Abstract
Rotarix® and RotaTeq® vaccines have led to a dramatic reduction in rotavirus disease worldwide. However, the detection of porcine circovirus type 1 (PCV-1) and 2 (PCV-2) DNA in these vaccines raised some safety concerns. Studies examining shedding of rotavirus in stool from rotavirus vaccine recipients have been performed but no published data exist regarding the shedding of PCV virus in stools of vaccinees. The goal of this study was to determine if PCV-1 and/or PCV-2 is shed in the feces of infants vaccinated with RotaTeq®. Using multiple PCR assays for detection of PCV DNA, we tested for PCV-1 and PCV-2 in 826 stool swab samples collected serially during the first 9 d after vaccination from 102 children vaccinated with RotaTeq®. Since the vaccine is recommended and uptake is high, we did not have samples from unvaccinated infants. A total of 235 (28.5%) samples from 59 vaccine recipients were positive for PCV-2 DNA by one or more assays used in this study. PCV-1 DNA was not detected in RotaTeq® or any of the stool swab extracts. Twenty-two of the 102 vaccine recipients (21.6%) shed RotaTeq® vaccine strain and 10 of these vaccinees (9.8%) were shedding both PCV DNA and rotavirus vaccine RNA. PCV DNA was detected up to 9 d post vaccination and was most frequently detected in the first 5 d after vaccination. This study demonstrated shedding of PCV-2 DNA by RotaTeq® vaccinees but we did not find evidence that this DNA was associated with viable PCV. Findings from this study support the continued use of current rotavirus vaccines.
Introduction
Group A rotaviruses are the most common cause of severe diarrheal disease in infants and children worldwide and are responsible for an estimated 453 000 deaths in children under five every yearCitation1 primarily in resource-poor countries of Africa and southeast Asia.Citation2 Two vaccines, Rotarix® (GlaxoSmithKline Biologicals) and RotaTeq® (Merck and Co, Inc.), are currently used in the US and other countries to reduce morbidity and mortality due to rotavirus.Citation3 The 2 live-attenuated vaccines are different in formulation and administration schemes and have been demonstrated to be highly efficacious in high- and middle-income countries and moderately effective in resource-poor countries of the world.Citation4-Citation6
Porcine circoviruses (PCV) are small (17–22 nm) isometric viruses that commonly infect pigs.Citation7 The PCV genome consists of circular, single stranded DNA molecules approximately 1900 bases in length.Citation8 While type 2 PCV (PCV-2) causes post-weaning multisystemic wasting syndrome (PWMS) in pigs, type 1 PCV (PCV-1) has not been associated conclusively with any porcine disease.Citation9 Neither PCV-1 nor PCV-2 is known to cause infection or illness in humansCitation8,Citation10,Citation11 but PCV DNA has been detected in 5% of stool samples from US adults, presumably a result of pork consumption. It is not known if PCV is able to replicate in the gastrointestinal tract of humans.Citation8
In early 2010, Rotarix® vaccine was found to contain PCV-1 DNA by microarray analysis and next generation sequencing.Citation11 In March 2010, the US Food and Drug Administration (FDA) released a recommendation for clinicians to temporarily suspend use of Rotarix® vaccine. In May 2010, the FDA reported findings that RotaTeq® also contained detectable levels of PCV-1 and PCV-2 DNA. Subsequent investigations suggested that PCV-containing material was most likely introduced into both rotavirus vaccines through porcine-derived materials such as trypsin, a reagent used during vaccine production in cell culture.Citation12,Citation13 Investigators have documented the presence of full length or nearly full-length sequence of PCV-1 DNA and infectious PCV-1 in Rotarix® vaccine.Citation11,Citation14-Citation17 In the RotaTeq® vaccine, short fragments of both PCV-1 and PCV-2 DNA were detected but no evidence of infectious virus has been reported.Citation15,Citation16
In May 2010, the FDA issued a recommendation for clinicians to resume the use of Rotarix® vaccine based on the observed benefits of rotavirus vaccines in preventing severe acute gastroenteritis among infants and available evidence that suggested only the theoretical possibility of PCV infection among humans. RotaTeq® use was never suspended by the FDA.
To date, no published data are available regarding shedding of PCV or PCV DNA in recipients of either Rotarix® or RotaTeq® vaccines. Increases in PCV DNA load in stools compared with the vaccine stocks would suggest that viral replication is taking place in vaccinees. This study reports efforts to detect PCV in stool samples from infants vaccinated with RotaTeq®.
