Abstract
The concept of biological containment was developed as a strategy to prevent environmental dissemination of engineered live vaccine or drug delivery vehicles. A mutation in the gene encoding thymidylate synthase (thyA), a key enzyme in the pyrimidine biosynthetic pathway, has previously been shown to limit growth of L. lactis vectors under restrictive conditions. We hypothesized that further mutations in the pyrimidine biosynthetic pathway might enhance the stability and safety of live L. lactis vectors. We show that a double mutation in the genes encoding ThyA and CTP synthase (PyrG) in L. lactis confers double auxotrophy for both thymidine and cytidine. However, the combination of two mutations failed to enhance the biological containment phenotype of the engineered strain. In the absence of thymine/thymidine, the thyA mutant exhibited a strong bactericidal phenotype. However, creation of the double mutant caused the loss of this phenotype, though survival in the mouse GI tract was enhanced. The implications for biological containment of live L. lactis based delivery vectors are discussed.
Keywords: :
Lactococcus lactis has many advantages as a potential vaccine and drug delivery vehicle including an established safety profile, the existence of a large number of available genetic tools, and the possibility of oral administration.Citation1-Citation12 Many applications have thus been developed which utilize lactic acid bacteria including anti-viral/anti-bacterial vaccines, treatment for autoimmune diabetes, and anticancer therapies.Citation11,Citation13-Citation15 Using L. lactis as a vaccine or immunotherapeutic vector involves the heterologous expression of the antigen or the therapeutic molecule by the microorganism. However, despite the GRAS (Generally regarded as safe) status of the original strain, the resulting genetically modified microorganism (GMM) must fulfill several safety requirements.Citation16 The ability of an engineered microorganism to establish or disseminate in the environment is considered to be an undesirable event.Citation16 As such, the use of containment strategies to prevent or reduce environmental dissemination of engineered vectors is therefore recommended.Citation17
To optimize the safety of their Actobiotic™ L. lactis strains, the Actogenix biopharmaceutical company replaced the essential thymidylate synthase gene (thyA) with genes encoding therapeutic molecules such as the human Trefoil Factor 1 in the AG013 product.Citation6 Thymidylate synthase is responsible for the conversion of dUMP into dTMP. If thyA is absent, an external source of thymine or thymidine is required for the microorganism to produce dTTP thereby limiting the strain from disseminating into the environment due to the lack of available thymidine. Thymine/thymidine auxotrophic microorganisms undergo cell death in response to thymidine deprivation (a phenomenon termed “thymineless death”).Citation18
We recently investigated whether the deletion of another gene with a role in pyrimidine metabolism (the pyrG gene) could provide a basis for biological containment.Citation2 The pyrG gene encodes CTP synthase which converts UTP into CTP in a unique step in the de novo synthesis of pyrimidines in L. lactis. The pyrG-deficient vaccine carrier became auxotrophic for cytidine and the effect of cytidine deprivation was found to be mainly bacteriostatic rather than bactericidal. Our work suggested the possibility of combining both biocontainment strategies to enhance the safety of the L. lactis based vaccine delivery system, a phenomenon that we investigate here.Citation2
