Abstract
Cryo-transmission electron microscopy (cryoTEM) is a powerful characterization method for assessing the structural properties of biopharmaceutical nanoparticles, including Virus Like Particle-based vaccines. We demonstrate the method using the Human Papilloma Virus (HPV) VLPs in GARDASIL®. CryoTEM, coupled to automated data collection and analysis, was used to acquire images of the particles in their hydrated state, determine their morphological characteristics, and confirm the integrity of the particles when absorbed to aluminum adjuvant. In addition, we determined the three-dimensional structure of the VLPs, both alone and when interacting with neutralizing antibodies. Two modes of binding of two different neutralizing antibodies were apparent; for HPV type 11 saturated with H11.B2, 72 potential Fab binding sites were observed at the center of each capsomer, whereas for HPV 16 interacting with H16.V5, it appears that 60 pentamers (each neighboring 6 other pentamers) bind five Fabs per pentamer, for the total of 300 potential Fab binding sites per VLP.
Introduction
Human papillomavirus (HPV) vaccines based on major capsid protein L1 are licensed in over 100 countries to prevent HPV infections that result in cervical cancer and warts. The yeast-derived recombinant quadrivalent HPV L1 (Types 6, 11, 16, and 18) vaccine GARDASIL® and the Baculovirus-derived bivalent vaccine (Types 16 and 18) CERVARIX® have played an important role in reducing cervical cancer since their introduction for human use in 2006 and 2007.Citation1 The L1 proteins expressed either in yeast or Baculovirus systems self-assemble into virus-like particles (VLPs) that when properly adjuvanted, elicit protective immune responses by mimicking the authentic epitopes of virions. Routine biochemical techniques can be applied to confirm the primary structure of the proteins in these recombinant vaccines. However, other tools are needed to visualize critical morphological characteristics of these VLP vaccine intermediates, including shape, particle integrity, and aggregation. CryoTEM provides an excellent visualization tool to directly determine these properties of the VLP particles and can also be used to observe the VLPs when absorbed onto aluminum adjuvants. These data can be combined with the results from orthogonal methods to provide information that is important for process development, process optimization, and comprehensive characterization of pivotal vaccine lots.
Results
VLP Morphology
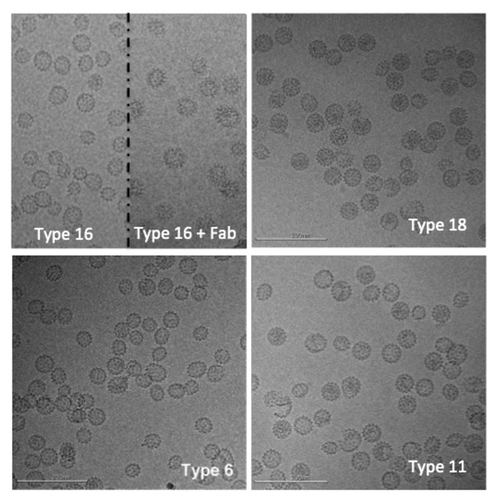
CryoTEM images for each of the four HPV L1 serotypes (L1 types 6, 11, 16 and 18) are shown in . Most of the VLPs appear to be fully assembled, are observed to have a range of sizes, and are predominantly spherical or ellipsoidal in shape, with no evidence of filaments or other large aggregates. Similar ranges in apparent morphology have been observed in other EM studies of L1 type 16,Citation2,Citation3 and type 11Citation4 VLPs as well as in unfractionated preparations of rabbit papillomavirus virus.Citation5
Figure 1. Representative cryo-electron microscopy images of human papillomavirus virus-like particles (L1 types 16, 18, 6, and 11). In the top left panel, the image is split to show type 16 particles on the left and type 16 particles decorated with an antibody fragment on the right. Scale bar is 200 nm.
