Abstract
Immune responses to cross-conserved T cell epitopes in novel H1N1 influenza may explain reports of diminished influenza-like illnesses and confirmed infection among older adults, in the absence of cross-reactive humoral immunity, during the 2009 pandemic. These cross-conserved epitopes may prove useful for the development of a universal H1N1 influenza vaccine, therefore, we set out to identify and characterize cross-conserved H1N1 T cell epitopes. An immunoinformatic analysis was conducted using all available pandemic and pre-pandemic HA-H1 and NA-N1 sequences dating back to 1980. Using an approach that balances potential for immunogenicity with conservation, we derived 13 HA and four NA immunogenic consensus sequences (ICS) from a comprehensive analysis of 5 738 HA-H1 and 5 396 NA-N1 sequences. These epitopes were selected because their combined epitope content is representative of greater than 84% of pre-pandemic and pandemic H1N1 influenza strains, their predicted immunogenicity (EpiMatrix) scores were greater than or equal to the 95th percentile of all comparable epitopes, and they were also predicted to be presented by more than four HLA class II archetypal alleles. We confirmed the ability of these peptides to bind in HLA binding assays and to stimulate interferon-γ production in human peripheral blood mononuclear cell cultures. These studies support the selection of the ICS as components of potential group-common H1N1 vaccine candidates and the application of this universal influenza vaccine development approach to other influenza subtypes.
Introduction
Despite intense efforts at epidemiological tracking and computational modeling, the timing and severity of influenza outbreaks remain difficult to predict, as illustrated by the last two influenza seasons. On average, influenza-like illness (ILI) reports exceed the Centers for Disease Control and Prevention’s (CDC) national baseline for a period of 12 weeks. During the 2011–2012 season, there was only one week of ILI reports above baseline. In contrast, the 2012–2013 season saw the earliest outbreak since 2003–2004, and roughly 15 weeks of ILI reports above baseline were documented.Citation1
Vaccination is the only public-health means for reducing the impact of influenza morbidity and mortality by offsetting the uncertainties of timing and virulence arising from uncontrollable complexities of population, behavioral, viral, and environmental factors. Vaccination is also considered a cornerstone approach for pandemic preparedness. The principle approach to influenza vaccine design focuses on raising antibodies that prevent hemagglutination. Generally, vaccination does induce hemagglutinating antibodies but these antibodies are neither cross-reactive with other strains, nor persistent. Furthermore, vaccination against influenza is only moderately effective. A meta-analysis using data from randomized, controlled trials conducted over 12 seasons and published between 1967 and 2011 demonstrated that trivalent influenza vaccination (TIV) in adults aged 18–65 years provided only moderate protection (59%) over eight seasons and significantly lower levels in other seasons.Citation2 Live attenuated influenza vaccine (LAIV), which stimulates both cellular and humoral immunity, showed higher efficacy (83%) in children aged 6 to 17 years, but not in adults. To improve on the shortcomings of existing influenza vaccination approaches, novel vaccine approaches that aim to provide universal protection are needed.
We became interested in the concept of cross-reactive T cell epitopes for influenza during the 2009 H1N1 pandemic. At that time, the Centers for Disease Control and Prevention reported that seasonal flu vaccines did not elicit cross-reactive neutralizing antibodies against the emerging pandemic (H1N1) 2009.Citation3 When early clinical reports released during the 2009 A(H1N1) pandemic suggested the novel influenza was more virulent among children and adults under 65 years than the elderly, we hypothesized that cellular responses to cross-reactive T cell epitopes might explain the unexpected disease distribution. The apparent lack of B cell epitope conservation in novel H1N1 and absence of cross-reactive antibodies raised by the seasonal vaccine H1N1 strain at the time supported this idea.
Thus, we set out to identify cross-conserved T cell epitopes in the pandemic and the 2008–2009 seasonal vaccine hemagglutinin (HA) and neuraminidase (NA) antigens, as soon as the first pandemic influenza sequences became available, using immunoinformatic methods.Citation4 The HLA class II epitope predictions were later confirmed experimentally using peripheral blood mononuclear cells from human donors not exposed to the pandemic virus,Citation5 illustrating that pre-existing CD4+ T cells elicit cross-reactive effector responses against the pandemic H1N1 virus. In addition, they demonstrated that the computational tools were 90% accurate in predicting CD4+ T cell epitopes and their HLA-DR-dependent response profiles in donors that were chosen at random for HLA haplotype. As HA and NA antigens are the principle components of seasonal trivalent inactivated and subunit influenza vaccines and CD4+ T cells support both humoral and cellular influenza immunity, we have now performed a significantly expanded immunoinformatic analysis of the H1-HA and N1-NA sequence space to identify HLA class II-restricted immunogenic consensus sequences covering isolates dating back to 1980 from the end of the 2009 pandemic. The novel antigens were validated in HLA binding and T cell assays in preparation for future vaccine efficacy studies in HLA transgenic mice. We provide a detailed report on the methods used to define these highly cross-conserved influenza vaccine epitopes. The method may be of interest for the design of future H7N9, H5N1, and H3N2 vaccines.
