizing antibodies, but also did not induce sterilizing immunity.Citation6,Citation7 There is also need for a vaccine against the Koala retrovirus. This virus induces fatal lymphomas and chlamydia infections on the background of a severe immunodeficiency in the infected animals.Citation8 Immunisation of rats with the TM protein of the KoRV resulted in neutralizing antibodies,Citation9 however higher titers of neutralizing antibodies were induced when the SU protein was used for immunisation (unpublished data). Both proteins may be used in combination as a vaccine to prevent the further spread of the KoRV infection in koalas in Australia and in international zoos.
Differences in the TM Proteins of Different Retroviruses
The family Retroviridae consists of two subfamilies, the Orthoretrovirinae and the Spumaretrovirinae. Orthoretroviruses are divided into different genera such as alpharetroviruses, betaretroviruses, gammaretroviruses, deltaretroviruses, epsilonretroviruses and lentiviruses (). The general structure of the retroviral envelope proteins is very similar, however there are considerable differences in the size, in the pattern of glycosylation and number of cysteine-cysteine loops ( and ). Also the mechanism of infection is similar for all retroviruses.Citation10 The SU protein, which is gp120 in the case of HIV-1 and gp70 in the case of gammaretroviruses, interacts with the receptor molecule(s) on the cell surface. The CD4 molecule and the chemokine receptors CXCR5 or CCR4 are the receptors for gp120 of HIV-1; different transporter molecules including amino acid transporters are the receptors for gp70 of gammaretroviruses.Citation11,Citation12 After interaction of the SU proteins with their receptors, the fusion peptide at the N-terminus of the TM protein intercalates into the cellular membrane, and conformational changes in the TM protein, including an interaction of the C-helical and N-helical regions of the TM proteins, bring the membrane proximal external region (MPER) close to the fusion peptide proximal region (FPPR) and the viral membrane close to the cell membrane. This promotes fusion between viral and cellular membranes and internalisation of the virus into the target cell.Citation10,Citation12
Figure 1. Unrooted Pol sequence neighbor joining (NJ) dendrogram of the retroviral genera: α-, β-, gamma-, delta-, epsilon-, lenti- and spuma retroviruses.Citation76 The viruses used in this comparative study are indicated.
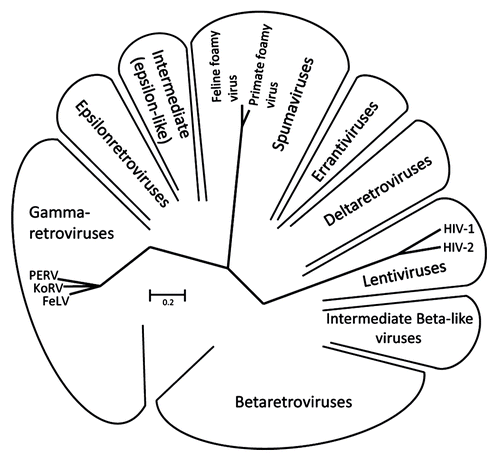
Figure 2. Main functional domains of the retroviral TM proteins (NHR - N-terminal helical region, C - C - Cys-Cys-loop, CHR - C-terminal helical region). The TM proteins of PERV, FeLV, HIV and FFV are presented. C indicates a cysteine, a star indicates a potential N-linked glycosylation site.
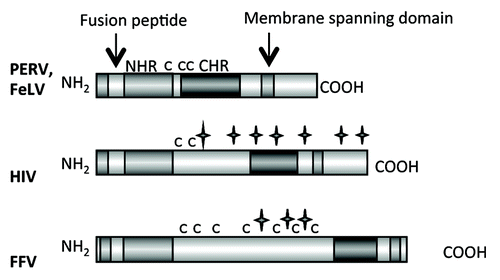
Table 1. The transmembrane envelope proteins of different retroviruses
Special Expression and Purification Methods for Retroviral TM Proteins
To induce neutralizing antibodies against the TM proteins, different steps are required, starting with a clone of the sequence of the protein (). Due to the membrane spanning domain and the hydrophobic fusion peptide the TM proteins are expressed at a low level and they are poorly soluble. Therefore special methods are required to express these proteins.Citation13 In most cases the ectodomain without the fusion peptide and without the membrane spanning domain were used for immunisation. Despite this, an optimization of the expression of retroviral TM proteins is beneficial in order to facilitate the subsequent purification steps. Optimization was performed by testing different fusion partners, the bacterial host strain, the medium, as well as the expression conditions such as temperature, inducer concentration and time point of induction. To speed up the optimization of the expression, a powerful procedure was developed based on simultaneous screening in 96 deep well plates.Citation13 In one plate four bacterial strains, three different growth media, three IPTG concentrations and two transformed bacterial clones as well as the empty vector as control could be analyzed and compared. Using such effective methods, the TM proteins of the FFV and PFV were expressed and purified.Citation13 To increase protein solubility during the purification process, various detergents were analyzed and the best condition were selected for large scale production and purification.