We originally created a ΔpyrG mutant in L. lactis through allelic replacement with the hly gene (encoding the listeriolysin antigen [LLO]) from L. monocytogenes (designated L. lactis ΔpyrG [P23:SEC-LLO]).Citation2 In the current study we created a thyA deletion mutant as well as a double ΔthyAΔpyrG mutant using gene replacement with a truncated thyA. To create a thyA mutant in L. lactis MG1363 and to create the L. lactis ΔthyAΔpyrG (P23:SEC-LLO) double mutant strain we utilized the pORI280/pVE6007 system.Citation19,Citation20 The chromosomal DNA region located upstream from the promoter of gene thyA(llmg_0964) was amplified using primers PRE-thyAFor-XbaI(5′- CCAATGGTTC TAGATTCACT AAGTCCAGC -3′) and PRE-ThyARev-BamHI(5′- ATACGACTTG GAAGGATCCA CACCACCTC -3′), resulting in a 928bp amplicon (PRE). The region located downstream from thyA gene was amplified using primers POST-thyAFor-XhoI (5′- TTCCAAGTCG TATCTCGAGT AACGACAAG -3′) and POST-thyARev-BglII (5′- TTTCCAGAAG ATCTTTCCAA GTTCCAAC -3′), resulting in a 719bp amplicon (POST).The complementary region (underlined sequence) in primers PRE-ThyARev-BamHI and POST-thyAFor-XhoI allowed the use of a mix of both amplicons to amplify the whole region with the primers PRE-thyAFor-XbaI and POST-thyARev-BglII. The resulting PCR product was thus flanked by XbaI and BglII restriction sites (bold characters in primer sequences) and the amplicon was subsequently digested by XbaI and BglII and ligated into a dephosphorylated and similarly digested plasmid, pORI280.Citation19 The ligation reaction was used to transform electro-competent E. coli EC101 (RepA+)Citation21 cells as described by Dower et al.Citation22 Ligation was performed using the Fast-link™ DNA ligation kit (Epicenter Biotechnologies). After electroporation, cells were plated onto LB agar containing 64 µg/ml X-Gal and 200 µg/ml Erythromycin and incubated at 37 °C for 24h. Plasmid pORI-PRE-POST was then extracted from E. coli EC101 (pORI-PRE-POST) using the QIAprep Spin Miniprep Kit (Qiagen) and sequenced to confirm the DNA sequence (GATC Biotech).
Electrocompetent L. lactis MG1363 (pVE6007) and L. lactis ΔpyrG (P23:SEC-LLO) RifR (pVE6007) were prepared, transformed with plasmid pORI-PRE-POST as described by Geertsma and Poolman, 2007Citation23 and Gerber and Solioz, 2007Citation24 and plated onto M17 agar supplemented with 0.5% (w/v) glucose, 0.5 M sucrose, 5 μg/ml erythromycin, and 64 μg/ml X-Gal (and 4 mM cytidine where required). Chromosomal integration and double crossover of pORI280 was performed by temperature shift as described previously.Citation2 20 µM thymidine and 4 mM cytidine were added as necessary. Subsequently, all strains were made rifampicin-resistant to permit analysis in complex microbial environments and all subsequent experiments were performed using the rifampicin-marked strains. In all cases the generation of spontaneous rifampicin-resistance did not impact the growth characteristics of the strains (data not shown).
Mutants and plasmids generated and utilized in this study, as well as growth and assay conditions are described in . Deletion of pyrG is predicted to result in the disruption of the only pathway available for the de novo synthesis of CTP. Strains where pyrG is deleted were able to grow only when broth was supplemented with cytidine (). Similarly, the thyA gene encodes thymidylate synthase which is responsible for the conversion of dUMP into dTMP. When thyA is absent, production of dTTP requires thymine or thymidine for the salvage pathway to be functional. Our ΔthyA mutants were able to grow only when broth was supplemented with thymidine (). The double ΔthyAΔpyrG mutant was auxotrophic for both cytidine and thymidine ().
Table 1. Strains and plasmids generated and used in this study
Figure 1. Growth curves of the L. lactis variants in presence/absence of cytidine and thymidine. Strains L. lactisMG1363 (closed circles), MG1363 ΔpyrG (P23:SEC-LLO) (open circles), MG1363 ΔthyA (closed triangles), and MG1363 ΔthyAΔpyrG (P23:SEC-LLO) (open triangles) were grown in broth (A) SA, (B) SA + cytidine 4mM, (C) SA + thymidine 20µM, or (D) SA + cytidine 4mM + thymidine 20 µM. The results are expressed as the mean ± SD (n = 3).
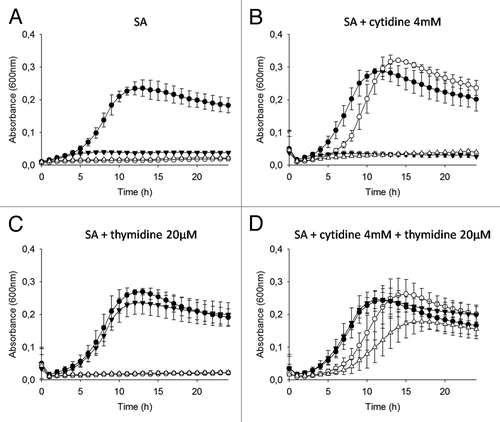
Whereas ΔpyrG and ΔthyA mutations have already been shown to have bacteriostatic or bactericidal effects respectively in non-supplemented broths,Citation2,Citation8 the consequences of combining ΔpyrG and ΔthyA mutations on long-term survival in broth or in the environment have not yet been investigated.