Results
Multiplexed TaqMan assay, testing of vaccines and clinical samples
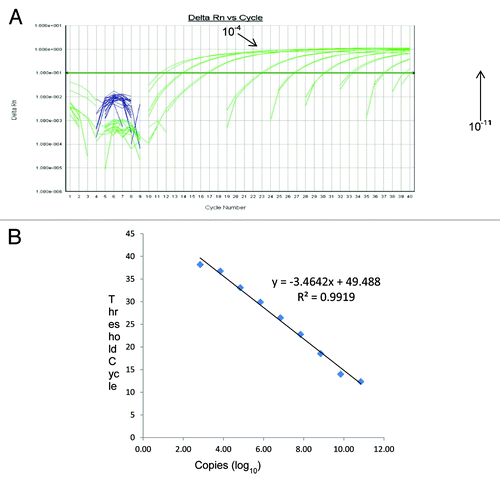
Assay sensitivity was determined by amplifying serial 10-fold dilutions of the PCV-1 and PCV-2 ssDNA oligonucleotide positive control standards of known concentration. The analysis of the calibration curves indicated that for PCV-1, the range of quantitative analysis covered an interval from 6.3 × 107 to 6 copies/reaction, corresponding to Ct values in the range of 12.2 to 38.4. The calibration curve indicated a linear correlation (R2 = 0.9966, slope = –3.4083) with an amplification efficiency of 96.8% (). Similarly, for PCV-2, the range of quantitative analysis covered an interval from 6.9 × 108 to 7 copies/reaction, corresponding to Ct values in the range of 12.35 to 39.2. The calibration curve indicated a linear correlation (R2 = 0.9919, slope = −3.4642) with an amplification efficiency of 94.5% (). Based on the range of Ct values obtained for both PCV-1 and PCV-2 control standards, samples with a Ct value ≤38.4 (PCV-1) and ≤39.2 (PCV-2) were considered positive.
Figure 1. (A) Amplification curves of 10-fold dilutions of PCV-1 positive control standards and (B) the linear relationship between threshold cycle (C) and log of copy number. Serial 10-fold dilutions were made using PCV-1 ssDNA oligonucleotide positive control standards. Dilutions used ranged from 10−3 to 10−11, corresponding to 6.3 × 107 to 6 copies/reaction, are included. The regression line and slope of the Ct value vs. log copy number graph were used for calculation of assay efficiency.
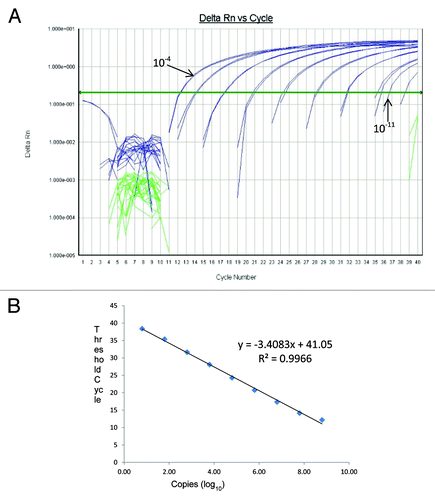
Figure 2. (A) Amplification curves of 10-fold dilution of PCV-2 positive control standards and (B) the linear relationship between threshold cycle (C) and log of copy number. Serial 10-fold dilutions were made using PCV-2 ssDNA oligonucleotide positive control standards. Dilutions used ranged from 10−3 to 10−11, corresponding to 6.9 × 107 to 7 copies/reaction, are included. The regression line and slope of the Ct value vs. log copy number graph were used for calculation of assay efficiency.
When testing vaccine stocks, PCV-1 DNA was detected at approximately 2.2 × 105 viral copies per reaction volume for Rotarix® vaccine stock and PCV-2 DNA was detected at approximately 8.4 × 103 viral copies per reaction volume for RotaTeq® stock, corresponding to 1.1 × 107 and 4.2 × 105 copies/vial of vaccine for Rotarix® and RotaTeq®, respectively. In stool samples, viral DNA was detected in the range of 1.26 × 103 to 3.24 × 106 (mean, 1 × 104; standard deviation, 3.6) copies per reaction volume of TNA. Using this assay to screen for PCV-1 and PCV-2 DNA in stool samples from vaccine recipients, none of the 55 RotaTeq® positive stool samples from vaccine recipients were positive for PCV-2 DNA, but 235 of 771 (30.5%) RotaTeq® negative stool samples from vaccinees were positive for PCV-2 DNA. PCV-1 DNA was not detected in any of the TNA extracts from stool samples ().