Summary of the Immunisation Experiments with the TM Proteins of Gammaretroviruses
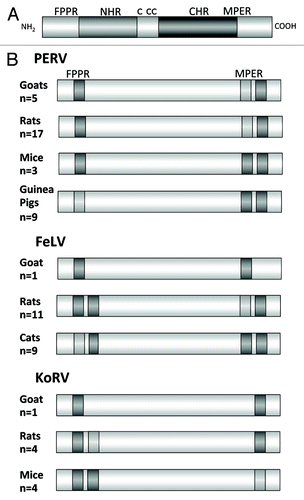
The cloning, expression and purification of the TM proteins of PERV,Citation14-Citation16 FeLVCitation6,Citation7,Citation17,Citation18 and the KoRVCitation9 have been described in great detail and recently summarized.Citation19 Immunising with the ectodomains of these gammaretroviruses, neutralizing antibodies against the corresponding virus were induced and epitopes were found in two regions of the TM antigen, in the MPER and FPPR (). Interestingly, the localization of the epitopes in the MPER was similar to the localization of the epitopes of antibodies broadly neutralizing HIV-1 isolated from infected patients such as 2F5, 10E8, Z13e1 and 4E10 in the TM protein gp41 if HIV-1.Citation20-Citation22 Despite the evolutionary difference between the lentivirus HIV-1 and the gammaretrovirus PERV, a limited sequence homology of the epitopes was observed. The epitope FEGWFN in p15E corresponds in localization and sequence to the epitope NWFN(D)IT in gp41 of HIV-1 recognized by the broadly neutralizing antibody 4E10 (identical amino acids in bold). Despite the sequence homology, the monoclonal antibody 4E10, broadly neutralizing HIV-1, and antibodies neutralizing PERV and binding to the MPER of p15E of PERV do not cross-react and do not cross-neutralize (unpublished).
Figure 4. (A) Schematic presentation of the ectodomain of p15E and (B) localization of the epitopes recognized by sera induced by immunisation with the ectodomain of the TM protein of three gammaretroviruses, PERV, FeLV and KoRV in different species. FPPR — fusion peptide proximal region, MPER — membrane proximal external region. Epitopes strongly recognized by sera from all immunised animals are in dark gray, epitopes recognized less and only by sera from some immunised animals are in light gray. The number of sera analyzed by epitope mapping are indicated.
It is important to note that the p15E-specific sera were not only neutralizing in vitro. At least in the case of FeLV, in in vivo studies, immunisation with the ectodomain of p15E prevented antigenemia and disease development in 50% of the cats immunised with the ectodomain of p15E of FeLV.Citation6
Although antibodies against epitopes in the FPPR and MPER have been found when immunising with the TM protein p15E of PERV, only the antibodies directed against the MPER were able to neutralize the virus (unpublished). This was shown isolating antibodies against the FPPR and MPER using affinity chromatography with immobilized recombinant proteins or synthetic peptides corresponding to the MPER and FPPR.
Increased Immune Response After Immunisation with the TM Protein and the Surface Envelope Protein of Two Gammaretroviruses
Both the surface envelope (SU) protein and the TM protein are involved in infection and represent ideal targets for neutralizing antibodies. Recently numerous potent antibodies neutralizing HIV-1 were isolated from HIV-1 infected individuals, which reacted with the surface envelope protein.Citation23-Citation29 Since the SU protein is larger and forms the top of the viral knob, consisting of three TM and three SU proteins, higher titers and a stronger neutralizing antibody response may be expected in comparison to the TM protein, which is partially hidden in the viral knob and protected by the glucan sheet of the SU protein.
To examine this probability, immunisations were performed with the SU protein of FeLV,Citation7 PERVCitation15 and KoRV (unpublished). In all cases a stronger neutralizing activity was induced compared with the immunisation with the TM protein alone. Moreover, when immunising with a combination of the SU and TM protein the induced neutralizing activity was stronger when compared with the immunisation with each of the envelope proteins alone.Citation7,Citation30
Whether the strategy to immunise with a combination of the SU and TM proteins is useful in the case of HIV-1 remains unclear since the SU protein of HIV-1 is highly variable in contrast to the SU protein of the gammaretroviruses.
Immunisation with the TM Proteins of HIV-1 And HIV-2 Produced in Bacteria
In contrast to the situation with the gammaretroviruses, immunisation with the ectodomain of HIV-1 and HIV-2 produced in bacteria failed to induce neutralizing antibodies (ref. Citation31 and data unpublished). Although some epitopes in the FPPR and the MPER were recognized (), neutralizing activity was not detected. The same was true when a DNA immunisation using a vector allowing expression of the TM protein on the cell surface to imitate the localization near a lipid membrane was performed.Citation32 This is in accordance with numerous other reports using gp41 or parts of it for immunisation. No or only a very weak neutralizing activity was found.Citation33-Citation40
Figure 5. Location of the epitopes recognized by immune sera from rats immunised with the recombinant ectodomain of gp41 of HIV-1 and gp36 of HIV-2. Epitopes strongly recognized by sera from all immunised animals are in dark gray, epitopes recognized less and only by sera from some immunised animals are in light gray.