Results
9-mer conservation and HLA binding potential
Influenza H1N1 HA and NA sequences dating back to 1980 were computationally screened in a step-wise process to identify conserved and potentially immunogenic epitopes (). 5 738 influenza A H1-HA sequences were collected, comprising 4 110 (71.6%) pandemic 2009 and 1 628 (28.3%) pre-pandemic sequences. A total of 3 200 273 9-mers were parsed from these sequences and duplicates were removed, leaving 16 247 unique 9-mers (0.5%). Of the unique HA 9-mers, 3 396 were predicted by EpiMatrix to bind to at least one of eight archetypal HLA class II alleles (20.1%). One hundred and sixty-eight 9-mers were found in more than 75% of the input HA proteins (), covering an average of 99% and 95% of pandemic and pre-pandemic sequences, respectively. A set of 5 396 influenza A N1-NA sequences was also assembled, including 3 574 (66.2%) pandemic and 1 822 (33.8%) pre-pandemic, post-1980 sequences. From these N1-NA sequences, 2 488 564 9-mers were parsed, 11 065 of which were classified as unique (0.4%). Of the unique NA 9-mers, 2 147 (19.4%) were predicted to bind to at least one HLA class II allele. One hundred and twenty-two 9-mers were found in more than 75% of the input NA proteins (), covering 99% of pandemic and 96% of pre-pandemic sequences.
Figure 1. Informatic-driven identification of influenza H1N1 hemagglutinin and neuraminidase immunogenic consensus sequences. Results of the step-wise computational process of screening H1-HA and N1-NA sequences for conserved and potentially immunogenic epitopes and constructing immunogenic consensus sequences are shown.
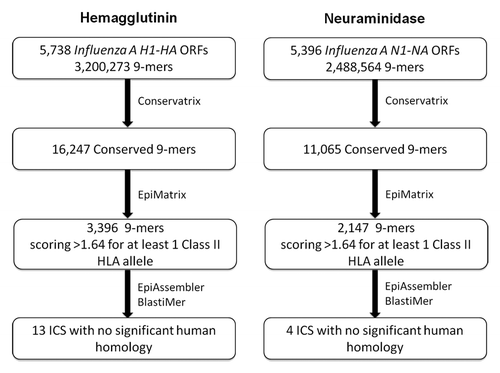
Figure 2. Conservation distribution of influenza H1N1 hemagglutinin and neuraminidase 9-mers. Pre-pandemic and pandemic H1-HA (A) and N1-NA (B) sequences were parsed into overlapping nine amino acid frames with a one amino acid frameshift, and unique 9-mers were evaluated for percent coverage of source antigens at 100% sequence identity using the Conservatrix algorithm. Inset: Close-up of the 75–100% coverage range.
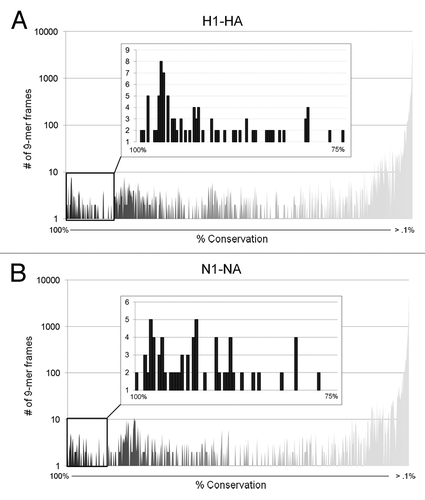
Interestingly, a second cluster of “high-range” conserved 9-mers was observed in the conservation distribution of both protein sets (). Three hundred 87 HA 9-mers (2.3%) and 339 NA 9-mers (3.1%) were found in 50–74% of input sequences. These sequences may represent regions that tolerate limited variability on the level observed in antigenic shifts. The remaining 15 692 HA (96.5%) and 10 604 NA peptides (95.8%) were found in <50% of input sequences with 14 671 HA and 9 744 NA peptides conserved in <1% of unique 9-mers, as is expected for these highly variable antigens.
ICS construction and selection
Drawing from the set of 9-mer sequences with > 75% conservation, a total of 19 immunogenic consensus sequences (ICS) derived from HA and eight ICS from NA were constructed by the EpiAssembler algorithm. This algorithm has been used to identify consensus sequences from highly variable proteins that are balanced for immunogenicity and conservation.Citation6,Citation7 One drawback to the approach is that given the large sampling of protein isolates, identifying multiple similar, though not identical, 9-mers that derive from the same region of HA or NA and retain significant HLA binding potential is highly likely. Indeed, 6 ICS from the HA set were eliminated due to redundant core peptide sequences. In this case, we selected the sequence that had the highest conservation from the set of redundant sequences. A final set of 13 HA and four NA ICS epitopes was retained based on the previously defined characteristics: high influenza strain coverage (> 85% conservation among input proteins), excellent immunogenicity scores (EpiMatrix cluster scores >10), and promiscuous HLA binding potential (, ). All HA and NA ICS are homologous to published HLA ligands and/or HLA DR-restricted T cell epitopes, according to a BLAST screen of the Immune Epitope Database at 90% coverage.Citation8
Figure 3. Immunogenic consensus sequence influenza H1N1 strain coverage and HLA binding potential. ICS construction yielded 13 H1-HA and four N1-NA sequences with > 85% coverage of input proteins and EpiMatrix cluster scores > 10, representing significant HLA binding potential. Bars represent EpiMatrix cluster score and open circles percent strain coverage.