Table 1. Results of stool sample testing for PCV viruses by TaqMan and SYBR green assays
SYBR Green assay, testing of vaccines and clinical samples
The SYBR Green assay permits verification of amplified DNA as PCV-2 without opening the test plate by melt curve analysis of PCR products. A specific peak which melted at 83.5–84.5 °C was always observed for PCV-2 positive materials (). Out of the 55 RotaTeq® positive stool samples and 771 RotaTeq® negative stool samples, a melt peak corresponding to that of PCV-2 DNA, was detected in 8 and 65 samples, respectively ().
Conventional hemi-nested PCR
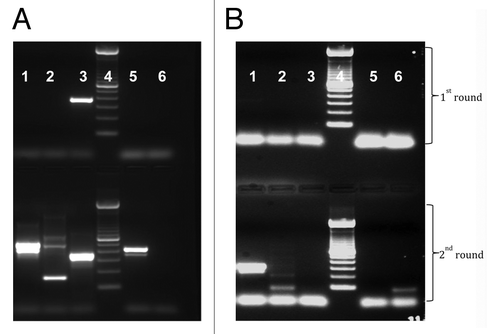
We tested for the presence of PCV-1 and PCV-2 DNA in vaccine stocks of Rotarix® and RotaTeq® (). We detected a PCV-1 product of the expected size in the Rotarix® vaccine and a PCV-2 amplicon of the correct size in RotaTeq®. PCV-1 was not detected in RotaTeq® vaccine stock and PCV-2 was not detected in Rotarix® vaccine stock. The amplicons obtained for PCV-1 and PCV-2 were sequenced and aligned with PCV sequences in GenBank. The approximately 300 bp sequence obtained for PCV-1 was 100% identical with PCV-1, strain CCL33-Urgent (accession number JN133303) previously isolated from PK-15 cell culture and 99% identical to previously reported PCV-1, strain PCV3_Rotarix_con1 (accession number HM143844) isolated from Rotarix vaccine A41XA799A.Citation11 The 236 bp DNA sequence obtained for PCV-2 was 99% identical to wild type PCV-2, strain 08p19B06_630–2 (accession number JN575645) detected from a swine sample collected in 2011 in the USA.
Figure 4. Presence of short fragments of PCV-1 and PCV-2 DNA in Rotarix® and RotaTeq® vaccines, respectively. (A) Conventional PCR using sequence specific primers for PCV-1. (B) Conventional PCR using sequence specific primers for PCV-2. For PCV-1, expected amplicon size for 1st and 2nd round PCR is 349 bp and 317 bp, respectively, and for PCV-2, 294 bp and 262 bp, respectively. For both gel A and B, samples were loaded in the following order: lane 1 = RotaTeq®; Lane 2 = Rotarix®; lane 3 = 100 bp molecular DNA ladder; lane 4 = stool sample, and lane 5 = negative control.
Using these two hemi-nested assays to detect PCV-1 and PCV-2 DNAs in stool TNA extracts, no amplicons of sizes specific for neither PCV-1 nor PCV-2 were obtained (data not shown).
Collective data analysis
Out of a total of 826 samples tested, 235 (28.5%) were found to contain fragments of PCV DNA. For samples positive by either the multiplexed TaqMan and/or SYBR Green assays, PCV-2 DNA was excreted throughout all 9 post-vaccination days with the peak detection of PCV-2 DNA on days 3 (34.07%) and 4 (36.79%) (). Of the 22 RotaTeq®-shedding vaccinees tested, only 10 were excreting both PCV and RotaTeq® (). Though PCV positive samples were found on all 9 post-vaccination days, they were most frequently detected in specimens collected on days 1–5 post-vaccination. On the other hand, RotaTeq® shedding was mostly detected in samples collected on days 6–9 post-vaccination. While 10 vaccinees excreted both rotavirus vaccine and PCV-2 over the 9 d post-vaccination period, both PCV-2 and RotaTeq® were never detected concurrently (). In these PCV-2 positive samples, the number of PCV-2 genomic copies ranged between approximately 900 and 5000 per reaction and was relatively constant over the 9 d study period ().
Figure 5. Percentage of stool samples positive for PCV-2 by multiplexed TaqMan and SYBR Green assays (black dotted shading) and multiplexed TaqMan or SYBR Green assay (solid gray shading).
