Abstract
A new avian-origin influenza virus emerged near Shanghai in February 2013, and by the beginning of May it had caused over 130 human infections and 36 deaths. Human-to-human transmission of avian-origin H7N9 influenza A has been limited to a few family clusters, but the high mortality rate (27%) associated with human infection has raised concern about the potential for this virus to become a significant human pathogen. European, American, and Asian vaccine companies have already initiated the process of cloning H7 antigens such as hemagglutinin (HA) into standardized vaccine production vehicles. Unfortunately, previous H7 HA-containing vaccines have been poorly immunogenic. We used well-established immunoinformatics tools to analyze the H7N9 protein sequences and compare their T cell epitope content to other circulating influenza A strains as a means of estimating the immunogenic potential of the new influenza antigen. We found that the HA proteins derived from closely related human-derived H7N9 strains contain fewer T cell epitopes than other recently circulating strains of influenza, and that conservation of T cell epitopes with other strains of influenza was very limited. Here, we provide a detailed accounting of the type and location of T cell epitopes contained in H7N9 and their conservation in other H7 and circulating (A/California/07/2009, A/Victoria/361/2011, and A/Texas/50/2012) influenza A strains. Based on this analysis, avian-origin H7N9 2013 appears to be a “stealth” virus, capable of evading human cellular and humoral immune response. Should H7N9 develop pandemic potential, this analysis predicts that novel strategies for improving vaccine immunogenicity for this unique low-immunogenicity strain of avian-origin influenza will be urgently needed.
Introduction
Between February and May 2013, avian-origin H7N9 emerged as a significant cause of human morbidity and mortality in China, with pandemic potential. As of May 2013, the new influenza virus has caused more than 130 infected patients in China to seek hospital care, of which 27% have died and many remain hospitalized.Citation1,Citation2 Infections are sporadic, usually associated with exposure to birds such as chickens and pigeons, and the extent of human-to-human transmission appears to be very low at present. Sampling methods may not be entirely accurate since viral titers are low in upper respiratory samples, thus some cases of asymptomatic infection and transmission may have been missed.Citation3
Despite the fact that human-to-human transmission seems to be rare, the ongoing outbreak of H7N9 influenza is of great concern to infectious disease experts for four important reasons. First, individual cases of nearly identical H7N9 virus infections have been identified in a wide range of locations within China in a very short period of time, which strongly suggests that the virus is already widespread in an as-yet-unidentified reservoir population.4 Second, an H7 subtype influenza has never before infected humans to any extent, meaning the human population has little or no immunity, which could lead to a very high mortality rate.Citation5 H7N9 infection has led to death in approximately 27% of reported cases, and many infected patients are still hospitalized in serious condition.2 Third, viral sequences suggest the possibility of resistance to certain antiviral medications, rendering some current treatments ineffective.Citation1,Citation6 And fourth, viral isolates bear hemagglutinin receptor binding site mutations that better enable this avian virus to infect mammals, and only a few additional mutations might be needed for full adaptation for human-to-human transmission.Citation6
The source of the viral infections appears to be contact with poultry in the majority of cases. Despite these reports, approximately 40% of patients have had no contact with birds of any type.Citation7 Furthermore, the number of infections among chickens in market-based samples is very low, an observation that has puzzled epidemiologists. Despite these reports of low infection rates, the genomic sequences of the virus suggests that the H7N9 sequence is closest to poultry-origin H7 viruses,Citation3 and as a result of this association, a mass culling of the poultry population has been implemented in Shanghai, Nanjing, and other regions across China.Citation33
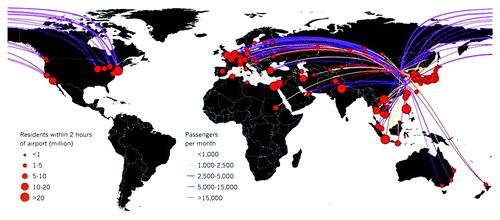
Alarmingly, human H7N9 cases have already spread outside of China; a business man who had traveled to one of the affected provinces was recently hospitalized in Taiwan and remains in serious condition as of May.Citation8 According to experts cited by Nature, 70% of the global population outside of China lives within two hours of an airport linked to the outbreak regions by either a direct flight or a single connection ().Citation9 Consequently, should genetic changes occur that permit facile human-to-human transmission, the H7N9 outbreak will rapidly become a global pandemic.