Table 1. H1N1 immunogenic consensus sequences
By sequence comparison, we evaluated the potential for the selected H1N1 ICS to trigger immune responses from pre-existing T cells that bear TCR specific for epitopes contained in host or commensal antigens. We normally eliminate any such cross-reactive epitopes because activation of T cells cross-reactive with self or with commensals in the context of immunization may lead to unexpected immune responses that limit efficacy. For example, activation of cross-reactive host-specific natural regulatory T cell activation may dampen vaccine responses. Alternatively, T cells that express T cell receptors (TCRs) trained on commensal antigens may lead to induction of cross-reactive T cells with effector phenotypes, an event that may perturb immune homeostasis. Fortunately, by BLAST analysis, no homology between influenza and human sequences was identified in any of the final 17 ICS selections. Even though 12 ICS were observed to be homologous to murine, commensal, and other pathogen (non-influenza) sequences, they were not eliminated from the final set selected for experimental validation. None of these ICS bear TCR-facing sequences that are fully conserved with their murine and commensal homologs and preserve MHC binding potential, suggesting a low probability for cross-reactivity. Using a new tool that evaluates TCR facing residues, JanusMatrix,Citation9 we found no ICS shares a T cell receptor-facing pattern with predicted mouse MHC-binding sequences, despite H1-1 and N1-1 containing 9-mers with seven out of nine identities. Potential cross-reactivity between H1-7, H1-9, N1-1, and N1-4 ICS and human commensal sequences was also considered to be negligible, using JanusMatrix. In contrast, a screen of non-influenza pathogens uncovered multiple 9-mers with 7 out of 9 identities, although few were well matched by HLA allele and TCR-facing side chains. The “other pathogen” that matched most often to ICS peptides was Salmonella Typhi, with similar sequences observed in H1–2, -3, -4, -5, -8, and -13. Notably, in a screen of H5N1 influenza HA and NA sequences, five HA and four NA ICS were found to be homologous. This suggests that this methodology has identified influenza sequences that are both highly conserved and potentially immunogenic, enhancing their utility as universal influenza vaccine candidate epitopes.
HLA binding
ICS peptides were assayed in vitro for their capacity to bind multiple HLA types, including DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*1101, and DRB1*1501. Of the 108 ICS peptide-HLA binding interactions assayed, 3% bound with very high affinity, 23% bound with high affinity, 26% bound with moderate affinity, 30% bound with low affinity, 2% with very low affinity, and 15% had no affinity for the HLA tested ().
Figure 4. ICS peptide – HLA DR binding affinities. Peptide identifiers and sequences are noted in the first and second columns, respectively. IC50 values in μM units were calculated from curves fitted to dose-dependence competition binding data for each peptide-HLA DR allele pair. Peptide binding affinity is shown according to the following classification: IC50 < 0.1 µM (black), 0.1 µM < IC50 < 1 µM (dark gray), 1 µM < IC50 < 10 µM (gray), 10 µM < IC50 < 100 µM (light gray), IC50 > 100 µM (lightest gray). IC50 values too high to accurately measure under binding conditions tested are considered non-binders (NB; shown in white cells).
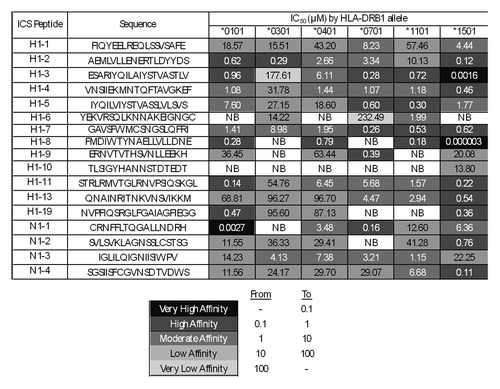
All (100%) of the peptides bound as predicted to at least three HLA alleles, 89% to at least four HLA alleles, 72% to at least five, and 39% to all six. A non-binder was defined as a confirmed (true negative) prediction if the peptide had an EpiMatrix Z-score that was lower than the defined cut-off (1.64) for its associated HLA allele. Positive predictions were defined as epitopes scoring ≥ 1.64 on the EpiMatrix Z-scale and binding HLA at any affinity. The concordance of computational predictions and binding assay results was evaluated with classification of peptide-HLA binding pairs as either true positive, false positive, true negative, or false negative. Overall, the concordance with predictions (both positive and negative) was 83%. With respect to each allele assayed, the values are 88% for DRB1*0101, 76% for DRB1*0301, 88% for DRB1*0401, 78% for DRB1*0701, 82% for DRB1*1101, and 82% for DRB1*1501. These HLA-binding and epitope prediction results are consistent with previously published studies using the same algorithms and assay conditions.Citation10,Citation11
Discrepancies between computational predictions and experimental results are expected, in part, because immunoinformatic algorithms are not 100% accurate. A recent retrospective evaluation of epitope mapping algorithm accuracy showed EpiMatrix was >75% accurate across all the HLA class II alleles studied here.Citation12 In comparison with other major prediction tools, EpiMatrix compared favorably, with equal or greater accuracy both overall and for individual alleles. Additionally, prediction/experimental discrepancies are introduced in binding assays by epitope-specific factors, including peptide design and the unique physical and chemical properties of individual peptides that, for example, may contribute to aggregation.
T cell reactivity
To further support the choice of these immunogenic consensus sequences as potential vaccine candidates, we stimulated human peripheral blood mononuclear cells (PBMCs) with ICS peptides and measured cytokine production to demonstrate that the peptides are immunoreactive. While HLA binding assays establish that a sequence can be presented to T cells, cytokine measurements demonstrate epitope antigenicity. Over a nine-day period, we expanded antigen-specific T cells from five young adult subjects, with unknown influenza infection and vaccination history, by stimulation with a pool of HA and NA ICS peptides. Cells were then re-stimulated with pooled or individual ICS peptides for measurement of interferon-γ (IFNγ) production by ELISpot assay. Cultured ELISpot assays were performed because ex vivo responses were not robust (data not shown), suggesting that antigen-specific T cell precursor frequencies were too low to observe without amplification. Cultured ELISpot responses were significantly greater than ex vivo responses and were considered positive when (1) the number of IFNγ spot-forming cells exceeded 50 per million PBMCs cultured, (2) spot counts were at least twice background, and (3) spot counts were statistically different from “no stimulus” measurements (p < 0.05). All subjects responded to the pool of ICS peptides (, ). Apart from Subject 844, all pooled peptide re-stimulations elicited robust numbers of IFNγ-producing cells, ranging from 1127 to 4737 per million PBMCs with stimulation index values at least three times the cutoff value of two. It is possible Subject 844 did not respond strongly to the ICS peptides because of no prior history of H1N1 exposure or vaccination. These data show that H1N1 cross-reactive memory T cell precursors do exist and have the potential to be substantially expanded by vaccination with universal H1N1 antigens.