Three-dimensional (3D) reconstruction of HPV VLP and VLP:Fab structure
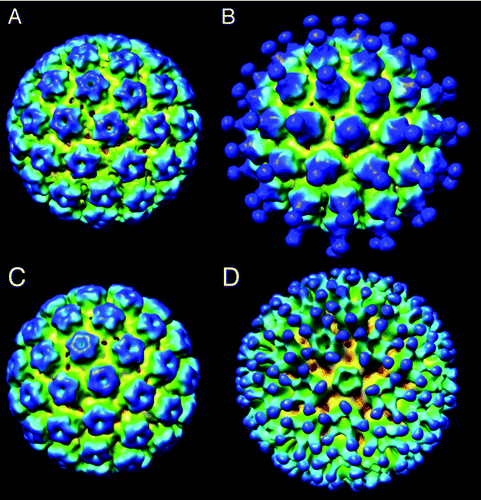
One of the unique advantages of the cryoTEM method is that a 3D map of the structure can be reconstructed by combining particles of the same morphology and conformation but in different relative orientations.Citation20 A set of particles of similar diameter (54 ± 3 nm) were selected from images of VLPs of type 11 and type 16, and single particle analysis methodsCitation6 were used to reconstruct 3D maps of each serotype, as shown in . The reconstructed volumes show that the capsids are constructed of 72 pentameric capsomers arranged in a T = 7 icosahedral lattice that closely resembles that of the native virions of human or bovine papillomavirus.Citation7,Citation8 These results are in agreement with an earlier cryoTEM study of vaccinia virus-produced L1 type 1 capsids that appeared similar (at a resolution of ~3.5 nm) to native HPV type 1.Citation9 Measurements obtained from a central section of the HPV11 map also provide an independent measure of the particle diameter (~55 nm).
Figure 2. Three-dimensional reconstructions of (A) HPV11 used 5135 particle images in the reconstruction and the resolution based on the FSC0.5 criteria was 1.3 nm and the EMDB deposition number is 28367; (B) HPV11 decorated with antibody fragment H11.B2 B2 (6009 particles, FSC0.5 = 2.0 nm, EMDB 28368); (C) HPV16 (5135 particles, FSC0.5 = 1.3 nm, EMDB 28369) (d) HPV16 decorated with antibody fragment H16.V5 (6009 particles, FSC0.5 = 2.0 nm, EMDB 28370).
During the manufacturing process for GARDASIL, some types of the purified VLP are subjected to a disassembly/reassembly step that optimizes the structure and stability of the final VLPs. The main benefit of the disassembly/reassembly process was to improve the stability of the VLP preparations since fully closed and discrete VLPs are less prone to aggregation 15, 31). This treatment was observed to increase the surface antigenicity for HPV 16 of clinically relevant epitopes such as those of mAb H16.V5 – particularly those known to be neutralizing in pseudovirion neutralization assayCitation10-Citation12 and to be immuno dominant when analyzed by competition assay using human sera of naturally infected individuals.Citation13 Epitope-specific antigenicity is important to our understanding of the VLP structures, particularly the presence of the neutralizing epitopes and resemblance to authentic virions.Citation14,Citation15 Furthermore, antibodies to these epitopes have been incorporated into the potency assay for the individual VLP antigens in GARDASIL.Citation16 The binding activities of these neutralizing mAbs are surrogate markers for the vaccine efficacy in eliciting neutralizing titers in vivo.Citation16
VLPs can be observed directly interacting with antibody fragments (see , top left inset) and 3D volumes () can be reconstructed from these decorated particles using the methods as described above. We reconstructed antibodies decorating two different HPV serotypes: Type 11 interacting with antibody fragment H11.B2 and Type 16 interacting with H16.V5. The reconstructed density maps are shown in . The antigenic site of interaction is quite different for the two serotypes. For Type 11+ H11.B2 the antigenic site is located at the center of the capsomer (), providing 72 potential binding sites per capsid. In the case of Type 16 + H16.V5, the antigenic site is located off to the side at the outer edge of the capsomers (), and it also appears that the antibody binds to the capsomers at the located 3-fold axes of symmetry, and not to those located at the 5-fold axes of symmetry,, possibly due to a more restricted spacing between capsomers at the 5-fold axis as compared with those at the 3-fold axis.Citation17 From these 3D maps, we deduce that there are 300 potential binding sites on the H16 serotype for the H16.V5 antibody.