Figure 1. The rapid emergence of H7N9 in several locations in China reflects wide-spread distribution in the animal population and potential for even greater spread. H7N9 is a virus that deserves serious consideration. Should a traveler get infected in China, flight routes from the outbreak regions would quickly carry any human-transmissible virus to huge population centers in Europe, North America and Asia. An estimated 70% of the world population resides within two hours’ travel time of destination airports (calculated using gridded population-density maps and a data set of global travel times, map supplied by A. J. Tatem, Z. Huang and S. I. Hay (2013). Unpublished data. (A.J.T., University of Southampton, UK; Z.H., University of Florida, Gainesville; S.I.H., University of Oxford, UK.) Reprinted by permission from Macmillan Publishers Ltd: Nature News 2013.Citation9
Analysis of H1N1 (2009) demonstrated cross-conservation of T cell epitopes.
The current H7N9 outbreak is somewhat reminiscent of March 2009, when pandemic H1N1 (A/California/07/2009) emerged in Mexico. At that time, CDC studies showed that most individuals lacked pre-existing humoral immunity that was cross-protective against the pandemic strain (A/California/07/2009), and that vaccination with the 2008/2009 seasonal trivalent influenza vaccine (TIV) containing H1N1 A/Brisbane/59/2007 rarely elicited neutralizing antibody responses against 2009 H1N1 isolates.Citation10,Citation11 Despite this concern, during the course of the 2009 pandemic, the new H1N1 was not associated with significant morbidity or mortality among older adults, a population normally at high risk for complications following seasonal influenza infection.Citation12,Citation13
At that time, the importance of T cell epitope responses to cross-protective immunity (against different strains of influenza) was already well established. Accordingly, we proposed that cross-reactive T memory responses with prior H1N1 viruses might protect against severe H1N1 disease, despite the absence of cross-reactive humoral immunity, and we published T cell epitopes in the hemagglutinin (HA) and neuraminidase (NA) proteins that were predicted by immunoinformatic analysis to be cross-conserved with HA and NA sequences present in the trivalent inactivated influenza vaccine administered that season.Citation14 This hypothesis was later validated in animal and human studies, and the impact of the H1N1 pandemic was, in fact, much less severe than had been expected. The lack of B cell epitope cross-reactivity did not appear to lead to high morbidity and mortality rates during the pandemic.
Unfortunately, our prediction is very different for H7N9: we find (1) very low T cell epitope content in the HA sequences, and (2) very poor potential for cross-reactivity with T cells specific for currently circulating influenza strains. Coupled with the absence of any prior exposure to H7 in human populations (meaning established humoral immunity may be entirely absent), these findings suggest that the H7N9 influenza virus, should it acquire the ability to be transmitted from person to person, has the potential to have a much more serious impact on human populations than did pandemic H1N1 influenza in 2009.
Application of in silico analysis to H7N9 demonstrates low T cell epitope content.
Employing the EpiMatrix immunoinformatics suite, we compared the protein sequences of emerging H7N9 strains to the sequences of other recent strains of influenza and previous H7 strains. We also provide a detailed analysis of the T cell epitopes (both class I cytotoxic T cell epitopes and class II T helper epitopes) in the H7N9 strains. We find: (1) HA from the 2013 outbreak H7N9 and a wide range of avian H7 strains have low immunogenic potential (based on T cell epitope analysis, as previously published)Citation15 when compared with HA from seasonal influenza strains that are known to be immunogenic; (2) H7-HA conservation analysis suggests limited potential for T cell epitope cross reactivity with previously and currently circulating seasonal strains; and (3) T cell epitopes that have cross-reactive potential are found among internal proteins. A full T cell epitope analysis for novel avian H7N9 and a list of cross-conserved epitopes is compiled in this report. This prediction of low immunogenic potential and poor epitope conservation has important implications for the development of effective H7N9 vaccines.