Figure 5. Antigen-specific human IFNγ ELISpot responses to computationally identified influenza HA and NA immunogenic consensus sequences. ICS were assayed for T cell reactivity by IFNγ ELISpot assay using PBMCs isolated from normal human donors (n = 5). An ELISpot response was considered positive if three criteria were met: (1) spot-forming cells (SFC) per million PBMC were at least 50 over background; (2) SFC per million PBMC were at least 2-fold over background; and (3) antigen-stimulated SFC numbers were statistically different (p < 0.05) from non-stimulated counts. (A) The numbers of SFC over background per million PBMCs that secrete IFNγ in response to individual and pooled influenza HA and NA ICS are presented. Individual subject responses are represented by dots and the average response across subjects by horizontal lines. The 50 SFC over background per million PBMCs cutoff is denoted by the dotted line. (B) The ELISpot response stimulation index, representing the ratio of antigen-stimulated SFC counts to non-stimulated counts, is presented. Stimulation index values per individual subject are represented by dots and the average values across subjects by horizontal lines.
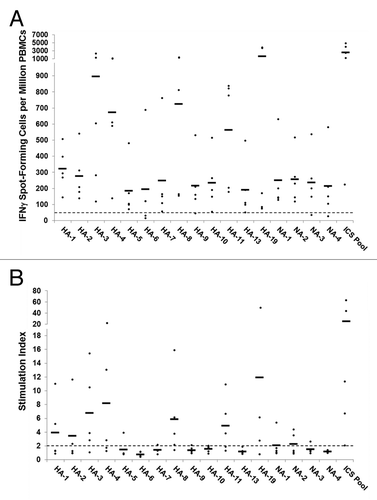
Table 2. Cultured human IFNγ ELISpot responses to influenza H1N1 ICS peptides
Individual ICS peptides stimulated significant IFNγ production in all subjects but Subject 844 (). Excluding this subject, positive responses per subject ranged from 24% to 65% over all 17 ICS peptides. By source antigen, positive responses ranging from 31% to 69% per subject were observed for the 13 HA ICS peptides and 0% to 50% for the four NA ICS peptides. Thirteen out of the 17 ICS peptides (76%) stimulated positive responses in at least one subject. By source antigen, 10 out of 13 HA ICS (77%) and 3 out of 4 NA ICS (75%) peptides were positives. Two ICS peptides, HA-3 and HA-8, were immunoreactive in all cases except Subject 844, suggesting that T cells specific for these sequences may be immunodominant. Unexpectedly, four ICS peptides stimulated no responses in any subjects. The small cohort size of the study with limited HLA diversity may explain this result. Alternatively, these sequences may stimulate type 1 helper T cell cytokines other than IFNγ, such as interleukin-2 and tumor necrosis α. Additionally, vaccination may increase the numbers of precursor cells to the level needed for detection by cultured ELISpot. Thus, positive responses may be observed in a larger cohort with broader HLA coverage, comparing multiple cytokine responses in samples drawn before and after vaccination. Future studies will address these factors.
Discussion
Cross-reactive T cell epitopes such as the ones identified here may have played a significant role in containing the human impact of the 2009 influenza H1N1 pandemic. Despite studies showing pandemic H1N1 was highly pathogenic in laboratory animals and shared few B cell epitopes with most seasonal H1N1 viruses,Citation13 the virus triggered only mild symptoms in middle-aged and elderly adults and, fortunately, failed to cause widespread morbidity and mortality.Citation14 One explanation for this unexpected observation is that pre-existing influenza-specific CD4+ T cells generated cross-reactive cellular responses against the virus that were capable of limiting disease severity and virus spread in individuals lacking cross-protective humoral immunity. This hypothesis is supported by a number of in vitro and in vivo studies: independent studies demonstrated cytotoxic T lymphocytes (CTLs) and CD4+ T cells raised against the seasonal H1N1 viruses, A/Brisbane/59/2007 and A/New Caledonia/20/99, respectively, were capable of responding against whole protein antigens from the pH1N1 virus.Citation5,Citation15-Citation17 In mice and in humans, memory T cells to conserved epitopes have been shown to confer protection to heterotypic infection.Citation18,Citation19 In addition, cross-reactive human T helper cell responses were observed for HLA-DR4 epitopes.Citation20 Moreover, ferrets infected with seasonal H1N1 influenza, though lacking sterilizing immunity, were protected from disease upon subsequent pH1N1 infection.Citation21 During the pandemic, a lower hospitalization rate and lower reports of H1N1 infection among recent seasonal vaccinees was observed in a case-control study in Mexico.Citation22 And finally, a T cell-driven influenza vaccine was recently shown to be protective against influenza challenge in human studies.Citation23 Taken together, these studies support the usefulness of including influenza antigens that can elicit cross-strain T helper cell responses in a universal influenza vaccine.
The activation of helper T cells is also critically important to the magnitude, quality, and kinetics of antibody response.Citation24 In the absence of functional (memory) CD4+ T cells, mouse studies have shown that the rate of viral clearance upon secondary infection slows considerably, beyond the degree seen in the primary response.Citation25-Citation27 Also in mice, cross-reactive memory T helper cells have been shown to contribute to cross-strain antibody responses.Citation28 In human populations, cross-reactive T cell responses have been observed between circulating strains of influenza and epidemic strains (such as H5N1) in the absence of cross-reactive antibodies.Citation29 Both cross-reactive CTLs and T helper cells have been identified by a number of investigators.Citation30,Citation31 T cell responses to conserved epitopes may be particularly important when new strains of influenza emerge.