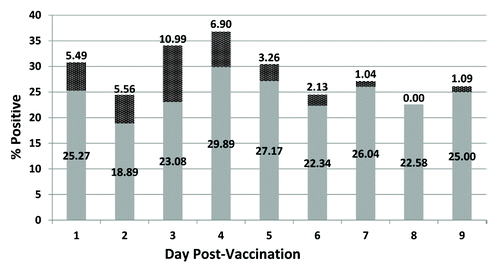
Table 2. Temporal shedding of PCV-2 and rotavirus in RotaTeq® vaccinees*
Discussion
In 2010, the finding of PCV components in Rotarix® and RotaTeq® vaccines caused great concern about the safety of current rotavirus vaccines. Though extensive clinical data supports the safety of both vaccines,Citation10,Citation23 there is still concern among expectant mothers or parents with children eligible for vaccination.Citation23 The shedding of rotavirus vaccine strains in RotaTeq® and Rotarix® vaccinees has been documentedCitation18,Citation24 hence the question remained whether or not these vaccinees were also excreting PCV.
In this study, we have investigated the excretion of PCV DNA among infants vaccinated with their first dose of RotaTeq®. This study reveals evidence of PCV-2 DNA excretion among the vaccinees but no evidence of PCV-1 DNA excretion. Out of the 826 stool swabs samples collected from 102 RotaTeq® vaccinees, a total of 235 out of 771 (30.5%) RotaTeq® negative stools and none of the 55 RotaTeq® positive stools samples from 59 vaccine recipients were positive for PCV-2 DNA by one or more assays used in this study. Though, we were able to detect PCV-2 DNA in multiple samples by qPCR, all attempts to obtain amplicons by conventional PCR for sequence confirmations were unsuccessful. This suggests that PCV-2 DNA is present in small fragments and/or at very low levels.
In the group of 10 vaccinees that were excreting both RotaTeq® and PCV, PCV-2 DNA was not detected in samples that were RotaTeq® positive and vice visa. Given these findings, we hypothesize that the presence of rotavirus dsRNA interferes with the detection of PCV ssDNA, perhaps by competition between the two types of nucleic acid during the extraction process. During the TNA extraction, the magnetic beads could become saturated with rotavirus nucleic acid, which is usually present at a very high concentration in positive samples, thus reducing or eliminating binding sites for PCV DNA which is apparently present at a relatively low copy number.
Furthermore, we have confirmed the presence of PCV-1 and PCV-2 DNA in single lots of Rotarix® (R41CA817A) and RotaTeq® vaccines (1660Z) at a concentration of 1.1 × 107 copies per vial of vaccine and 4.2 × 105 copies per vial of vaccine, respectively. These values are within the range that has been reported by other investigators.Citation11,Citation14-Citation16,Citation25 In this study, the PCV-1 DNA level found in the Rotarix® vaccine is high, but correlates with a previous report that the PCV-1 DNA load in Rotarix® exceeds 105 DNA molecules per dose.Citation11 PCV-1 DNA titers slightly lower or similar to what was found in this study have also been reported.Citation11,Citation14,Citation15 For PCV-2 in RotaTeq®, the value obtained here is also slightly higher than what has been reported.Citation15,Citation16,Citation25 Gilliland and colleagues reported only traces of PCV-2 DNA in RotaTeq® which could not be quantified.Citation15 Although the presence of PCV-1 DNA has been detected and documented in different materials used at various stages in the production of RotaTeq® vaccine and in several lots of final container of the RotaTeq® vaccine, albeit at low concentrations,Citation15,Citation16,Citation25 we could not detect or confirm the presence of PCV-1 DNA in RotaTeq® vaccine stock by using conventional PCR and qRT-PCR. After several attempts to amplify the full length PCV-1 and PCV-2 genomes from RotaTeq® failed, we concluded that PCV-1 and PCV-2 DNA detected in the rotavirus vaccines lots used in this study either existed as small fragments or that if entire PCV-1 or PCV-2 genomes were present, their concentrations are too low to be detected by our conventional PCR.