Results
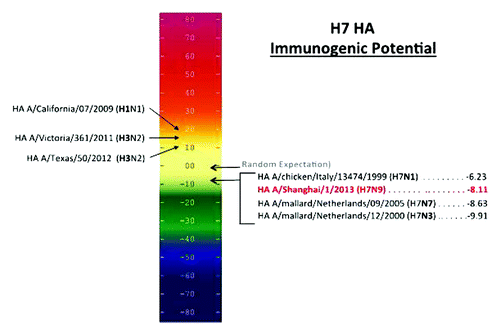
Overall HLA class II epitope content. The immunogenicity potential score of H7N9 antigens was derived by summing EpiMatrix scores over the entire protein and normalizing for protein length. Immunogenicity potential has been directly correlated with antibody titers in several published studies.Citation16,Citation18 The immunogenicity score for H7N9 HA is remarkably low (): H7-HA scores -8.11, well below the expectation for a random sequence, and also significantly below the scores of H1-HA (+18.22) and H3-HA (+14.30). The current H7 also scores within the same range as other H7-HA strains, some of which were part of poorly immunogenic monovalent inactivated vaccines.Citation19,Citation20 Other low-immunogenicity proteins that score in the negative range on this immunogenicity scale include albumin, transferrin, and the immunoglobulin G Fc domain, observations that have been previously published by our group.Citation21
Figure 2. Potential immunogenicity of emerging influenza A (H7N9) HA and HA from the current seasonal vaccine and H7 vaccines. The number of HLA ligands (putative T cell epitopes) per unit protein is plotted on an Immunogenicity Scale. This scale is correlated with observed immunogenicity in retrospective and prospective studies.Citation16,Citation17 The numbers used in this scale reflect the difference between the number of predicted T cell epitopes we would expect to find in a protein of any given size by chance alone (based on an evaluation of more than 10,000 random protein sequences) and the number of putative epitopes predicted by the EpiMatrix System for a given protein. The EpiMatrix Protein Score of an “average” protein is zero. EpiMatrix Protein Scores above zero indicate the presence of excess MHC ligands and denote a higher potential for immunogenicity, while scores below zero indicate the presence of fewer potential HLA ligands than expected and a lower potential for immunogenicity. H7 has fewer HLA ligands than expected, which is reflected in its negative immunogenicity score. Seasonal strains of influenza are scored for potential immunogenicity on the left; their scores are much higher than H7N9.
HLA class II cluster analysis.
We have published our approach to finding highly immunogenic epitopes that are ‘promiscuous’ and highly conserved in multiple strains of the same pathogen.Citation22 We call this approach the ‘immunogenic consensus sequence (ICS) approach.’ In our own work, we have found that these clustered epitopes are the most effective ones to include in vaccines to promote antibody response and vaccine efficacy.Citation23 Accordingly, we derived a total of 101 ICS epitopes for H7N9, containing multiple 9-mer frames predicted to bind to many HLA alleles and conserved in a minimum of three of the four human-derived avian-origin H7N9 sequences (). ICS were derived from among all 11 H7N9 antigens. Of the 101 ICS class II epitopes, 16 were found to contain 9-mer sequences that were previously published as T cell epitopes or class II MHC binders. None of these 16 published 9-mers have been identified in the H7N9 influenza surface HA and NA antigens, but rather all are found in internal proteins: six in M1, eight in NP, and two in PB1 (see ‘Published in IEDB’ column, ).
Table 1. Numbers of putative class II ICS constructed from the four emerging avian H7N9 strains and numbers of those epitopes conserved (at least 80% amino acid similarity) in recently circulating and vaccine backbone influenza strains
Next we checked for conservation among circulating strains of influenza and vaccine backbone proteins. In this case, we considered all epitopes sharing at least 80% amino acid similarity as defined by Kesmir et al. “conserved”.Citation34 Only 11 promiscuous T cell epitopes were found in H7N9-HA, and only four of these are somewhat cross-conserved in other strains. In contrast, we found 16 promiscuous helper T cell epitopes in the 2009 H1N1-HA sequence, of which nine (56%) were 100% conserved in the 2008–2009 influenza vaccine strain; 81% were either identical or had one conservative amino acid substitution. Thus the emerging H7N9 is predicted to be less immunogenic, based on our analysis, and is also shown to be less conserved in other influenza strains. Table S1 provides detailed information on the H7N9 class II clustered (promiscuous) epitopes, their conservation in other influenza strains, and information on published epitopes if cross conservation is present.