Immunisation with Hybrid Proteins
It has been shown that during infection an interaction between the MPER and the FPPR takes place.Citation41-Citation45 In addition we had shown that the binding of 2F5 is enhanced when in addition to the peptide containing its epitope a peptide corresponding to the FPPR is present, suggesting that the interaction between both peptides induces a conformation allowing a better binding of 2F5.Citation46 Whether this conformation is also required to induce broadly neutralizing antibodies remains unclear. Based on our positive results with the TM proteins of the gammaretroviruses inducing antibodies against the MPER and FPPR of p15E, based on our negative results with the TM proteins of HIV-1 (see below) and being aware that there is an interaction between the MPER and FPPR during retroviral infection, we developed the strategy of hybrid proteins (). In p15E of PERV the regions corresponding to the epitopes of the antibodies induced by immunisation with p15E were substituted by the FPPR and MPER domains of gp41 of HIV-1. The idea behind this was that an interaction of the helical regions NHR and CHR of p15E from PERV will bring the transplanted FPPR and MPER domains of gp41 from HIV-1 in close proximity and this may induce a conformation able to induce neutralizing antibodies such as 2F5 and 4E10. Different hybrids were designed, the first containing the fusion peptide (FP) and the membrane spanning domain (MSD) derived from gp41 of HIV-1, the second containing the FP and MSD from p15E of PERV, the third without any FP and MSD and the fourth with a changed localization of the FPPR domain (). All hybrids were recognized by the monoclonal antibodies 2F5 and 4E10 and hybrid 3 and 4 were producing trimers and multimers. Whereas the first two hybrid antigens induced only antibodies binding the FPPR of gp41, the third and fourth hybrid antigens induced antibodies binding exactly the epitope of 2F5 (). Therefore systematic changes in the surrounding and localization of the transplanted domains allowed induction of antibodies binding the 2F5 epitope. Despite this, all induced sera did not neutralize HIV-1, at least in the neutralization assay based on TZM-bl cells. Nevertheless these results suggest that further modifications of the hybrid antigens may induce neutralizing antibodies.
Figure 6. Generation of hybrid proteins consisting of a backbone derived from the TM protein p15E of PERV and the FPPR and MPER of gp41 of HIV-1, which replace the FPPR and MPER of p15E. The FPPR and MPER of p15E were removed and the FPPR and MPER (containing the epitopes of the broadly neutralizing antibodies 2F5 and 4E10) of gp41 of HIV-1 were inserted. NHR - N-terminal helical region, C - C or C - CC - Cys-Cys-loop, CHR - C-terminal helical region, MSD - membrane spanning domain.
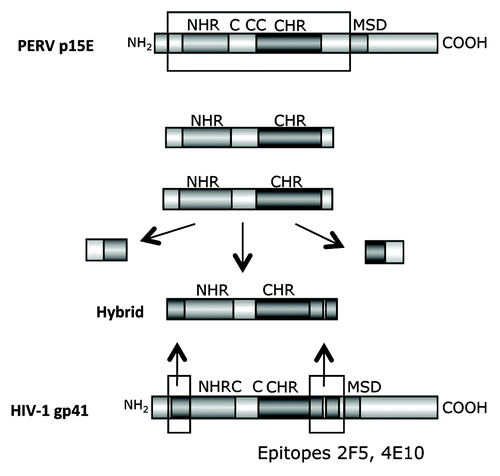
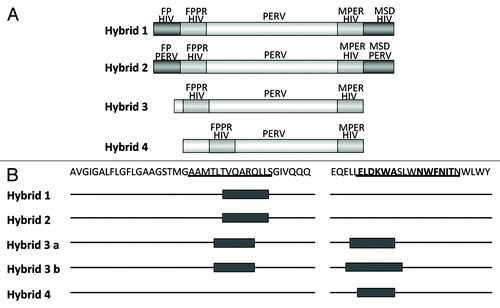
Figure 7. (A) Structure of the hybrid protein used for immunisation. The hybrids were generated as described in . For abbreviation see and . The origin of the fusion peptide (FP) and the membrane spanning domain (MSD), either from gp41 of HIV-1 or from p15E of PERV are indicated. Hybrid 3 and 4 lack the FPPR and MSD, in hybrid 4 the FPPR region was shifted. (B) Localization of the epitopes recognized by sera from animals immunised with the different hybrid proteins. The first part of the sequence corresponds to the FP/FPPR sequence, the second part to the MPER. The sequences of the epitopes of 2F5 and 4E10 are in bold, the sequences of the peptides shown to enhance the binding of 2F5 and 4E10 to their epitopes when interacting are underlined.
Immunisation with the TM Protein of HIV-1 Produced in Human Cells
In contrast to gp41 produced in bacteria (see above), gp41 produced in human cells is glycosylated,Citation47 with a molecular mass of 41kDa. Due to the use of the exportation signal of gp120, this protein was released from the producing cell. Only the fully glycosylated form was released; non-glycosylated and partially glycosylated proteins remained in the cells. The released proteins were able to trimerise and were recognized by the monoclonal antibodies 2F5 and 4E10. When immunising rats with two forms of the gp41, one containing a part of the cytoplasmatic tail, the other presenting only the ectodomain similar to the antigen used in the case of the gammaretroviruses, a strong antibody response was induced. The sera recognized several epitopes in gp41, among them the 2F5 epitope (unpublished). Studies on the neutralizing activity of these antibodies are in progress.