Survival of the different mutant strains were thus compared in a cytidine and thymidine-deprived defined broth (SA)Citation25 (). Strain L. lactis MG1363 was able to grow in SA though the resulting acidification of broth (reaching pH 3 by day 3) prevented us from carrying on the experiment with that strain after 3 d. However, this phenomenon was not encountered with the mutant strains due to reduced growth potential. The ΔpyrG mutant exhibited a decline in the number of viable cells from 1×108 to 3×105 CFU/ml over a 10 d period. As expected the ΔthyA mutant showed a more dramatic loss of viability over this time period. Interestingly the long-term survival of the double ΔthyAΔpyrG mutant in SA broth was consistently similar to that of the single ΔpyrG mutant suggesting no enhanced loss of viability over time due to the double mutation. Repeat experiments (data not shown) gave similar results. While the potential mechanisms are currently unclear, our results suggest that the presence of the ΔpyrG mutation prevents thymine-less deathCitation18 in these double mutation cells.
Figure 2. Survival of the different L. lactis strains in a cytidine and thymidine deprived broth and in environmental samples. Strains L. lactis MG1363 (closed circles), MG1363 ΔpyrG (P23:SEC-LLO) (open circles), MG1363 ΔthyA (closed triangles), and MG1363 ΔthyAΔpyrG (P23:SEC-LLO)(open triangles) were used to inoculate SA broth (A), soil (B), or river water (C).The results are expressed as the mean ± SD of at least 3 independent experiments.
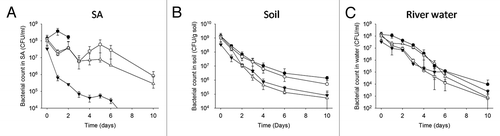
To mimic natural environmental conditions, survival of the different mutant strains was evaluated in autoclaved soil. No significant difference in survival was observed between the cytidine auxotroph and the wild type strain. However, we show that both the thymidine auxotroph and the double ΔthyAΔpyrG mutant were significantly more sensitive to persistence in soil than the wild type and the ΔpyrG mutant (0.033 < P value < 0.045 at Day 4, for instance) (). The double auxotrophic strain actually displays the greatest log reduction during the 10 d period in soil (−4.27 log compared with −2.98, −3.39, and −3.61 log for the wild type, the ΔpyrG and the ΔthyA mutant, respectively). Although combining the deletion of pyrG and thyA genes appears to provide a better containment of bioengineered L. lactis strains in soil, no significant differences between the different containment strategies was observed in river water (). We performed other experiments using sea water as a support medium (data not shown) and did not detect significant differences between the different strains and the wild-type. Overall therefore, we found rather minimal benefits to the biological containment of L. lactis provided by the various approaches outlined when strains were examined in complex environments. We determined that double mutation of the thyA and pyrG alleles in L. lactis failed to provide significantly enhanced bio-containment properties over the thyA mutant alone.
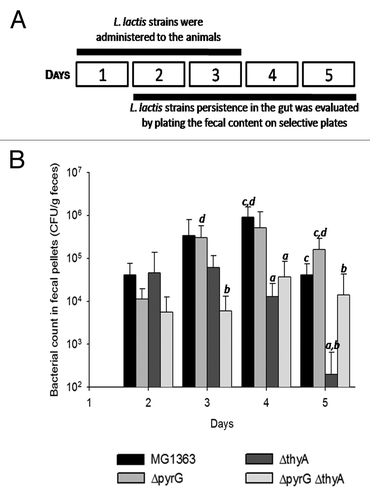
As engineered L. lactis strains are optimized for oral delivery and gastrointestinal applications, we evaluated the in vivo viability of the ΔpyrG and/or ΔthyA strains in orally inoculated mice. 109 L. lactis cells were administered to mice by gavage for 3 consecutive days and bacteria collected from the fecal pellets were enumerated from the second to the fifth day (). The number of viable wild type and ΔpyrG cells increased until day 4 before declining at day 5 (). Strains ΔthyA and ΔthyAΔpyrG failed to accumulate during the experiment. At day 5, strain ΔthyA was undetectable in 4 out of 5 analyzed fecal samples, whereas strain ΔthyAΔpyrG was still present with an average 1.4 × 104 CFU/g feces. Thus, after 5 d, all of the strains reach similar levels in the feces, with the exception of the single ΔthyA mutant which is almost cleared from the gastrointestinal tract (). Although a thyA-deficiency is deleterious for the viability of L. lactis in vivo, the deletion of thyA combined with the replacement of pyrG by the LLO-encoding gene hly enhances the ability of the strain to survive within the gastrointestinal tract.