Cross reactivity—class II.
As shown in , a total of 96 of the 101 putative epitopes are conserved in other H7N9 strains. Of the 11 putative H7-HA epitopes, only four have minimal cross-reactivity to the current 2012–2013 TIV; three are found in A/Victoria/361/2011 (H3N2), one was found in A/California/7/2009 (H1N1), and no conserved sequences were identified in B/Wisconsin/1/2010. Greater homology with H3-HA is expected, as H7 and H3 are group 2 influenza A viruses whereas H1 is a group 1 virus. Similarly, only one of the six N9-NA putative epitopes was found to be conserved in the current TIV, with 86% similarity to the 2012–2013 vaccine strain A/Victoria/361/2011 (H3N2) as well as the 2013–2014 vaccine recommendation A/Texas/50/2012 (H3N2). Conversely, 83 of the 84 putative epitopes derived from internal H7N9 proteins are conserved in at least one of the currently circulating influenza A strains. Fourteen H7N9 ICS derived from internal proteins are conserved in influenza B/Wisconsin/1/2010; high conservation of internal antigen epitopes is consistent in vaccine backbone strains. Still, previously circulating strains of influenza had more T cell epitopes than were found in the current H7N9.
Epitope content—class I.
As shown in , a total of 1,242 putative class I epitopes were found by EpiMatrix, spanning the 11 H7N9 antigens across the six supertype class I alleles. Of those 1,242 sequences, a total of 48 were found to be identified as either published T cell epitopes or MHC binders. None of these 48 included epitopes from the external HA and NA antigens; instead, the epitopes conserved with published epitopes were found in the M1 (17), NP (9), NS1 (1), PA (6), and PB1 (15) proteins, as shown in . Table S2 provides detailed information on the H7N9 epitopes, their conservation in other influenza strains, and information on published epitopes if cross-conservation is present.
Table 2. Numbers of putative class I epitopes discovered in four emerging avian H7N9 strains and numbers of those epitopes conserved (at least 80% amino acid similarity) in recently circulating and vaccine backbone influenza strains
Cross reactivity—class I.
A total of 967 (77.9%) of the 1,242 putative epitopes were found to be > 90% conserved in other H7N9 strains. Only 4/150 and 1/146 HA and NA class I putative epitopes, respectively, were found to be conserved in the current 2012–2013 TIV, which is not surprising since this is an entirely new strain of influenza for the human population. A similar trend of low conservation can be seen when H7N9 is compared with the LAIV backbone strain and the strains recommended for 2013–2014 vaccination. A total of 609 of the remaining 946 putative class I epitopes, spanning the internal proteins, were found to be conserved in one or more of the circulating 2012–2013 influenza A strains. Similar conservation trends can be seen when comparing the epitopes derived from internal antigens to the LAIV backbone strain and the strains recommended for 2013–2014 vaccination.
The above analysis is based on in silico predictions using a well-validated immunoinformatics toolkit.Citation24 Based on historical performance, we expect these epitope predictions to be 93-99% accurate; in addition, correlations with published H1N1 and H3N2 epitopes are provided in Tables S1 and S2. Our findings suggest that overall H7-HA immunogenicity will be poor, and that immune responses to H7N9 will be driven by T cell epitopes found in the internal proteins. Given the poor immunogenicity of previous H7 vaccines,Citation25 whole attenuated or inactivated vaccines containing a greater number of T cell epitopes in their internal proteins (or subunit vaccines that incorporate conserved internal protein T cell epitopes) may be more effective immunogens than subunit HA/NA for H7N9.