VLPs maintain their spherical morphology on aluminum adjuvant
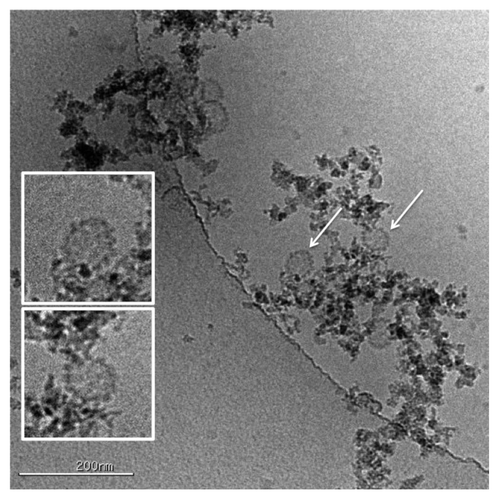
The quadrivalent HPV VLP vaccine, GARDASIL, is a sterile liquid suspension that is prepared by combining the adsorbed VLPs of each HPV type. All of the VLPs were adsorbed onto an aluminum-containing adjuvant (Amorphous Aluminum Hydroxyphosphate Sulfate or AAHS) and the final formulation buffer. It is of interest to understand whether interaction with the aluminum adjuvant surface alters the VLP morphology, so HPV VLPs adsorbed onto the aluminum adjuvant were directly visualized using cryoTEM in the vitrified state (see ). There was no apparent change in shape upon adsorption onto the amorphous adjuvants and the VLPs retained their spherical shape.
Discussion
Biophysical characterization of VLP-based vaccines is an integral part of the characterization package, along with the results from biochemical and functional analyses. TEM, cryoTEM, AFM, dynamic light scattering, size exclusion chromatography, and sedimentation experiments are among the most popular techniques that are employed for VLP-based vaccine characterization. In this report, the application of cryoTEM was demonstrated for generating qualitative, semi-quantitative, and quantitative data using HPV16 VLPs as an example. Importantly, a 3D reconstruction of the recombinant protein based VLPs closely resembles the authentic capsids of similar viruses.
The functionality of the recombinant VLPs was also probed using the Fab fragments of neutralizing mAbs. Among different mAbs tested, H11.B2 and H16.V5 showed discrete particles when the VLP-Fab complexes were examined with cryoTEM, making the data amenable for 3D reconstruction of the complex structure. While linking L1 molecules or aggregating virions was demonstrated to be an important mechanism for several neutralizing mAbs, certain mAbs, with H11.B2 and H16.V5 included, showed similar neutralization efficiencies when compared with their parental antibodies.Citation8,Citation12 The mAbs relying on cross-linking virions for neutralization, Fab fragments could show lower neutralization efficiencies by 2- to 3-orders of magnitude. On the contrary, monovalent Fab fragments of H11.B2 and H16.V5 showed only approximately 3-, and 5-fold reduction, respectively in neutralizing efficiency, when compared with respective bivalent IgGs.Citation8,Citation12 Moreover, the Fab binding locations for H11.B2 or for H16.V5 are consistent with their corresponding epitopes, DE loops for H11.B2, and FG loop (with some contribution from HI loop) for H16.V5.Citation12,Citation18,Citation19
The binding mode for H16.V5 was observed to be near the center of the pentamer, similar to that previously reported for a neutralizing mAb again bovine papillomavirus.Citation20 In that paper, a further binding mode for virus neutralization was illustrated for a mAb binding to the valley in-between pentamers. Therefore, the binding to the area near the central openings in the pentamers, the putative binding sites for L2 molecules, for a neutralizing mAb is a novel finding using 3D reconstruction method.