A prerequisite for any infectious virus is the presence of intact viral genomes.Citation25 Our inability to detect full-length or nearly full-length PCV-1 or PCV-2 sequences in stool samples from vaccinees suggests the absence of any infectious particles. Detection of a full-length or nearly full-length PCV-1 DNA genome has been reported in Rotarix® stocks but not in RotaTeq®.Citation14,Citation25 Also, the infectivity of PCV-1 in Rotarix® vaccine lots and non-infectious nature of PCV-2 detected in RotaTeq® vaccine lots has been reported.Citation16
Three limitations of this study should be noted. Because the samples used for these analyses were collected for a RotaTeq® rotavirus vaccine strain study,Citation18 the data reported here does not include those from samples collected from children pre-vaccination. Since the vaccine is recommended and uptake is high, we did not have samples from unvaccinated infants. Therefore, it is not possible to definitively state that these infants did not come into contact with PCV from some other source. Also, no internal positive control was included in these analyses. Another limitation of this study was the lack of a PCV-positive control that could be spiked into negative stool, to show that the extraction process does not inhibit PCV DNA detection post-extraction by qRT-PCR and PCR. Finally, this study only focuses on RotaTeq® vaccine, hence we do not have similar data for the Rotarix vaccine which has been shown to contain viable PCV-1 virus.Citation16 A similar study investigating the excretion of Rotarix rotavirus vaccine strain in stool collected from children vaccinated with Rotarix is underway.
This study reports for the first time the excretion of PCV-2 DNA in stool samples from RotaTeq® vaccinees. We also describe the detection and confirmation of PCV-1 and PCV-2 in Rotarix® and RotaTeq® vaccine stocks, respectively. PCV-1 and PCV-2 were found at low levels in vaccines and do not appear to cause human infections. Hence these findings support the continued use of current rotavirus vaccines.
Materials and Methods
Samples, controls, and study population
Between the period of May 2008 and February 2009, a total of 826 stool swab samples were collected from 102 infants, 6 to 12 weeks of age vaccinated with their first dose of RotaTeq®. Study participants were recruited at the Broadway Practice of the Ambulatory Care Network of New York-Presbyterian Hospital, located in the borough of Manhattan, New York City, NY. Stool samples were collected using double-ended swabs once daily during the 9 d following vaccination, as described previously.Citation18 All swabs were identified only by a unique study number, which was not linked to date of specimen collection, patient name, medical record number, or any other individual identifying information. As part of a previously-reported RotaTeq® shedding study, one end of each swab was processed and tested by rotavirus EIA and genotyping.Citation18
Commercially available RotaTeq® (1660Z) and Rotarix® (R41CA817A) were used as positive controls. Each dose of RotaTeq® vaccine is supplied as a 2 mL liquid for oral administration, while Rotarix® vaccine is provided as a lyophilized pellet that is reconstituted before administration using 2 mL diluent provided by manufacturer.
PCV-1 and PCV-2 standard curve preparation
PCV-1 (PCV1PC, Mfg. ID 92894593, IDT) and PCV-2 (PCV2PC, Mfg. ID 92894594, IDT) single-stranded DNA (ssDNA) oligonucleotide positive control standards were used to generate the standard curves use for calculating the copy numbers. The sequences of the PCV1PC and PCV2PC were as follows; 5′-TACTGTTCCT TTTTTGGCCC GCAGTATTTT GATTACCAGC AATCAGGCCC CCCAGGAATG GTACTCCTCA ACTGCTGTCC CAGCTGTAGA AGCTCTCTAT-3′ and 5′-AACTGTACCT TTTTTGGCCC GCAGTATTTT GATTACCAGC AATCAGACCC CGTTGGAATG GTACTCCTCA ACTGCTGTC CCAGCTGTAG AAGCTCTCTA T-3′, respectively. Each of the positive control standards was diluted serially from 10−1 to 10−14 in nuclease-free water (Ambion®, Inc.) containing 100 µg/µL of yeast RNA (Bio-Rad Laboratories).
Total nucleic acid extraction from stool samples and vaccines
Fecal material from each stool swab was eluted into 500 µL of phosphate buffered saline (PBS) to yield an approximately 20% stool suspension. Total nucleic acid (TNA) from each of the 826 specimens previously screened for rotavirus antigen using EIA (55 were confirmed as RotaTeq® rotavirus vaccine strain by RT-PCR and nucleotide sequencing and 771 rotavirus vaccine negative)Citation18 was extracted using the MagNA Pure 96 instrument and the DNA and Viral NA Small Volume kit (Roche Applied Science), while vaccine lots were extracted using the MagNA Pure Compact instrument and NA Isolation Kit I (Roche Applied Science). To avoid any possible cross-contamination, the RotaTeq® and Rotarix® vaccines were extracted in separate runs. With the exception of Rotarix®, in which diluent was used as negative control, each extraction run included a PBS blank as negative control. All extractions were performed according to the manufacturer’s instructions. TNA extracts were either analyzed immediately by real-time PCR or stored at −80 °C until tested.