The comprehensive approach to defining highly conserved H1N1 sequences described here builds on our initial analysis and identification of cross-conserved H1-HA and N1-NA T-cell epitopes that was published during the 2009 influenza A(H1N1) pandemic. A comparison of the results from these two independent analyses shows that the wider net cast for H1N1 sequences in the present study yielded a set of sequences, some of which are similar and some different from the original screen. Nine of 13 ICS derived from this collection of HA antigens and three of four NA ICS newly derived in the present work are similar to those identified in the initial analysis. Additionally, we previously identified three cross-conserved immunogenic sequences from each antigen, which were not sufficiently conserved in the larger sequence data set to be represented in the final selections made here. This comprehensive set of more highly conserved H1N1 sequences will be further tested in in vivo studies with HLA transgenic mice, before moving them forward into formulation studies for vaccine development purposes.
We used HLA binding as a proxy for immunogenicity, although binding is not an absolute indicator of immunogenicity potential. The results showed that the ICS peptides are promiscuous binders, suggesting they may be broadly immunoreactive. As well, they showed that the immunoinformatic predictions were highly (83%) accurate. Furthermore, in vitro T cell responses to 76% of the individual ICS peptides were observed in a small cohort of healthy subjects. In addition, the results confirmed epitopes that have been previously published, based on a search of the IEDB. While our report focuses on HLA class II-restricted CD4+ T helper epitopes, CTLs are also required for viral clearance, some of which may be CD4+.Citation32 Taken together, the combination of epitope prediction and HLA binding and immunoreactivity data here shows that these cross-conserved influenza sequences have promiscuous HLA binding and antigenicity properties required to identify broadly reactive, cross-protective H1N1 influenza-specific T cells raised in infection or vaccination and to serve as immunogens in a universal H1N1 influenza vaccine.
The level of conservation of these sequences among H1N1 sequences extending from 1980 to 2011 is quite remarkable and illustrates the capacity of epitope-mapping tools to discover high-value sequences for vaccine design. The immunoinformatics approach described here allows for the identification and characterization of cross-conserved T cell epitopes for any set of source sequences. Notably, this universal influenza vaccine development approach may be applied to other influenza subtypes (H7, H5, for example). We have also developed tools that identify highly cross-conserved CD8+ T cell responses and have validated their accuracy.Citation33-Citation35 Highly efficient algorithms, such as these, may be useful for accelerated development of vaccines against emerging infections in the context of newly emerging infections or bioterror events.Citation36
An important safety feature of the vaccine design approach described here is that T cell epitopes that have a high degree of cross-conservation with human genome are taken into consideration and eliminated from the list of epitopes to be tested and included in vaccine constructs, as there is at least initial evidence that such epitopes may be immunopathogenic (refs. Citation9,Citation37,Citation38 and Losikoff P et al, in preparation), or tolerated by the immune system, or they may stimulate regulatory T cell responses.Citation9
Accumulating evidence suggests that the sequences identified here may stimulate influenza-specific T helper cells that can limit disease through activation of cellular and humoral immune mechanisms reported to be critical for immunity. Not only do CD4+ T cells play a role in the rate of viral clearance,Citation25 but memory helper T cells specific to a previous influenza strain contribute to distinct cross-strain antibody responses.Citation28 Thus, influenza vaccine strategies that focus the T cell response on cross-reactive sequences may harness cellular and humoral mechanisms with the potential to provide group-common protection against disease.
Materials and Methods
Immunoinformatics
Sequence collection
Hemagglutinin and neuraminidase sequences were obtained from the NIAID Influenza Research Database (http://www.fludb.org).Citation39 H1N1 HA and NA sequences isolated from human hosts and deposited between January 1980 and June 2011 were downloaded and annotated by origin. Swine origin viral isolates were designated “Pandemic,” whereas all other isolates were designated “non-Pandemic H1N1,” according to the SOP for New Pandemic (H1N1) Classification (http://www.fludb.org/brcDocs/documents/2009H1N1vSOP.pdf).
Conservation analysis
Because the HLA binding groove accepts 9-mer peptides, input HA and NA sequences were parsed into overlapping 9-mer frames, irrespective of protein alignment, using the Conservatrix algorithm. All resulting 9-mers were ranked by their conservation within each antigen set.
EpiMatrix analysis
All unique 9-mers resulting from Conservatrix analysis were scored for binding potential against a panel of eight representative class II HLA alleles using the EpiMatrix algorithm.Citation40 The peptide-binding preferences of these alleles, HLA DRB1*0101, *0301, *0401, *0701, *0801, *1101, *1301 and *1501, are expected to cover over 95% of human populations worldwide.Citation41 EpiMatrix scores for each allele are normalized on a Z-scale; peptides scoring >1.64 are called “hits.” These sequences represent the top 5% of any normally distributed set of 9-mer peptides and are highly likely to be true HLA ligands.
Construction of immunogenic consensus sequences
The EpiAssembler algorithm was employed to create immunogenic consensus sequences (ICS) using data collected during conservation analysis and EpiMatrix analysis.Citation7 ICS are class II epitope-length peptides of 20–25 residues each, whose composition has been enriched for both HLA binding potential and pathogen strain coverage using 9-mers from multiple isolates aligned by position in their native protein sequence. By definition, an ICS is seeded with a core 9-mer, and overlapping regions are subsequently screened for high-scoring, highly conserved 9-mer candidates to append until the optimal peptide length is reached. The resulting ICS peptides are compact clusters of putative T cell epitopes, each offering the advantage of targeting multiple strains and widely variable individual hosts using a single sequence. A conservation threshold was set such that the constituent 9-mer frames of each resulting ICS were required, in aggregate, to cover a minimum of 75% of input protein sequences.