Figure 3. Survival of the different L. lactis strains after passing through the gastrointestinal tract. (A) Schematic representing the experimental procedure. (B) Graph expressing the survival of the lactococcal strains following administration to mice by gavage and transition through the gastrointestinal tract. Bacteria were recovered from the faeces at the indicated CFU count. Results are expressed as the mean ± SD (n = 5). a, b, c, or d indicate that the value is significantly different (P value ≤ 0.05) from that affected to strains MG1363, ΔpyrG, ΔthyA, or ΔthyAΔpyrG, respectively.
ThyA-deficient strains have been shown to exhibit a high spontaneous mutagenesis rate in conditions deprived of thymine.Citation18 Although the occurrence of those spontaneous mutations may partly explain the thymineless death effect in ΔthyA mutants, the level of this phenomenon has not yet been evaluated for our double mutant ΔthyAΔpyrG strain. Future work will examine complementation of thyA and pyrG genes in our system to determine the possible influence of secondary mutations upon mutant phenotypes. Indeed the estimation of mutation rate in such strains would be an important consideration prior to their use in therapeutic settings.
Overall, we confirm that deletion of thyA in L. lactis promotes a bactericidal effect in the absence of thymine (thymineless death), but show that a double mutation in both thyA and pyrG compromises this effect. The increased survival potential of the double mutation has the effect of increasing survival of the mutant in the mouse GI tract and therefore may enhance vector delivery, but reduces the efficacy of biological containment. To our knowledge, significant analyses of the biological containment phenomenon for L. lactis vaccine vectors in natural environments are lacking. In our experiments, the lack of a dramatic containment phenotype in soil or natural water model systems suggests that much further work is necessary to determine the true efficacy of these biological containment approaches in the natural environment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors’ work was supported by grants from Science Foundation Ireland in the form of a center grant (Alimentary Pharmabiotic Centre; Grant Numbers SFI/12/RC/2273 and 12/RC/2273).
References
- Bahey-El-Din M, Casey PG, Griffin BT, Gahan CGM. Lactococcus lactis-expressing listeriolysin O (LLO) provides protection and specific CD8(+) T cells against Listeria monocytogenes in the murine infection model. Vaccine 2008; 26:5304 - 14; http://dx.doi.org/10.1016/j.vaccine.2008.07.047; PMID: 18691625
- Bahey-El-Din M, Casey PG, Griffin BT, Gahan CG. Efficacy of a Lactococcus lactis ΔpyrG vaccine delivery platform expressing chromosomally integrated hly from Listeria monocytogenes.. Bioeng Bugs 2010; 1:66 - 74; http://dx.doi.org/10.4161/bbug.1.1.10284; PMID: 21327128
- Bahey-El-Din M, Casey PG, Griffin BT, Gahan CGM. Expression of two Listeria monocytogenes antigens (P60 and LLO) in Lactococcus lactis and examination for use as live vaccine vectors. J Med Microbiol 2010; 59:904 - 12; http://dx.doi.org/10.1099/jmm.0.018770-0; PMID: 20488938
- Bahey-El-Din M, Gahan CGM. Lactococcus lactis-based vaccines: current status and future perspectives. Hum Vaccin 2011; 7:106 - 9; http://dx.doi.org/10.4161/hv.7.1.13631; PMID: 21263226
- Bahey-El-Din M. Lactococcus lactis-based vaccines from laboratory bench to human use: an overview. Vaccine 2012; 30:685 - 90; http://dx.doi.org/10.1016/j.vaccine.2011.11.098; PMID: 22154771
- Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, Vanhoenacker P, Corveleyn S, Watkins B, Sonis S, Coulie B, Rottiers P. AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil Factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol 2010; 46:564 - 70; http://dx.doi.org/10.1016/j.oraloncology.2010.04.008; PMID: 20542722