Testing for PCV-1 and PCV-2 in stool samples from RotaTeq®vaccinees
Four different assays, including two real-time PCR-based assays (a multiplexed TaqMan PCV-1 and PCV-2 assayCitation19 and a SYBR Green PCV-2 real-time PCRCitation20) and two conventional hemi-nested PCR assays (one specific for PCV-1Citation21 and the other for PCV-2), were used to screen stool TNA extracts for PCV-1 and PCV-2. The multiplexed TaqMan assay was chosen as the primary assay because of its sensitivity, 6.5 and 43.6 copies/reaction for PCV-1 and PCV-2, respectively,Citation19 and because it was designed to detect and quantify short fragments of PCV-1 and PCV-2 DNA in clinical (serum) specimens.Citation19 The SYBR Green assay works on the principle of melt curve analysis and was used as a confirmatory assay for PCV-2. Samples found to contain short fragments of PCV-2 DNA by either qRT-PCR or SYBR Green analysis were further evaluated for the presence of longer fragments of PCV DNA using the PCV-1 specific conventional hemi-nested PCR or PCV-2 specific conventional hemi-nested PCR.
Multiplexed TaqMan assay
The multiplexed TaqMan assay used the PCV-1 and PCV-2 specific primers and probes designed to target a highly conserved region in the ORF1 of all PCV strains as described by Puvanendiran et al.Citation19 with the following modifications: (1) the detector and quencher dyes in the PCV-1 and PCV-2 probes were changed to 5′-FAM-CCAGCAATCA GGCCCCCCAG GAAT-BHQ1-3′ and 5′-Quasar670-CCA GCAATCAGAC CCCGTTGGAA TG-BHQ3-3′, respectively; (2) The reaction composition and conditions were changed: Each real-time PCR reaction contained 1 × TaqMan Universal PCR master Mix No AmpErase® UNG (Applied Biosystems), 600 nM each of forward (PCV-F; 5′-TGGCCCGCAG TATTTTGATT-3′) and reverse (PCV-R; 5′-CAGCTGGGAC AGCAGTTGAG-3′) primers, PCV-1 and PCV-2 probes in a final concentration of 300 nM and 150 nM, respectively, and 5 µl of TNA extract (or water for no template controls [NTCs]) in a 25 µl reaction volume. Each sample was tested in duplicate. Thermal cycling, fluorescent data collection, and data analysis were performed using the ABI 7500 Fast Real-Time PCR System (Applied Biosystems) with the 7500 Fast SDS software v1.4.0. The PCR program used was 10 min at 95 °C (1 cycle), followed by 40 cycles of amplification that consisted of denaturation at 95 °C for 15 s, and annealing and extension at 58 °C for 1 min. NTCs and positive controls were included in each run. On each plate, 10−2 to 10−14 dilutions of both PCV-1 and PCV-2 positive control standards were run in duplicate.
SYBR Green PCV-2 specific real-time PCR
A SYBR Green real-time PCR assay was designed using previously described PCV-2 specific primers CF8 and CR8.Citation20 Each 25 µl reaction contained 12.5 µl 2X iQTM SYBR® Green Supermix (Bio-Rad Laboratories), 500 nM of each primer, and 5 µl of TNA extract or water. Thermal cycling, fluorescent data collection, and data analysis were performed using the ABI 7500 Fast Real-Time PCR System (Applied Biosystems) with the 7500 Fast SDS software v1.4.0. The PCR program used was initial denaturation at 95 °C for 3 min (1 cycle), followed by 40 cycles that consisted of denaturation at 94°C for 30 s, annealing at 59 °C for 30 s, and extension at 72 °C for 30 s (fluorescent signal was collected after the end of each extension step). A final melting curve analysis of the PCR products was performed after amplification with heating from 60 °C to 95 °C in 0.5 °C steps with fluorescence recorded after holding for 10 s at each step. Negative and positive controls were included in each run.