The functional and neutralizing mAbs, H11.B2 and H16.V5, studied in this report were also used in the approved in vitro relative potency tests for GARDASIL lot release and stability.Citation16,Citation21 These assays have been shown to correlate to in vivo mouse potency as well as to clinical serological results. The observation of two different binding modes of these two neutralizing mAbs also sheds some light on the mechanism of how HPV virions are neutralized during in vitro studies, as well as in biological systems. In general, the neutralizing epitopes are more exposed or better formed when the VLPs have undergone the disassembly and reassembly treatment. This treatment likely causes aggregated recombinant VLP proteins to dissociate and re-oligomerize into virion-like assemblies.Citation14 This is the desired outcome during vaccine production, as it maximizes the content of neutralizing epitopes and enhances product stability after formulation and filling.Citation22
Aluminum adjuvants are commonly used in vaccines to enhance immunogenicity. Gardasil VLPs 6, 11, 16 and 18 formulated with Merck’s proprietary aluminum adjuvant, AAHS, demonstrated significantly higher antibody levels in rhesus macaques compared with the VLPs alone (12.7 to 41.9-fold higher after four weeks and 4.3 to 26.7 higher levels at 52 wk).Citation23 Particle integrity after adjuvant adsorption could be confirmed using cryoTEM by visualizing whole vaccine mixtures containing VLP antigen adsorbed to amorphous adjuvants. The AAHS particles also keep the VLPs from aggregating without deforming the VLP morphology or their key epitopes. The morphology of the individual VLPs were confirmed to be unaffected by the adjuvant by both visual inspection and by using tomographic methodsCitation24 to obtain 3D reconstructions of the structures (results not shown). Furthermore, the antigenicity is recovered completely after adjuvant dissolution.
In conclusion, cryoTEM can be a highly informative complementary tool for VLP characterization. The characterization of a recombinant VLP-based vaccine requires the application of an extensive panel of analytical procedures to the product, and the characterization data set is included in regulatory filings to demonstrate an understanding of the product. A comprehensive characterization data set is important for supporting selection of the potency assay, justifying specifications for the product and the impurity profile, characterizing the process, and establishing the “historical” comparability and therapeutic/clinical equivalence of the product. Structural characterization, along with functional characterizationCitation14,Citation16,Citation21 is an important aspect of this analytical assessment.
The 3D maps shown in have been deposited to the EMDB database; accession numbers are: EMDB28367, EMDB28368, EMDB28369, and EMDB28370.
Materials and Methods
HPV 6, 11, 16 and 18 L1 VLPs
Full-length L1 proteins of HPV types 6, 11, 16 and 18 were overexpressed in yeast using previously described procedures.Citation25-Citation27 Purification of the recombinant VLPs, and the disassembly/reassembly process was previously described.Citation28,Citation29 The protein concentration was determined with the Pierce bicinchoninic acid (BCA) protein assay method, and then verified with UV absorbance at 280 nm.
Anti-HPV 16 L1 mouse mAbs (IgGs) and Fab of H16.V5 and H11.B2
Mouse mAb IgGs
Mouse mAbs, H16.V5 and H11.B2,Citation10-Citation12 were produced in cell culture or ascites using the hybridoma cell lines provided by Professor Neil Christensen at Penn State University (Hershey, PA). Standard Protein A chromatography was used to purify IgGs. The concentration was determined with UV absorbance at 280 nm. Purity of the IgGs (> 95% pure) was determined by size-exclusion HPLC and the isotype of each mAb was verified with an IsoStrip test strip (Roche).
H16.V5 and H11.B2 Fab preparation
To prevent cross-linking of the VLPs during cryoTEM experiments (with both IgG and Fab), Fab antibody fragments were prepared from the H16.V5 IgG2b. Reduction of disulfide bonds and specific cleavage at the hinge region were achieved by adding a solution of H16.V5 or H11.B2 (1.0 mg/mL) in 20 mM l-cysteine hydrochloride monohydrate to immobilized papain on agarose beads (Pierce), followed by gentle mixing over the course of two days at ambient temperature for complete digestion. Supernatant containing the Fab fragments was purified via gentle mixing with immobilized Protein A (Repligen) for one hour. Fab fragments in solution were concentrated using a 10 kDa filter (Centriprep YM-10, Millipore). The completeness of the cleavage of IgG and purity of purified Fab was assessed with a size-exclusion HPLC. Concentrations of the H16.V5 or H11.B2 Fab fragments were determined before and after filtration by UV absorbance measurements at 280 nm using an Agilent 8453 UV-Visible spectrophotometer.