Discussion
In 2009, we performed a similar analysis of H1N1, following emergence of the H1N1 2009 virus. Using EpiMatrix, a T cell epitope prediction and comparison tool, we compared the sequences of the three HA and NA proteins contained in 2008–2009 TIV to their counterparts in A/California/04/2009 (H1N1), looking for cross-conserved T cell epitope sequences. We found > 50% conservation of T helper and cytotoxic T lymphocyte (CTL) epitopes between novel swine-origin influenza virus and TIV HA for selected HLA.Citation14 At that time, we suggested that seasonal influenza vaccination or infection might protect against pandemic H1N1. We then extended the analysis to define core T cell epitopes in H1N1 2009, and to define the extent of cross-reactivity between the pandemic virus and the 2009-2010 seasonal H1N1 vaccine strain, A/Brisbane/59/2007. The 2009 H1-HA protein contained 16 class II (T helper) peptide sequences that were both highly conserved between the pandemic and A/Brisbane/59/2007 H1N1 strains and likely to be presented in the context of multiple HLA alleles.Citation14 Schanen et al. (VaxDesign), in collaboration with EpiVax, then confirmed that human influenza-specific T helper cells from donors not previously exposed to H1N1 2009 were capable of generating cross-reactive effector responses against the 2009 H1N1 virus.Citation26 These studies helped support the hypothesis that existing influenza-specific CD4+ T cells generated cross-protective cellular responses that limited disease severity and virus spread in individuals lacking cross-reactive humoral immunity, and was substantiated by subsequent animal studies demonstrating that seasonal H1N1 exposure protected ferretsCitation27 and mice (Ted Ross, unpublished) from disease in pandemic H1N1 infection.
Our previous study contradicted predictions that H1N1 would cause serious illness in human populations. Even though H1N1 spread rapidly (presumably due to the absence of any memory B cell response), hospitalizations were limited to younger individuals and pregnant women who might have had limited prior exposure to influenza and absent T memory responses.
The lack of antibody response to the 2009 H1N1 is similar to the situation with the current H7N9, but from the T cell perspective, the two are markedly different. If H7N9 becomes easily transmissible between humans, widespread transmission (due to the absence of memory B cell responses and antibody that might protect against H7, which has never circulated in human populations) and widespread illness (due to the absence of cross-reactive T cell potential) are probable. The mortality rate following establishment of H7N9 disease is greater than 27%; thus concern about the potential for this virus to cause significant morbidity and mortality in human populations, should the virus adapt to spread from human to human, are entirely justified.
In addition, due to the poor predicted immunogenicity of H7N9, inactivated vaccines based on the H7-HA and N9-NA antigens, whether produced in eggs or mammalian cells, are likely to be poorly immunogenic based on the analysis described here. Previous studies of influenza vaccines have clearly demonstrated the link between T cell response and antibody titers,Citation28 thus the absence of T helper epitopes in the HA antigen of the current circulating strain of H7N9 can be expected to lead to very poor immunogenicity, particularly for subunit vaccines containing HA alone. New strategies that will improve protection against H7N9 are needed, as the virus has the potential to become a worldwide threat.
Alternative vaccine approaches for H7N9 would include: (1) addition of adjuvants to boost immune responses against the weaker T cell epitopes; (2) addition of cross-conserved epitopes to the HA in a subunit vaccine (as proposed by our team to NIAID); (3) enhancement of the HA with cross-conserved epitopes (a vaccine design currently under production at EpiVax); (4) development of T cell vaccines for influenza that contain all of the most immunogenic T cell epitopes derived from H7, including those that are cross conserved with H7N9 (a DNA-string-of beads design is currently in production); and (5) use of two doses of vaccine rather than one.
In contrast with 2009 H1N1, 2013 H7N9 is a significant concern for global health. As described above, H7N9 has fewer epitopes than would normally occur in a random protein sequence. As this virus has not, by current reports, circulated in humans previously, it is unclear why the virus would evolve to have fewer T cell epitopes than predicted. Those T cell epitopes that we predict might be of use for vaccine design are included in the supplemental tables (Tables S1 and S2) as a service to the influenza vaccine development community. Whether ‘evolutionary’ in nature or not, we postulate that the relative paucity of T cell epitopes (both novel and conserved) in the key antigens of emerging H7N9 will have serious consequences for human immune response and vaccine design.
Materials and Methods
Sequence collection.