Conventional hemi-nested PCR assay for PCV-1 and PCV-2
For PCV-1, a hemi-nested PCR assay was performed using previously described primers and conditions.Citation21 These primers were designed to amplify 349 bp and 317 bp regions of the ORF1 for the first and hemi-nested second reactions, respectively. For PCV-2, a new, conventional hemi-nested PCR assay was developed using primers designed by an alignment of multiple PCV-2 strains from the GenBank sequence database.Citation22 The PCV-2 forward primer spanned nucleotides 151 to 172 (CDC-PCV2F1: 5′-CGCACCTTCG GATATACTGT CA-3′), the PCV-2 reverse primer spanned nucleotides 423–445 (CDC-PCV2R2: 5′-TGTAAACTAC TCCTCCCGCC ATA-3′), and the reverse primer for the hemi-nested PCR spanned nucleotides 394 to 413 (CDC-PCV2R1: 5′-AAGGCCACAG CCCWAACYTA-3′). The oligonucleotide primers were designed to amplify 294 bp and 262 bp regions of ORF1 in the first round and hemi-nested second round reactions, respectively.
The extracted TNAs from the vaccines and stool swabs were used as PCR templates for both PCV-1 and PCV-2 in the first reaction, and 5 µL of the amplified product from both PCV-1 and PCV-2 assays, was used for the second reaction. The hemi-nested amplification was performed in a 50 µL reaction mixture containing 1.25 mM MgCl2, 1 × PCR buffer, 0.2 mM of each dNTP, 1 µM of each primer, and 2.5 U of Platinum Taq DNA polymerase (Perkin ElmerBoth reactions were run in a GeneAmp PCR System 9600 Thermocycler (Perkin Elmer-Cetus) under the following conditions: denaturation at 95 °C for 1 min, primer annealing at 65 °C for 1 min, and extension at 72 °C for 1 min (35 cycles total) with a final extension step at 72 °C for 10 min. Amplified products were visualized by electrophoresis on a 3% agarose gel. DNA fragments of specific sizes were located by UV transillumination after staining with GelRedTM (Biotium, Inc.). Negative and positive controls were included in each run.
| Abbreviations: | ||
| PCV-1 | = | porcine circovirus type 1 |
| PCV-2 | = | porcine circovirus type 2 |
| FDA | = | US Food and Drug Administration |
| TNA | = | total nucleic acid |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:136 - 41; http://dx.doi.org/10.1016/S1473-3099(11)70253-5; PMID: 22030330
- WHO. Global and national estimates of deaths under age five attributable to rotavirus infection: 2004. http://wwwwhoint/immunization_monitoring/burden/Global_national_estimates_2004_deaths_under_age_five_attributable_to_rotavirus_infection_2004pdf 2006.
- de Zoysa I, Feachem RG. Interventions for the control of diarrhoeal diseases among young children: rotavirus and cholera immunization. Bull World Health Organ 1985; 63:569 - 83; PMID: 3876173
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 2010; 376:606 - 14; http://dx.doi.org/10.1016/S0140-6736(10)60889-6; PMID: 20692030
- Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira B, Victor JC, Gillard PH, Cheuvart BB, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 2010; 362:289 - 98; http://dx.doi.org/10.1056/NEJMoa0904797; PMID: 20107214
- Zaman K, Dang DA, Victor JC, Shin S, Yunus M, Dallas MJ, Podder G, Vu DT, Le TP, Luby SP, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet 2010; 376:615 - 23; http://dx.doi.org/10.1016/S0140-6736(10)60755-6; PMID: 20692031
- Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest 2000; 12:3 - 14; http://dx.doi.org/10.1177/104063870001200102; PMID: 10690769
- Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, et al. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 2010; 84:1674 - 82; http://dx.doi.org/10.1128/JVI.02109-09; PMID: 20007276
- Allan G, Krakowka S, Ellis J, Charreyre C. Discovery and evolving history of two genetically related but phenotypically different viruses, porcine circoviruses 1 and 2. Virus Res 2012; 164:4 - 9; http://dx.doi.org/10.1016/j.virusres.2011.09.013; PMID: 21945213
- Kuehn BM. FDA: Benefits of rotavirus vaccination outweigh potential contamination risk. JAMA 2010; 304:30 - 1; http://dx.doi.org/10.1001/jama.2010.863; PMID: 20606142
- Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, Delwart EL. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol 2010; 84:6033 - 40; http://dx.doi.org/10.1128/JVI.02690-09; PMID: 20375174
- Ranucci C. RotaTeq® (Rotavirus Vaccine, Live Oral, Pentavalent): Update on Porcine Circovirus (PCV). Presented at: Advisory Committee on Immunization Practices; 2010 Atlanta, GA; http://www.cdc.gov/vaccines/recs/ acip/downloads/mtg-slides-oct10/12-6-rota-PCVupdate.pdf.