- Wells JM, Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat Rev Microbiol 2008; 6:349 - 62; http://dx.doi.org/10.1038/nrmicro1840; PMID: 18345021
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, Cox E, Remon JP, Remaut E. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 2003; 21:785 - 9; http://dx.doi.org/10.1038/nbt840; PMID: 12808464
- Steidler L, Rottiers P. Therapeutic drug delivery by genetically modified Lactococcus lactis.. Ann N Y Acad Sci 2006; 1072:176 - 86; http://dx.doi.org/10.1196/annals.1326.031; PMID: 17057198
- Vandenbroucke K, Hans W, Van Huysse J, Neirynck S, Demetter P, Remaut E, Rottiers P, Steidler L. Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology 2004; 127:502 - 13; http://dx.doi.org/10.1053/j.gastro.2004.05.020; PMID: 15300583
- Wang Z, Gao J, Yu Q, Yang Q. Oral immunization with recombinant Lactococcus lactis expressing the hemagglutinin of the avian influenza virus induces mucosal and systemic immune responses. Future Microbiol 2012; 7:1003 - 10; http://dx.doi.org/10.2217/fmb.12.69; PMID: 22913358
- Wells J. Mucosal vaccination and therapy with genetically modified lactic acid bacteria. Annu Rev Food Sci Technol 2011; 2:423 - 45; http://dx.doi.org/10.1146/annurev-food-022510-133640; PMID: 22129390
- Bolhassani A, Zahedifard F. Therapeutic live vaccines as a potential anticancer strategy. Int J Cancer 2012; 131:1733 - 43; http://dx.doi.org/10.1002/ijc.27640; PMID: 22610886
- Ribelles P, Benbouziane B, Langella P, Suárez JE, Bermúdez-Humarán LG. Protection against human papillomavirus type 16-induced tumors in mice using non-genetically modified lactic acid bacteria displaying E7 antigen at its surface. Appl Microbiol Biotechnol 2013; 97:1231 - 9; http://dx.doi.org/10.1007/s00253-012-4575-1; PMID: 23212671
- Takiishi T, Korf H, Van Belle TL, Robert S, Grieco FA, Caluwaerts S, Galleri L, Spagnuolo I, Steidler L, Van Huynegem K, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J Clin Invest 2012; 122:1717 - 25; http://dx.doi.org/10.1172/JCI60530; PMID: 22484814
- EU Council Directive 2009/41/EC on the contained use of genetically modified micro-organisms [Internet]. Official Journal L 125, 21/05/2009 P. 0075 - 0097; Available from: http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2009:125:0075:01:EN:HTML - Last accessed May, 21st 2012.
- Lee P. Biocontainment strategies for live lactic acid bacteria vaccine vectors. Bioeng Bugs 2010; 1:75 - 7; http://dx.doi.org/10.4161/bbug.1.1.10594; PMID: 21327129
- Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol 1998; 52:591 - 625; http://dx.doi.org/10.1146/annurev.micro.52.1.591; PMID: 9891809
- Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 1996; 253:217 - 24; http://dx.doi.org/10.1007/s004380050315; PMID: 9003306
- Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol 1992; 174:5633 - 8; PMID: 1324906
- Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 1995; 177:7011 - 8; PMID: 8522504
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 1988; 16:6127 - 45; http://dx.doi.org/10.1093/nar/16.13.6127; PMID: 3041370
- Geertsma ER, Poolman B. High-throughput cloning and expression in Lactococcus lactis. Protocol Exchange [Internet] 2007; Available from: http://www.nature.com/protocolexchange/protocols/287
- Gerber SD, Solioz M. Efficient transformation of Lactococcus lactis IL1403 and generation of knock-out mutants by homologous recombination. J Basic Microbiol 2007; 47:281 - 6; http://dx.doi.org/10.1002/jobm.200610297; PMID: 17518422
- Jensen PR, Hammer K. Minimal Requirements for Exponential Growth of Lactococcus lactis.. Appl Environ Microbiol 1993; 59:4363 - 6; PMID: 16349136