Four human H7N9 influenza sequences were downloaded from GISAID (http://platform.gisaid.org/). The downloaded sequences were from A/Hangzhou/1/2013, A/Anhui/1/2013, A/Shanghai/1/2013, and A/Shanghai/2/2013. The 25 remaining non-human H7N9 strains available on GISAID were also downloaded to confirm if epitopes found within the four human-sourced strains were conserved in H7N9 as a whole, which would indicate functional dependency and low probability of mutation. The novel H7N9 sequences analyzed were selected from sequences that were available on GISAID on April 6th, 2013. Three additional influenza strains found in the current 2012–2013 TIV [A/California/07/2009(H1N1), A/Victoria/361/2011(H3N2), and B/Wisconsin/01/2010] were also downloaded for evaluation of conservation and cross-protection with EpiMatrix-identified H7N9 T cell epitopes.
The following accessions were utilized for epitope cross-conservation analysis: A/California/7/2009 (H1N1): GISAID EPI_ISL_77637; A/Victoria/361/2011 (H3N2): GISAID EPI_ISL_134450; B/Wisconsin/01/2010: GISAID EPI_ISL_115174; A/Christchurch/16/2010 (H1N1): GISAID EPI_ISL_79722; A/Texas/50/2012 (H3N2): GISAID EPI_ISL_136503; A/Ann Arbor/7/1967 (H2N2): GISAID EPI_ISL_130404; A/Puerto Rico/8/34 (H1N1): Genbank AAM75164.1; AAM75162.1; AAM75610.1; AAM75158.1; AAM75156.1; AAM75163.1; AAM75161.1; AAM75159.1; AAM75157.1; AAM75155.1; ADX99593.1.
Conservatrix.
The Conservatrix algorithmCitation29 was employed to parse and compare 9-mers and 10-mers (class I analysis only) for highly conserved segments among the four human H7N9 influenza sequences, organized by protein of origin: HA, M1, M2, NA, NEP, NP, NS1, PA, PB1, PB1-F2, or PB2. Highly conserved epitope sequences were then analyzed for putative binding to eight common HLA class II (DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, DRB1*1301, DRB1*1501) and six common HLA class I alleles (A0101, A0201, A0301, A2402, B0702, and B4403).
EpiMatrix.
Using the EpiMatrix algorithm,Citation30 epitopes were selected from the output of highly conserved epitopes produced by Conservatrix. Each amino acid was scored for predicted affinity to eight common HLA class II and six common HLA class I alleles, covering > 90% of the human population.Citation31
EpiAssembler.
Sets of overlapping, conserved, and immunogenic class II epitopes were identified and assembled into extended immunogenic consensus sequences (ICS) with the EpiAssemblerCitation32 tool. Processing and presentation of these sequences would allow for presentation of the highly conserved peptides in the context of more than one class II MHC. The resulting peptide is not a “pseudo-sequence” as such, since each constituent epitope occurs in its corresponding position in the native protein but in different variants of the pathogen. These epitopes are of high value for vaccine design as they are both promiscuous and highly conserved.
Additional material
Download Zip (189.9 KB)Disclosure of Potential Conflicts of Interest
ADG and WDM are senior officers and majority shareholders at EpiVax, Inc., a privately owned immunoinformatics and vaccine design company located in Providence, RI, USA. MA, FT, LL, and LM are employees at EpiVax, in which LM holds stock options. TR is currently collaborating with EpiVax on an influenza SBIR proposal to the National Institutes of Health. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company.