Homology analysis
To eliminate peptide candidates unlikely to stimulate effector T cell responses, H1N1 ICS sequences were screened for homology against a set of host and commensal protein databases using the BLAST algorithm and parameters established as standards for short (peptide-length) sequences. Given that viral epitopes may stimulate regulatory T cell responses, ICS were screened against the complete human genome and the complete murine genome. Sequences were also screened against human commensals cataloged as part of the Human Microbiome Project (www.hmpdacc.org/resources/data_browser.php),Citation42 as well as the larger GenBank non-redundant protein database—excluding influenza—to identify homologous sequences in other pathogens. Standards have been established for the annotation and finishing of microbial genome sequences.Citation43,Citation44 All genomes at Finishing Level 3 (High Quality Draft) and above as of April 2011 were used to construct a local database of human microbiome sequences against which to screen H1N1 peptides. Finally, to establish whether any H1N1 ICS might stimulate cross-reactive T cell responses in other potential pandemic influenza outbreaks, peptides were screened against all H5N1 sequences isolated from humans and available at the GISAID database for influenza sequence sharing (http://platform.gisaid.org) as of June 2011. As a general practice, any ICS sharing more than seven amino acid identities per 9-mer frame was tagged; results of homology analysis against these five databases were reviewed on a case-by-case basis.
ICS selection
ICS peptides were manually reviewed to eliminate redundancy of core sequences due to similar, though not identical, motifs across isolates. Ambiguity due to sequences containing unidentifiable residues at certain positions was corrected. ICS with significant homology to host genomes were excluded, whereas sequence similarity to other organisms would not necessarily remove a peptide from the list of candidates. In one case, a highly hydrophobic ICS was hand modified by adding charged residues to the N-terminal flank of the peptide to increase the probability of successful peptide synthesis.
Peptide synthesis
Synthetic peptides were manufactured using 9-fluoronylmethoxycarbonyl (Fmoc) chemistry by 21st Century Biochemicals (Marlboro, MA). Peptide purity was >80% as ascertained by analytical reversed phase HPLC. Peptide mass was confirmed by tandem mass spectrometry.
HLA binding assay
Class II HLA binding assays were performed to screen predicted epitope sequences for binding to multiple HLA alleles. A competition-based HLA binding format was adapted from Steere et al.Citation45 Briefly, in racks of 1.1 ml tubes, non-biotinylated test peptides over a wide range of concentrations competed for binding to soluble class II molecules (2.5 nM) against a biotinylated standard peptide at a fixed concentration (25 nM) at 37°C for 24 h to reach equilibrium. Class II molecules were then captured on ELISA plates using pan anti-Class II antibodies (L243, anti-HLA-DR). Plates were washed and incubated with Europium-labeled streptavidin for one hour at room temperature. Europium activation buffer was added to develop the plates for 15–20 min at room temperature before they were read on a Time Resolved Fluorescence (TRF) plate reader. All assays were performed in triplicate. Dose dependence curves were generated by fitting data using the four-parameter logistic equation, and IC50 values were calculated in SigmaPlot 11.0 (Systat, Chicago, IL). Based on the IC50 values, peptide binding to a given HLA allele was classified as very high affinity (< 100 nM), high affinity (100–1000 nM), moderate affinity (1000–10 000 nM), low affinity (10 000–100 000 nM), or very low affinity (> 100 000 nM). Peptides that did not inhibit the binding of the biotinylated reference peptide at any concentration were considered non-binders. Binding assays were performed for six alleles: DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*1101 and DRB1*1501, providing a broad representation of class II HLA allele binding pockets.Citation41
PBMC collection and characterization
Frozen PBMCs donated by five healthy adults, ages 18 to 65 y, were generously provided by VaxDesign. No information on influenza vaccination or exposure was provided. Donor HLA class II types were determined using the One Lambda Micro SSPTM High Resolution HLA class II kit at the Hartford Hospital Transplant Immunology Laboratory. Human subject studies were performed in accordance with NIH regulations and with the approval of the Ethical and Independent Review Services institutional review board.
PBMC culture
Thawed whole PBMC populations were rested overnight and then expanded by antigen stimulation over nine days at 37°C under a 5% CO2 atmosphere. In a 48-well plate, 5 × 106 cells in 150 μl Iscove's Modified Dulbecco's Medium (IMDM) were stimulated with a pool of HA and NA ICS peptides at 10 μg/ml on Day 1. Three days later, IL-2 was added to 10 ng/ml and the culture volume raised to 300 μl. On Day 7, cells were supplemented with 10 ng/ml IL-2 by half media replacement. Finally, two days later, PBMCs were collected and washed in preparation for antigen re-stimulation to measure cytokine secretion measurements by enzyme-linked immunospot (ELISpot) assay.
ELISpot assay
Interferon-gamma (IFNγ) ELISpot assays were performed using kits purchased from Mabtech and performed according to the manufacturer’s specifications. Target peptides were added individually at 10 μg/ml and pooled at 1.25 μg/ml to triplicate wells containing 100 000 PBMCs in IMDM supplemented with 10% human AB serum. Cells were incubated for 48 h at 37°C under a 5% CO2 atmosphere. Triplicate wells were plated with PHA (10 μg/ml) as a positive control, and six wells with no peptide were used for background determination. Raw spot counts were recorded by ZellNet Consulting, Inc. using a Zeiss high-resolution automated ELISpot reader system and companion KS ELISpot software. Results were calculated as the average number of spots in the peptide wells, adjusted to spots per one million cells. A response was considered peptide-specific if the number of spots was at least twice background, greater than 50 spot forming cells per well (1 response per 20 000 PBMCs), and statistically different (p < 0.05) from that of the control wells by the Student’s t-test.