TM Protein Associated Immunosuppression
HIV-1 and many other retroviruses induce immunodeficiencies in the infected host. Meanwhile there is evidence that the TM proteins of the retroviruses contribute to this immunosuppression. The TM proteins of numerous retroviruses including gp41 of HIV-1 as well as synthetic peptides corresponding to a highly conserved domain in the retroviral TM proteins, the so called immunosuppressive domain, are able to inhibit mitogen-driven activation of immune cells and to modulate their cytokine release and gene expression (for review see refs. Citation48–Citation50). The TM protein gp41 of HIV-1 was shown to induce interleukin - 10 (IL-10) and Il-6 release and to increase the expression of genes such as matrix metallopeptidase 1 (MMP-1) and triggering receptor expressed on myeloid cells 1 (TREM1). Genes involved in innate immunity such as ficolin (FCN1), selenoprotein P, plasma, 1 (SEPP1) and chemokine (C-X-C motif) ligand 9 (CXCL9) were found downregulated.Citation47,Citation51 Single mutations in the immunosuppressive domain abrogated these immunosuppressive activities.Citation47 This result had also consequences for the vaccine development against retroviruses. When rats were immunised with the wild-type gp41 produced in human cells a good immune response was observed. However, when immunising with gp41 mutated in the immunosuppressive domain, a much higher antibody response were achieved.Citation47
Immunisation with the TM Protein of Foamy Viruses
Despite typically cytopathic in vitro, foamy viruses are considered apathogenic in their natural host and after interspecies transmission. For instance, chronic infections without any symptoms are induced when simian foamy viruses (SFV) are transmitted to humans by bush-meat preparation, biting etc.Citation52-Citation54 The zoonotic transmission of a chimpanzee foamy virus led to the primate (previously called human) FV isolate PFV.Citation52 It is important to note that the titers of foamy viruses in the natural host as well as in the new host appear to be considerably low, however the infection is not cleared even in the presence of a clear immune response.Citation55,Citation56 The apathogenicity and the very limited distribution of the virus in the human population is the basis for the development to use these viruses as vector for gene therapy and vaccination. In addition, secondary transmission of the PFV from humans to humans has not been detected so far.
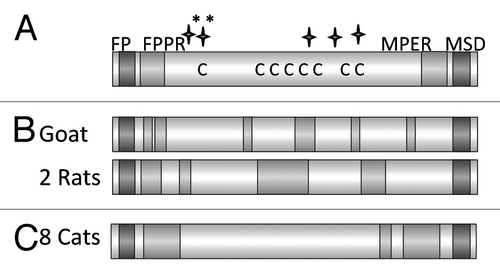
Using the 96 deep well screening strategy (see above) the TM proteins of FFV and PFV were produced in bacteriaCitation13 and characterized. As mentioned above, the TM protein of the foamy viruses contains numerous cysteines and several glycosylation sites in the sequence (). In addition to the immunisation studies the TM protein of the FFV was also used to establish a diagnostic tool based on the detection of TM protein-specific antibodies in infected animals.Citation57,Citation58 Furthermore, one goat and eight rats were immunised with these proteins and binding and neutralizing antibodies were analyzed. In all animals binding antibodies were induced recognizing epitopes located in the FPPR and in the central part of the molecule, but not in the MPER (). Correlating with the absence of antibodies directed against the MPER no neutralizing activity was observed in the sera. However, when sera from naturally FFV infected cats were analyzed, binding and neutralizing antibodies were found in the sera of all animals. It remains unclear, whether the neutralizing antibodies were directed against the TM protein or the SU protein, but interestingly antibodies targeting the MPER were detected (). The sera recognized mainly epitopes in the FPPR and MPER. No epitopes were not detected in the central part of the protein, possibly because in this region of the gylcosylated the viral TM the main glycosylation sites are located. Since the protein used for immunisation was produced in bacteria, and therefore was not glycosylated, antibodies against this domain of the protein were induced.
Figure 8. (A) Schematic presentation of the main domains of the TM protein of the feline foamy virus (FFV). Abbreviations see . C marks the cysteine residues, long stars the N glycosylation sites, long stars marked with * the potential N glycosylation sites. (B) Localization of the epitopes recognized by the immune sera induced by immunisation with the TM protein of FFV. Sera from one goat, and two rats which were immunised with the TM protein produced in bacteria were analyzed. (C) Localization of the epitopes recognized by the sera from 8 cats naturally infected by FFV.
Toward a Replication Competent Vaccine Virus Based on Foamy Viruses
Advantages of foamy virus vectors are in addition to the apathogenicity of the parental virusesCitation59 their high physical and genetic stability, their capacity for the uptake of larger foreign DNA sequences and their reduced dependence on replicating cells. Concerning pathogenicity, foamy virus infection of rabbits and mice however gave rise to a mild transient immunosuppressive effect, which regressed within 30 d.Citation60,Citation61 In transgenic mice expressing PFV, degeneration of the cerebellar granule cell layer was observed.Citation62,Citation63
Env proteins of the foamy viruses and virus budding are unique compared with other retroviruses: (1) particle budding strongly depends on co-expression of Env proteins (or compensatory changes in Gag)Citation64-Citation69 and (2) the about 125 aa long leader peptide of FV Env, designated Elp, is an important component of FFV and PFV particles.Citation68-Citation70 The C-terminus of Elp is exposed on the surface of virus particles and may be thus very much suited to display vaccine epitopes to the vaccines immune system.Citation69 Importantly, Env of PFV has been described to bud from cells,Citation65 a feature that could be used to create vaccine virus-like particles (VLPs) or a particulate protein vaccine.
In the past years, foamy virus-based vectors for gene therapy and vaccination have been successfully established and clearly demonstrated the potential of foamy viruses to deliver therapeutic genes or vaccine antigens in order to correct genetic defects or to induce (partially protective) immunity against heterologous pathogens.Citation71
The idea was to identify the epitopes in the TM protein of foamy viruses, to prepare hybrid proteins as in the case of the gammaretroviruses () and to create a replication-competent hybrid virus. Since already the first step of this strategy failed and no neutralizing antibodies against the TM protein of the foamy viruses were induced, a new approach was developed using the Bet protein as carrier for gp41-derived sequences corresponding to the MPER and the FPPR. This protein was already successfully grafted with a caliciviral antigen in a replication competent virus and an immune response against the calicivirus was induced.Citation69 Bet is an accessory protein, the corresponding bet gene is generated via splicing. Bet counteracts cellular APOBEC3-mediated restriction and may have a role in particle release and in establishing viral persistence.Citation72-Citation74 Using Bet-gp41 hybrids containing sequences corresponding to the MPER and/or FPPR of gp41 of HIV-1 for immunisation, antibodies against gp41 were induced.Citation75 These antibodies recognized epitopes in the FPPR and MPER of gp41, some overlapping with the epitope of 2F5, ELDKWAS, however no neutralizing activity was observed.