Acknowledgments
Funding to support the discussions leading to this work can be attributed to U19 (AI082642) and R21 (AI090359) grants awarded to ADG from the National Institutes of Health, NIAID, DAIT and DMID (respectively). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888 - 97; http://dx.doi.org/10.1056/NEJMoa1304459; PMID: 23577628
- World Health Organization. Global Alert and Response: Human infection with avian influenza A(H7N9) virus – update. 2013 May 17. Retrieved from: http://www.who.int/csr/don/2013_05_17/en/index.html
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 2013; 381:1916 - 25; http://dx.doi.org/10.1016/S0140-6736(13)60903-4; PMID: 23623390
- World Health Organization. Number of confirmed human cases for avian influenza A(H7N9) reported to WHO: Report 3 – data in WHO/HQ as of 01 May 2013, 14:45 GMT+1. 2013 May 01. Retrieved from: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/04_ReportWebH7N9Number.pdf
- Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, et al. Preliminary Report: Epidemiology of the Avian Influenza A (H7N9) Outbreak in China. N Engl J Med 2013; http://dx.doi.org/10.1056/NEJMoa1304617; PMID: 23614499
- Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 2013; 18:20453; PMID: 23594575
- Smith M. Infection Route of H7N9 Flu Remains Puzzling. MedPage Today 2013 Apr 18. Retrieved from: http://www.medpagetoday.com/InfectiousDisease/URItheFlu/38553
- Centers for Disease Control. R.O.C. (Taiwan). Press Release: In response to occurrence of first imported human infection with avian influenza A (H7N9) in Taiwan, Premier Jiang convenes 6th meeting held by CECC for H7N9 influenza to monitor progress in disease prevention efforts by each government agency. 2013 Apr 25. Retrieved from: http://www.cdc.gov.tw/english/info.aspx?treeid=BC2D4E89B154059B&nowtreeid=EE0A2987CFBA3222&tid=A14678FBB1632075
- Butler D. Mapping the H7N9 avian flu outbreaks: Where are the 104 human cases confirmed so far, and where might the virus go next? Nature News 2013 Apr 24; Retrieved from: http://www.nature.com/news/mapping-the-h7n9-avian-flu-outbreaks-1.12863
- Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009; 361:1945 - 52; http://dx.doi.org/10.1056/NEJMoa0906453; PMID: 19745214
- Centers for Disease Control and Prevention (CDC). Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep 2009; 58:521 - 4; PMID: 19478718
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003; 289:179 - 86; http://dx.doi.org/10.1001/jama.289.2.179; PMID: 12517228
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA 2004; 292:1333 - 40; http://dx.doi.org/10.1001/jama.292.11.1333; PMID: 15367555
- De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008-2009 conventional influenza vaccine. Vaccine 2009; 27:5740 - 7; http://dx.doi.org/10.1016/j.vaccine.2009.07.040; PMID: 19660593
- Weber CA, Mehta PJ, Ardito M, Moise L, Martin B, De Groot AS. T cell epitope: friend or foe? Immunogenicity of biologics in context. Adv Drug Deliv Rev 2009; 61:965 - 76; http://dx.doi.org/10.1016/j.addr.2009.07.001; PMID: 19619593
- De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol 2009; 131:189 - 201; http://dx.doi.org/10.1016/j.clim.2009.01.009; PMID: 19269256
- Jawa V, Cousens LP, De Groot AS. Immunogenicity of Therapeutic Fusion proteins: Contributory Factors and Clinical Experience. In: Schmidt SR, ed. Fusion Protein Technologies for Biopharmaceuticals: Applications and Challenges. Hoboken, NJ: John Wiley and Sons, Inc., 2013:75-90.