Disclosure of potential conflicts of interest
Two of the contributing authors, ASDG and WDM, are senior officers and majority shareholders at EpiVax, Inc., a privately owned biotechnology company located in Providence, RI. LM is employed by and holds stock options in EpiVax. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company.
Acknowledgments
We are grateful to Dr Donald Drake (VaxDesign) for providing PBMC samples and to Lauren Levitz for a thorough reading of the manuscript. This work was supported by NIH grant R21AI090359 (ADG).
References
- Centers for Disease Control and Prevention. 2012-2013 Influenza Season Week 8 ending February 23, 2013. Centers for Disease Control and Prevention. Retrieved from: http://www.cdc.gov/flu/weekly/.
- Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. [Erratum in: Lancet Infect Dis 2012; 12:655; PMID:22032844; 10.1016/S1473-3099] [11] [70295-X] Lancet Infect Dis 2012; 12:36 - 44; http://dx.doi.org/10.1016/S1473-3099(11)70295-X; PMID: 22032844
- Centers for Disease Control and Prevention (CDC). Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep 2009; 58:521 - 4; PMID: 19478718
- De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008-2009 conventional influenza vaccine. Vaccine 2009; 27:5740 - 7; http://dx.doi.org/10.1016/j.vaccine.2009.07.040; PMID: 19660593
- Schanen BC, De Groot AS, Moise L, Ardito M, McClaine E, Martin W, et al. Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4(+) T cells against the swine-origin H1N1 influenza virus. Vaccine 2011; 29:3299 - 309; http://dx.doi.org/10.1016/j.vaccine.2011.02.019; PMID: 21349362
- Koita OA, Dabitao D, Mahamadou I, Tall M, Dao S, Tounkara A, et al. Confirmation of immunogenic consensus sequence HIV-1 T-cell epitopes in Bamako, Mali and Providence, Rhode Island. Hum Vaccin 2006; 2:119 - 28; PMID: 17012903
- De Groot AS, Bishop EA, Khan B, Lally M, Marcon L, Franco J, et al. Engineering immunogenic consensus T helper epitopes for a cross-clade HIV vaccine. Methods 2004; 34:476 - 87; http://dx.doi.org/10.1016/j.ymeth.2004.06.003; PMID: 15542374
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, et al. The immune epitope database 2.0. Nucleic Acids Res 2010; 38:Database issue D854 - 62; http://dx.doi.org/10.1093/nar/gkp1004; PMID: 19906713
- Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Abdel Hady KM, et al. The two-faced T cell epitope: Examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother 2013; 9; In press http://dx.doi.org/10.4161/hv.24615; PMID: 23584251
- De Groot AS, Ardito M, Moise L, Gustafson EA, Spero D, Tejada G, et al. Immunogenic consensus sequence T helper epitopes for a pan-Burkholderia biodefense vaccine. Immunome Res 2011; 7:7; PMID: 22130150
- Moise L, McMurry JA, Pappo J, Lee DS, Moss SF, Martin WD, et al. Identification of genome-derived vaccine candidates conserved between human and mouse-adapted strains of H. pylori. Hum Vaccin 2008; 4:219 - 23; http://dx.doi.org/10.4161/hv.4.3.5394; PMID: 18376134
- De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol 2009; 131:189 - 201; http://dx.doi.org/10.1016/j.clim.2009.01.009; PMID: 19269256
- Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009; 361:1945 - 52; http://dx.doi.org/10.1056/NEJMoa0906453; PMID: 19745214
- Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, et al, 2009 Pandemic Influenza A (H1N1) Virus Hospitalizations Investigation Team. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N Engl J Med 2009; 361:1935 - 44; http://dx.doi.org/10.1056/NEJMoa0906695; PMID: 19815859
- Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A 2009; 106:20365 - 70; http://dx.doi.org/10.1073/pnas.0911580106; PMID: 19918065
- Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 2010; 185:4998 - 5002; http://dx.doi.org/10.4049/jimmunol.1001395; PMID: 20889549
- Tu W, Mao H, Zheng J, Liu Y, Chiu SS, Qin G, et al. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J Virol 2010; 84:6527 - 35; http://dx.doi.org/10.1128/JVI.00519-10; PMID: 20410263
- Boon AC, de Mutsert G, van Baarle D, Smith DJ, Lapedes AS, Fouchier RA, et al. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J Immunol 2004; 172:2453 - 60; PMID: 14764717
- Kreijtz JH, Bodewes R, van Amerongen G, Kuiken T, Fouchier RA, Osterhaus AD, et al. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 2007; 25:612 - 20; http://dx.doi.org/10.1016/j.vaccine.2006.08.036; PMID: 17005299
- Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol 2010; 84:3312 - 9; http://dx.doi.org/10.1128/JVI.02226-09; PMID: 20071564
- Ellebedy AH, Ducatez MF, Duan S, Stigger-Rosser E, Rubrum AM, Govorkova EA, et al. Impact of prior seasonal influenza vaccination and infection on pandemic A (H1N1) influenza virus replication in ferrets. Vaccine 2011; 29:3335 - 9; http://dx.doi.org/10.1016/j.vaccine.2010.08.067; PMID: 20840835
- Garcia-Garcia L, Valdespino-Gómez JL, Lazcano-Ponce E, Jimenez-Corona A, Higuera-Iglesias A, Cruz-Hervert P, et al. Partial protection of seasonal trivalent inactivated vaccine against novel pandemic influenza A/H1N1 2009: case-control study in Mexico City. BMJ 2009; 339:b3928; http://dx.doi.org/10.1136/bmj.b3928; PMID: 19808768