Outlook
The major advantage of neutralizing antibodies is their ability to prevent infection and subsequent integration of the provirus into the cellular genome where it may persist and if not expressed it remains undetectable by the immune system.
Retroviral transmembrane envelope (TM) proteins are a suitable target for the induction of neutralizing antibodies. Especially the membrane proximal external region (MPER), which is highly conserved, plays an important role during infection and antibodies binding to this region have been shown to neutralize HIV-1 and other retroviruses including PERV. The question how to induce such antibodies is still unsolved. Obviously only a distinct, still unknown conformation in the chain of the complex conformational changes during the infection process may represent the required target for neutralizing antibodies.
First immunisation studies with hybrid proteins based on the TM protein of PERV and transplanted MPER and FPPR of gp41 of HIV-1 failed, however by systematic changes in the localization of the grafted gp41 sequences a correct recognition of the 2F5 epitope was achieved. Further modifications of the hybrid proteins should be introduced, increasing the probability to induce neutralizing antibodies.
| Abbrevations: AIDS | = | acquired immunodeficiency syndrome |
| FeLV | = | feline leukemia virus |
| FFV | = | feline foamy virus |
| FPPR | = | fusion peptide proximal region |
| HAART | = | highly active antiretroviral therapy |
| HIV | = | human immunodeficiency virus |
| MPER | = | membrane proximal external region |
| PBMCs | = | peripheral blood mononuclear cells |
| PERV | = | porcine endogenous retroviruses |
| PFV | = | primate foamy virus |
| SU protein | = | surface envelope protein |
| TM protein | = | transmembrane envelope protein |
Acknowledgments
I would like to thank Dr. V. Morozov and M. Mühle for critical reading of the manuscript. Part of the studies was performed in the frame of the EuroNeut-41 project in the Seventh Framework Program (7–1517). Other parts were supported by the Volkswagen Foundation and the Berliner Saving Bank Foundation Medicine. This paper was presented at the Eight World Congress on Vaccines, Immunisation and Immunotherapy, Barcelona, Spain, 5–7 June 2010
Submitted
11/28/12
Accepted
12/04/12
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Jarrett O. Efficacy of recombinant feline leukemia virus vaccines. AIDS Res Hum Retroviruses 1996; 12:435 - 6; http://dx.doi.org/10.1089/aid.1996.12.435; PMID: 8882328
- Hoover EA, Mullins JI, Chu H-J, Wasmoen TL. Efficacy of an inactivated feline leukemia virus vaccine. AIDS Res Hum Retroviruses 1996; 12:379 - 83; http://dx.doi.org/10.1089/aid.1996.12.379; PMID: 8882314
- Hofmann-Lehmann R, Holznagel E, Aubert A, Ossent P, Reinacher M, Lutz H. Recombinant FeLV vaccine: long-term protection and effect on course and outcome of FIV infection. Vet Immunol Immunopathol 1995; 46:127 - 37; http://dx.doi.org/10.1016/0165-2427(94)07012-V; PMID: 7618252
- Hofmann-Lehmann R, Tandon R, Boretti FS, Meli ML, Willi B, Cattori V, et al. Reassessment of feline leukaemia virus (FeLV) vaccines with novel sensitive molecular assays. Vaccine 2006; 24:1087 - 94; http://dx.doi.org/10.1016/j.vaccine.2005.09.010; PMID: 16198454
- Hines DL, Cutting JA, Dietrich DL, Walsh JA. Evaluation of efficacy and safety of an inactivated virus vaccine against feline leukemia virus infection. J Am Vet Med Assoc 1991; 199:1428 - 30; PMID: 1666097
- Langhammer S, Hübner J, Jarrett O, Kurth R, Denner J. Immunization with the transmembrane protein of a retrovirus, feline leukemia virus: absence of antigenemia following challenge. Antiviral Res 2011; 89:119 - 23; http://dx.doi.org/10.1016/j.antiviral.2010.11.011; PMID: 21108970
- Langhammer S, Fiebig U, Kurth R, Denner J. Increased neutralizing antibody response after simultaneous immunization with leucogen and the feline leukemia virus transmembrane protein. Intervirology 2011; 54:78 - 86; http://dx.doi.org/10.1159/000318892; PMID: 20829603
- Hanger JJ, Bromham LD, McKee JJ, O’Brien TM, Robinson WF. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J Virol 2000; 74:4264 - 72; http://dx.doi.org/10.1128/JVI.74.9.4264-4272.2000; PMID: 10756041
- Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. Transspecies transmission of the endogenous koala retrovirus. J Virol 2006; 80:5651 - 4; http://dx.doi.org/10.1128/JVI.02597-05; PMID: 16699047
- Chan DC, Kim PS. HIV entry and its inhibition. Cell 1998; 93:681 - 4; http://dx.doi.org/10.1016/S0092-8674(00)81430-0; PMID: 9630213
- Tailor CS, Lavillette D, Marin M, Kabat D. Cell surface receptors for gammaretroviruses. Curr Top Microbiol Immunol 2003; 281:29 - 106; http://dx.doi.org/10.1007/978-3-642-19012-4_2; PMID: 12932075
- Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology 2008; 5:111; http://dx.doi.org/10.1186/1742-4690-5-111; PMID: 19077194
- Mühle M, Löchelt M, Denner J. Optimisation of expression and purification of the feline and primate foamy virus transmembrane envelope proteins using a 96 deep well screen. Protein Expr Purif 2012; 81:96 - 105; http://dx.doi.org/10.1016/j.pep.2011.09.006; PMID: 21964437
- Fiebig U, Stephan O, Kurth R, Denner J. Neutralizing antibodies against conserved domains of p15E of porcine endogenous retroviruses: basis for a vaccine for xenotransplantation?. Virology 2003; 307:406 - 13; http://dx.doi.org/10.1016/S0042-6822(02)00140-X; PMID: 12667808
- Kaulitz D, Fiebig U, Eschricht M, Wurzbacher C, Kurth R, Denner J. Generation of neutralising antibodies against porcine endogenous retroviruses (PERVs). Virology 2011; 411:78 - 86; http://dx.doi.org/10.1016/j.virol.2010.12.032; PMID: 21237477
- Denner J, Mihica D, Kaulitz D, Schmidt CM. Increased titers of neutralizing antibodies after immunization with both envelope proteins of the porcine endogenous retroviruses (PERVs). Virol J 2012; 9:260; http://dx.doi.org/10.1186/1743-422X-9-260; PMID: 23126255
- Langhammer S, Fiebig U, Kurth R, Denner J. Neutralising antibodies against the transmembrane protein of feline leukaemia virus (FeLV). Vaccine 2005; 23:3341 - 8; http://dx.doi.org/10.1016/j.vaccine.2005.01.091; PMID: 15837241
- Langhammer S, Hübner J, Kurth R, Denner J. Antibodies neutralizing feline leukaemia virus (FeLV) in cats immunized with the transmembrane envelope protein p15E. Immunology 2006; 117:229 - 37; http://dx.doi.org/10.1111/j.1365-2567.2005.02291.x; PMID: 16423059
- Denner J. Towards an AIDS vaccine: the transmembrane envelope protein as target for broadly neutralizing antibodies. Hum Vaccin 2011; 7:Suppl 4 - 9; http://dx.doi.org/10.4161/hv.7.0.14555; PMID: 21266839
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 2001; 75:10892 - 905; http://dx.doi.org/10.1128/JVI.75.22.10892-10905.2001; PMID: 11602729
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 1993; 67:6642 - 7; PMID: 7692082
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012; 491:406 - 12; http://dx.doi.org/10.1038/nature11544; PMID: 23151583
- Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol 2012; 86:4688 - 92; http://dx.doi.org/10.1128/JVI.07163-11; PMID: 22301150
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al, NISC Comparative Sequencing Program. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011; 333:1593 - 602; http://dx.doi.org/10.1126/science.1207532; PMID: 21835983
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011; 333:1633 - 7; http://dx.doi.org/10.1126/science.1207227; PMID: 21764753
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329:856 - 61; http://dx.doi.org/10.1126/science.1187659; PMID: 20616233
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al, Protocol G Principal Investigators. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011; 477:466 - 70; http://dx.doi.org/10.1038/nature10373; PMID: 21849977
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 2011; 85:9998 - 10009; http://dx.doi.org/10.1128/JVI.05045-11; PMID: 21795340
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al, Protocol G Principal Investigators. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009; 326:285 - 9; http://dx.doi.org/10.1126/science.1178746; PMID: 19729618
- Denner J, Mihica D, Kaulitz D, Schmidt CM. Increased titers of neutralizing antibodies after immunization with both envelope proteins of the porcine endogenous retroviruses (PERVs). Virol J 2012; 9:260; http://dx.doi.org/10.1186/1743-422X-9-260; PMID: 23126255
- Behrendt R, Fiebig U, Kurth R, Denner J. Induction of antibodies binding to the membrane proximal external region of gp36 of HIV-2. Intervirology 2012; 55:252 - 6; http://dx.doi.org/10.1159/000324483; PMID: 21454955
- Behrendt R, Fiebig U, Schmolke M, Kurth R, Denner J. Induction of Antibodies Specific for Gp41 of HIV-1 by Gene Gun DNA Vaccination. J Vaccines Vaccin 2012; http://dx.doi.org/10.4172/2157-7560.1000145
- McGaughey GB, Barbato G, Bianchi E, Freidinger RM, Garsky VM, Hurni WM, et al. Progress towards the development of a HIV-1 gp41-directed vaccine. Curr HIV Res 2004; 2:193 - 204; http://dx.doi.org/10.2174/1570162043484933; PMID: 15078183
- Hinz A, Schoehn G, Quendler H, Hulsik DL, Stiegler G, Katinger H, et al. Characterization of a trimeric MPER containing HIV-1 gp41 antigen. Virology 2009; 390:221 - 7; http://dx.doi.org/10.1016/j.virol.2009.05.015; PMID: 19539967