- Friedland L. Rotarix® (Rotavirus Vaccine, Live Oral): GSK’s PCV1 Investigation. Presented at: Advisory Committee on Immunization Practices; 2010; Atlanta, GA; http://www.cdc.gov/vaccines/recs/acip/ downloads/mtg-slides-jun10/13-3-rota.pdf.
- Baylis SA, Finsterbusch T, Bannert N, Blümel J, Mankertz A. Analysis of porcine circovirus type 1 detected in Rotarix vaccine. Vaccine 2011; 29:690 - 7; http://dx.doi.org/10.1016/j.vaccine.2010.11.028; PMID: 21093497
- Gilliland SM, Forrest L, Carre H, Jenkins A, Berry N, Martin J, Minor P, Schepelmann S. Investigation of porcine circovirus contamination in human vaccines. Biologicals 2012; 40:270 - 7; http://dx.doi.org/10.1016/j.biologicals.2012.02.002; PMID: 22402185
- McClenahan SD, Krause PR, Uhlenhaut C. Molecular and infectivity studies of porcine circovirus in vaccines. Vaccine 2011; 29:4745 - 53; http://dx.doi.org/10.1016/j.vaccine.2011.04.087; PMID: 21569811
- Ma H, Shaheduzzaman S, Willliams DK, Gao Y, Khan AS. Investigations of porcine circovirus type 1 (PCV1) in vaccine-related and other cell lines. Vaccine 2011; 29:8429 - 37; http://dx.doi.org/10.1016/j.vaccine.2011.07.123; PMID: 21835219
- Yen C, Jakob K, Esona MD, Peckham X, Rausch J, Hull JJ, Whittier S, Gentsch JR, LaRussa P. Detection of fecal shedding of rotavirus vaccine in infants following their first dose of pentavalent rotavirus vaccine. Vaccine 2011; 29:4151 - 5; http://dx.doi.org/10.1016/j.vaccine.2011.03.074; PMID: 21477676
- Puvanendiran S, Stone S, Yu W, Johnson CR, Abrahante J, Jimenez LG, Griggs T, Haley C, Wagner B, Murtaugh MP. Absence of porcine circovirus type 1 (PCV1) and high prevalence of PCV 2 exposure and infection in swine finisher herds. Virus Res 2011; 157:92 - 8; http://dx.doi.org/10.1016/j.virusres.2011.02.012; PMID: 21352865
- Larochelle R, Antaya M, Morin M, Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods 1999; 80:69 - 75; http://dx.doi.org/10.1016/S0166-0934(99)00032-4; PMID: 10403678
- Kim J, Han DU, Choi C, Chae C. Differentiation of porcine circovirus (PCV)-1 and PCV-2 in boar semen using a multiplex nested polymerase chain reaction. J Virol Methods 2001; 98:25 - 31; http://dx.doi.org/10.1016/S0166-0934(01)00348-2; PMID: 11543881
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res 2011; 39:Database issue D32 - 7; http://dx.doi.org/10.1093/nar/gkq1079; PMID: 21071399
- Payne DC, Humiston S, Opel D, Kennedy A, Wikswo M, Downing K, Klein EJ, Kobayashi A, Locke D, Albertin C, et al. A multi-center, qualitative assessment of pediatrician and maternal perspectives on rotavirus vaccines and the detection of Porcine circovirus. BMC Pediatr 2011; 11:83; http://dx.doi.org/10.1186/1471-2431-11-83; PMID: 21943237
- Rivera L, Peña LM, Stainier I, Gillard P, Cheuvart B, Smolenov I, Ortega-Barria E, Han HH. Horizontal transmission of a human rotavirus vaccine strain--a randomized, placebo-controlled study in twins. Vaccine 2011; 29:9508 - 13; http://dx.doi.org/10.1016/j.vaccine.2011.10.015; PMID: 22008819
- Ranucci CS, Tagmyer T, Duncan P. Adventitious Agent Risk Assessment Case Study: Evaluation of RotaTeq(R) for the Presence of Porcine Circovirus. PDA J Pharm Sci Technol 2011; 65:589 - 98; http://dx.doi.org/10.5731/pdajpst.2011.00827; PMID: 22294581