- Powell TJ, Peng Y, Berthoud TK, Blais ME, Lillie PJ, Hill AV, et al. Examination of influenza specific T cell responses after influenza virus challenge in individuals vaccinated with MVA-NP+M1 vaccine. PLoS One 2013; 8:e62778; http://dx.doi.org/10.1371/journal.pone.0062778; PMID: 23658773
- Kamperschroer C, Dibble JP, Meents DL, Schwartzberg PL, Swain SL. SAP is required for Th cell function and for immunity to influenza. J Immunol 2006; 177:5317 - 27; PMID: 17015717
- Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol 2002; 76:12388 - 93; http://dx.doi.org/10.1128/JVI.76.23.12388-12393.2002; PMID: 12414983
- Cardin RD, Brooks JW, Sarawar SR, Doherty PC. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med 1996; 184:863 - 71; http://dx.doi.org/10.1084/jem.184.3.863; PMID: 9064346
- Brooks JW, Hamilton-Easton AM, Christensen JP, Cardin RD, Hardy CL, Doherty PC. Requirement for CD40 ligand, CD4(+) T cells, and B cells in an infectious mononucleosis-like syndrome. J Virol 1999; 73:9650 - 4; PMID: 10516078
- Marshall D, Sealy R, Sangster M, Coleclough C. TH cells primed during influenza virus infection provide help for qualitatively distinct antibody responses to subsequent immunization. J Immunol 1999; 163:4673 - 82; PMID: 10528164
- Lee LY, Ha LA, Simmons C, de Jong MD, Chau NV, Schumacher R, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest 2008; 118:3478 - 90; PMID: 18802496
- Townsend AR, Skehel JJ. The influenza A virus nucleoprotein gene controls the induction of both subtype specific and cross-reactive cytotoxic T cells. J Exp Med 1984; 160:552 - 63; http://dx.doi.org/10.1084/jem.160.2.552; PMID: 6206181
- Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C, et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol 2008; 82:12241 - 51; http://dx.doi.org/10.1128/JVI.01563-08; PMID: 18842709
- McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, et al. T cell responses are better correlates of vaccine protection in the elderly. J Immunol 2006; 176:6333 - 9; PMID: 16670345
- Levitz L, Koita OA, Sangare K, Ardito MT, Boyle CM, Rozehnal J, et al. Conservation of HIV-1 T cell epitopes across time and clades: validation of immunogenic HLA-A2 epitopes selected for the GAIA HIV vaccine. Vaccine 2012; 30:7547 - 60; http://dx.doi.org/10.1016/j.vaccine.2012.10.042; PMID: 23102976
- De Groot AS, Levitz L, Ardito MT, Skowron G, Mayer KH, Buus S, et al. Further progress on defining highly conserved immunogenic epitopes for a global HIV vaccine: HLA-A3-restricted GAIA vaccine epitopes. Hum Vaccin Immunother 2012; 8:987 - 1000; http://dx.doi.org/10.4161/hv.20528; PMID: 22777092
- De Groot AS, Rivera DS, McMurry JA, Buus S, Martin W. Identification of immunogenic HLA-B7 “Achilles’ heel” epitopes within highly conserved regions of HIV. Vaccine 2008; 26:3059 - 71; http://dx.doi.org/10.1016/j.vaccine.2007.12.004; PMID: 18206276
- De Groot AS, Ardito M, Terry F, Levitz L, Ross TM, Moise L, et al. Low immunogenicity predicted for emerging avian-origin H7N9: Implication for influenza vaccine design. Hum Vaccin Immunother 2013; 9; In press http://dx.doi.org/10.4161/hv.24939; PMID: 23807079
- Elfaki ME, Khalil EA, Degroot AS, Musa AM, Gutiérrez Núñez A, Younis BM, et al. Immunogenicity and immune modulatory effects of in silico predicted L. donovani candidate peptide vaccines. Hum Vaccin Immunother 2012; 8:1769 - 74; http://dx.doi.org/10.4161/hv.21881; PMID: 22922767
- Poland GA. Vaccines against Lyme disease: What happened and what lessons can we learn?. Clin Infect Dis 2011; 52:Suppl 3 s253 - 8; http://dx.doi.org/10.1093/cid/ciq116; PMID: 21217172
- Squires RB, Noronha J, Hunt V, García-Sastre A, Macken C, Baumgarth N, et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi Viruses 2012; 6:404 - 16; http://dx.doi.org/10.1111/j.1750-2659.2011.00331.x; PMID: 22260278
- De Groot AS, Jesdale BM, Szu E, Schafer JR, Chicz RM, Deocampo G. An interactive Web site providing major histocompatibility ligand predictions: application to HIV research. AIDS Res Hum Retroviruses 1997; 13:529 - 31; http://dx.doi.org/10.1089/aid.1997.13.529; PMID: 9135870
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 1998; 160:3363 - 73; PMID: 9531296
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al, NIH HMP Working Group. The NIH Human Microbiome Project. Genome Res 2009; 19:2317 - 23; http://dx.doi.org/10.1101/gr.096651.109; PMID: 19819907
- Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, et al, Genomic Standards Consortium Human Microbiome Project Jumpstart Consortium. Genomics. Genome project standards in a new era of sequencing. Science 2009; 326:236 - 7; http://dx.doi.org/10.1126/science.1180614; PMID: 19815760
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, et al, Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science 2010; 328:994 - 9; http://dx.doi.org/10.1126/science.1183605; PMID: 20489017
- Steere AC, Klitz W, Drouin EE, Falk BA, Kwok WW, Nepom GT, et al. Antibiotic-refractory Lyme arthritis is associated with HLA-DR molecules that bind a Borrelia burgdorferi peptide. J Exp Med 2006; 203:961 - 71; http://dx.doi.org/10.1084/jem.20052471; PMID: 16585267