- Bianchi E, Joyce JG, Miller MD, Finnefrock AC, Liang X, Finotto M, et al. Vaccination with peptide mimetics of the gp41 prehairpin fusion intermediate yields neutralizing antisera against HIV-1 isolates. Proc Natl Acad Sci U S A 2010; 107:10655 - 60; http://dx.doi.org/10.1073/pnas.1004261107; PMID: 20483992
- Ho J, Uger RA, Zwick MB, Luscher MA, Barber BH, MacDonald KS. Conformational constraints imposed on a pan-neutralizing HIV-1 antibody epitope result in increased antigenicity but not neutralizing response. Vaccine 2005; 23:1559 - 73; http://dx.doi.org/10.1016/j.vaccine.2004.09.037; PMID: 15694508
- Joyce JG, Hurni WM, Bogusky MJ, Garsky VM, Liang X, Citron MP, et al. Enhancement of alpha -helicity in the HIV-1 inhibitory peptide DP178 leads to an increased affinity for human monoclonal antibody 2F5 but does not elicit neutralizing responses in vitro. Implications for vaccine design. [Erratum in: J Biol Chem 2003; 278:5492] J Biol Chem 2002; 277:45811 - 20; http://dx.doi.org/10.1074/jbc.M205862200; PMID: 12237296
- McGaughey GB, Citron M, Danzeisen RC, Freidinger RM, Garsky VM, Hurni WM, et al. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 2003; 42:3214 - 23; http://dx.doi.org/10.1021/bi026952u; PMID: 12641452
- Zwick MB. The membrane-proximal external region of HIV-1 gp41: a vaccine target worth exploring. AIDS 2005; 19:1725 - 37; http://dx.doi.org/10.1097/01.aids.0000189850.83322.41; PMID: 16227780
- Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 2008; 72:54 - 84; http://dx.doi.org/10.1128/MMBR.00020-07; PMID: 18322034
- Bellamy-McIntyre AK, Lay CS, Baär S, Maerz AL, Talbo GH, Drummer HE, et al. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J Biol Chem 2007; 282:23104 - 16; http://dx.doi.org/10.1074/jbc.M703485200; PMID: 17526486
- Noah E, Biron Z, Naider F, Arshava B, Anglister J. The membrane proximal external region of the HIV-1 envelope glycoprotein gp41 contributes to the stabilization of the six-helix bundle formed with a matching N’ peptide. Biochemistry 2008; 47:6782 - 92; http://dx.doi.org/10.1021/bi7023139; PMID: 18540633
- Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog 2010; 6:e1000880; http://dx.doi.org/10.1371/journal.ppat.1000880; PMID: 20463810
- Hager-Braun C, Katinger H, Tomer KB. The HIV-neutralizing monoclonal antibody 4E10 recognizes N-terminal sequences on the native antigen. J Immunol 2006; 176:7471 - 81; PMID: 16751393
- Lorizate M, Gómara MJ, de la Torre BG, Andreu D, Nieva JL. Membrane-transferring sequences of the HIV-1 Gp41 ectodomain assemble into an immunogenic complex. J Mol Biol 2006; 360:45 - 55; http://dx.doi.org/10.1016/j.jmb.2006.04.056; PMID: 16813835
- Fiebig U, Schmolke M, Eschricht M, Kurth R, Denner J. Mode of interaction between the HIV-1-neutralizing monoclonal antibody 2F5 and its epitope. AIDS 2009; 23:887 - 95; http://dx.doi.org/10.1097/QAD.0b013e3283292153; PMID: 19414989
- Morozov VA, Morozov AV, Semaan M, Denner J. Single mutations in the transmembrane envelope protein abrogate the immunosuppressive property of HIV-1. Retrovirology 2012; 9:67; http://dx.doi.org/10.1186/1742-4690-9-67; PMID: 22889273
- Oostendorp RAJ, Meijer CJLM, Scheper RJ. Immunosuppression by retroviral-envelope-related proteins, and their role in non-retroviral human disease. Crit Rev Oncol Hematol 1993; 14:189 - 206; http://dx.doi.org/10.1016/1040-8428(93)90009-S; PMID: 8397847
- Denner J. Immunosuppression by retroviruses: implications for xenotransplantation. Ann N Y Acad Sci 1998; 862:75 - 86; http://dx.doi.org/10.1111/j.1749-6632.1998.tb09119.x; PMID: 9928208
- Denner J. How does HIV induce AIDS? The virus protein hypothesis. J Hum Virol 2000; 3:81 - 2; PMID: 10850893
- Denner J, Eschricht M, Lauck M, Semaan M, Schlaermann P, Ryu H, et al. Modulation of cytokine release and gene expression by the immunosuppressive domain of gp41 of HIV-1. PLoS ONE 2012; In Press
- Heneine W, Schweizer M, Sandstrom P, Folks T. Human infection with foamy viruses. Curr Top Microbiol Immunol 2003; 277:181 - 96; http://dx.doi.org/10.1007/978-3-642-55701-9_8; PMID: 12908773
- Heneine W, Switzer WM, Sandstrom P, Brown J, Vedapuri S, Schable CA, et al. Identification of a human population infected with simian foamy viruses. Nat Med 1998; 4:403 - 7; http://dx.doi.org/10.1038/nm0498-403; PMID: 9546784
- Boneva RS, Switzer WM, Spira TJ, Bhullar VB, Shanmugam V, Cong ME, et al. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res Hum Retroviruses 2007; 23:1330 - 7; http://dx.doi.org/10.1089/aid.2007.0104; PMID: 18184074
- Cummins JE Jr., Boneva RS, Switzer WM, Christensen LL, Sandstrom P, Heneine W, et al. Mucosal and systemic antibody responses in humans infected with simian foamy virus. J Virol 2005; 79:13186 - 9; http://dx.doi.org/10.1128/JVI.79.20.13186-13189.2005; PMID: 16189020