- Koren E, De Groot AS, Jawa V, Beck KD, Boone T, Rivera D, Li L, Mytych D, Koscec M, Weeraratne D, et al. Clinical validation of the “in silico” prediction of immunogenicity of a human recombinant therapeutic protein. Clin Immunol 2007; 124:26 - 32; http://dx.doi.org/10.1016/j.clim.2007.03.544; PMID: 17490912
- Couch RB, Patel SM, Wade-Bowers CL, Niño D. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One 2012; 7:e49704; http://dx.doi.org/10.1371/journal.pone.0049704; PMID: 23239968
- Couch RB, Decker WK, Utama B, Atmar RL, Niño D, Feng JQ, Halpert MM, Air GM. Evaluations for in vitro correlates of immunogenicity of inactivated influenza a H5, H7 and H9 vaccines in humans. PLoS One 2012; 7:e50830; http://dx.doi.org/10.1371/journal.pone.0050830; PMID: 23239987
- De Groot AS, Goldberg M, Moise L, Martin W. Evolutionary deimmunization: an ancillary mechanism for self-tolerance?. Cell Immunol 2006; 244:148 - 53; http://dx.doi.org/10.1016/j.cellimm.2007.02.006; PMID: 17445787
- De Groot AS, McMurry J, Moise L. Prediction of immunogenicity: in silico paradigms, ex vivo and in vivo correlates. Curr Opin Pharmacol 2008; 8:620 - 6; http://dx.doi.org/10.1016/j.coph.2008.08.002; PMID: 18775515
- De Groot AS, Bishop EA, Khan B, Lally M, Marcon L, Franco J, Mayer KH, Carpenter CC, Martin W. Engineering immunogenic consensus T helper epitopes for a cross-clade HIV vaccine. Methods 2004; 34:476 - 87; http://dx.doi.org/10.1016/j.ymeth.2004.06.003; PMID: 15542374
- Levitz L, Koita OA, Sangare K, Ardito MT, Boyle CM, Rozehnal J, Tounkara K, Dao SM, Koné Y, Koty Z, et al. Conservation of HIV-1 T cell epitopes across time and clades: validation of immunogenic HLA-A2 epitopes selected for the GAIA HIV vaccine. Vaccine 2012; 30:7547 - 60; http://dx.doi.org/10.1016/j.vaccine.2012.10.042; PMID: 23102976
- Uyeki TM, Cox NJ. Global concerns regarding novel influenza A (H7N9) virus infections. N Engl J Med 2013; 368:1862 - 4; http://dx.doi.org/10.1056/NEJMp1304661; PMID: 23577629
- Schanen BC, De Groot AS, Moise L, Ardito M, McClaine E, Martin W, Wittman V, Warren WL, Drake DR 3rd. Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4(+) T cells against the swine-origin H1N1 influenza virus. Vaccine 2011; 29:3299 - 309; http://dx.doi.org/10.1016/j.vaccine.2011.02.019; PMID: 21349362
- Ellebedy AH, Ducatez MF, Duan S, Stigger-Rosser E, Rubrum AM, Govorkova EA, Webster RG, Webby RJ. Impact of prior seasonal influenza vaccination and infection on pandemic A (H1N1) influenza virus replication in ferrets. Vaccine 2011; 29:3335 - 9; http://dx.doi.org/10.1016/j.vaccine.2010.08.067; PMID: 20840835
- Galli G, Medini D, Borgogni E, Zedda L, Bardelli M, Malzone C, Nuti S, Tavarini S, Sammicheli C, Hilbert AK, et al. Adjuvanted H5N1 vaccine induces early CD4+ T cell response that predicts long-term persistence of protective antibody levels. Proc Natl Acad Sci U S A 2009; 106:3877 - 82; http://dx.doi.org/10.1073/pnas.0813390106; PMID: 19237568
- De Groot AS, Bosma A, Chinai N, Frost J, Jesdale BM, Gonzalez MA, Martin W, Saint-Aubin C. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine 2001; 19:4385 - 95; http://dx.doi.org/10.1016/S0264-410X(01)00145-1; PMID: 11483263
- De Groot AS, Jesdale BM, Szu E, Schafer JR, Chicz RM, Deocampo G. An interactive Web site providing major histocompatibility ligand predictions: application to HIV research. AIDS Res Hum Retroviruses 1997; 13:529 - 31; http://dx.doi.org/10.1089/aid.1997.13.529; PMID: 9135870
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 1998; 160:3363 - 73; PMID: 9531296
- De Groot AS, Bishop EA, Khan B, Lally M, Marcon L, Franco J, Mayer KH, Carpenter CC, Martin W. Engineering immunogenic consensus T helper epitopes for a cross-clade HIV vaccine. Methods 2004; 34:476 - 87; http://dx.doi.org/10.1016/j.ymeth.2004.06.003; PMID: 15542374
- Yiqi Y. Culls imposed to minimize contact. China Daily. 2013 Apr 18. Retrieved from: http://www.chinadaily.com.cn/cndy/2013-04/18/content_16417457.htm
- Calis JJ, de Boer RJ, Keşmir C. Degenerate T-cell recognition of peptides on MHC molecules creates large holes in the T-cell repertoire. PLoS Comput Biol 2012; 8:e1002412; http://dx.doi.org/10.1371/journal.pcbi.1002412; PMID: 22396638