- Alke A, Schwantes A, Zemba M, Flügel RM, Löchelt M. Characterization of the humoral immune response and virus replication in cats experimentally infected with feline foamy virus. Virology 2000; 275:170 - 6; http://dx.doi.org/10.1006/viro.2000.0537; PMID: 11017797
- Linial M. Why aren’t foamy viruses pathogenic?. Trends Microbiol 2000; 8:284 - 9; http://dx.doi.org/10.1016/S0966-842X(00)01763-7; PMID: 10838587
- Santillana-Hayat M, Rozain F, Bittoun P, Chopin-Robert C, Lasneret J, Périès J, et al. Transient immunosuppressive effect induced in rabbits and mice by the human spumaretrovirus prototype HFV (human foamy virus). Res Virol 1993; 144:389 - 96; http://dx.doi.org/10.1016/S0923-2516(06)80054-3; PMID: 8284516
- Hooks JJ, Detrick-Hooks B. Simian foamy virus-induced immunosuppression in rabbits. J Gen Virol 1979; 44:383 - 90; http://dx.doi.org/10.1099/0022-1317-44-2-383; PMID: 230289
- Lampe J, Marino S, Rethwilm A, Aguzzi A. Degeneration of the cerebellar granule cell layer in transgenic mice expressing genes of human foamy virus. Neuropathol Appl Neurobiol 1998; 24:36 - 43; http://dx.doi.org/10.1046/j.1365-2990.1998.00086.x; PMID: 9549727
- Tschopp RR, Brandner S, Marino S, Bothe K, Horak I, Rethwilm A, et al. Analysis of the determinants of neurotropism and neurotoxicity of HFV in transgenic mice. Virology 1996; 216:338 - 46; http://dx.doi.org/10.1006/viro.1996.0069; PMID: 8607263
- Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, et al. Foamy virus particle formation. J Virol 1998; 72:1610 - 5; PMID: 9445065
- Shaw KL, Lindemann D, Mulligan MJ, Goepfert PA. Foamy virus envelope glycoprotein is sufficient for particle budding and release. J Virol 2003; 77:2338 - 48; http://dx.doi.org/10.1128/JVI.77.4.2338-2348.2003; PMID: 12551971
- Lindemann D, Goepfert PA. The foamy virus envelope glycoproteins. Curr Top Microbiol Immunol 2003; 277:111 - 29; http://dx.doi.org/10.1007/978-3-642-55701-9_5; PMID: 12908770
- Kong XH, Yu H, Xuan CH, Wang JZ, Chen QM, Geng YQ. The requirements and mechanism for capsid assembly and budding of bovine foamy virus. Arch Virol 2005; 150:1677 - 84; http://dx.doi.org/10.1007/s00705-005-0518-9; PMID: 15834655
- Wilk T, Geiselhart V, Frech M, Fuller SD, Flügel RM, Löchelt M. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J Virol 2001; 75:7995 - 8007; http://dx.doi.org/10.1128/JVI.75.17.7995-8007.2001; PMID: 11483744
- Geiselhart V, Schwantes A, Bastone P, Frech M, Löchelt M. Features of the Env leader protein and the N-terminal Gag domain of feline foamy virus important for virus morphogenesis. Virology 2003; 310:235 - 44; http://dx.doi.org/10.1016/S0042-6822(03)00125-9; PMID: 12781711
- Geiselhart V, Bastone P, Kempf T, Schnölzer M, Löchelt M. Furin-mediated cleavage of the feline foamy virus Env leader protein. J Virol 2004; 78:13573 - 81; http://dx.doi.org/10.1128/JVI.78.24.13573-13581.2004; PMID: 15564468
- Schwantes A, Truyen U, Weikel J, Weiss C, Löchelt M. Application of chimeric feline foamy virus-based retroviral vectors for the induction of antiviral immunity in cats. J Virol 2003; 77:7830 - 42; http://dx.doi.org/10.1128/JVI.77.14.7830-7842.2003; PMID: 12829823
- Mühle M, Bleiholder A, Kolb S, Hübner J, Löchelt M, Denner J. Immunological properties of the transmembrane envelope protein of the feline foamy virus and its use for serological screening. Virology 2011; 412:333 - 40; http://dx.doi.org/10.1016/j.virol.2011.01.023; PMID: 21316070
- Bleiholder A, Mühle M, Hechler T, Bevins S, vandeWoude S, Denner J, et al. Pattern of seroreactivity against feline foamy virus proteins in domestic cats from Germany. Vet Immunol Immunopathol 2011; 143:292 - 300; http://dx.doi.org/10.1016/j.vetimm.2011.06.007; PMID: 21724269
- Alke A, Schwantes A, Kido K, Flötenmeyer M, Flügel RM, Löchelt M. The bet gene of feline foamy virus is required for virus replication. Virology 2001; 287:310 - 20; http://dx.doi.org/10.1006/viro.2001.1065; PMID: 11531409
- Löchelt M. Foamy virus transactivation and gene expression. Curr Top Microbiol Immunol 2003; 277:27 - 61; http://dx.doi.org/10.1007/978-3-642-55701-9_2; PMID: 12908767
- Russell RA, Wiegand HL, Moore MD, Schäfer A, McClure MO, Cullen BR. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J Virol 2005; 79:8724 - 31; http://dx.doi.org/10.1128/JVI.79.14.8724-8731.2005; PMID: 15994766
- Mühle M, Hoffmann K, Löchelt M,, Denner J. Immunisation with foamy virus Bet fusion proteins as novel strategy for HIV-1 epitope delivery. Clin Vaccine Immunol In Press
- Jern P, Sperber GO, Blomberg J. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology 2005; 2:50; http://dx.doi.org/10.1186/1742-4690-2-50; PMID: 16092962