CryoTEM of virions or VLPs in hydrated form
Samples were preserved in a thin layer of vitrified ice over C-Flat holey carbon films (Protochips, Inc.) supported on 400 mesh copper grids. Samples were not diluted prior to vitrification. A ~3 μL drop of the sample was placed onto an EM grid covered by a holey carbon support film. The bulk of the sample was removed to leave a thin film, which was then rapidly plunged into a cryogen (liquid ethane) capable of vitrifying the sample (i.e., freezing it without creating ice crystals). The sample preserved in this vitrified state was transferred into the electron microscope using a cryogenic stage that maintained the sample temperature below -170 °C. To limit damage to the specimen by the electron beam, images were acquired using a very low dose of electrons (typically ~10–20e-/Å2); the resulting images accurately reflected the structure of the VLPs but had a low signal to noise ratio (typically ~ < 1) and limited image contrast. Electron microscopy was performed using a Tecnai F20 electron microscope (FEI Co.) operating at 120KeV equipped with a Gatan 4kx4k digital camera. Images were acquired, using the Leginon data collection softwareCitation30 at a nominal magnification of 50,000×, corresponding to 0.226 nm/pixel at the specimen, using a dose of ~16 e-/Å2, and a nominal defocus of ~3 μm.
For the complexes of VLP:H16.V5 Fab or VLP:H11.B2 Fab, mixing of different ratios of VLP:Fab was performed to optimize binding and minimize aggregation of the VLP:Fab complexes.
3D reconstruction of T = 7 icosahedral VLPs and VLP:Fab complexes
Particle subimages were automatically selected from the images using a template based method described in.Citation31 The contrast transfer function of each image was estimated using ACE.Citation32 The measured defocus was then used to correct the phases in each image using the “applyctf” function in the EMAN software package.Citation33 Reconstructions were performed using a projection-matching algorithm in the EMAN softwareCitation33 beginning with a low-pass filtered map of a previously obtained structure as an initial model. Icosahedral symmetry was imposed and 8 rounds of refinement were used to obtain the final map. Images were binned by 2 prior to the refinement, resulting in an isotropic voxel size of 0.33 nm for the final 3D map, contained within an array size of 224x224x224 voxels. After each round of refinement, the map was filtered to the resolution as determined by the FSC0.5 criteria and this map was then used as a starting model for the next round of refinement. The final maps are shown in ; the number of particles contributing to the maps is indicated in the figure legend. The resolution of the final maps are ~15–25 Angstroms based on Fourier Shell Correlation method with a threshold of 0.5. The Chimera visualization packageCitation34 was used to produce all of the surface renderings shown in the figures.
Visualization of HPV VLPs on aluminum based adjuvants
HPV VLPs were adsorbed onto amorphous aluminum hydroxyphosphate sulfate adjuvant. The total protein concentration was 320 μg/mL with a total of aluminum content of 450 μg/mL. Sample preparation and microscopy was the same as described in the section “VLPs maintain their spherical morphology on aluminum adjuvant.”
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Professor Neil D. Christensen and Penn State University for providing us the antibodies or the cell lines for the monoclonal antibodies and Martha Brown for producing the antibodies in house used in this study. Support by the Chinese National Science Fund (81273327) and 863 Major Project (2012AA02A408) (to Q.Z.) are acknowledged.
References
- Dunne EF, Datta SD, E Markowitz L. A review of prophylactic human papillomavirus vaccines: recommendations and monitoring in the US. Cancer 2008; 113:Suppl 2995 - 3003; http://dx.doi.org/10.1002/cncr.23763; PMID: 18980283
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A 1992; 89:12180 - 4; http://dx.doi.org/10.1073/pnas.89.24.12180; PMID: 1334560
- Deschuyteneer M, Elouahabi A, Plainchamp D, Plisnier M, Soete D, Corazza Y, Lockman L, Giannini S, Deschamps M. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix™, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum Vaccin 2010; 6:407 - 19; http://dx.doi.org/10.4161/hv.6.5.11023; PMID: 20953154
- McCarthy MP, White WI, Palmer-Hill F, Koenig S, Suzich JA. Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. J Virol 1998; 72:32 - 41; PMID: 9420197
- Finch JT, Klug A. The structure of viruses of the papilloma-polyoma type 3. Structure of rabbit papilloma virus, with an appendix on the topography of contrast in negative-staining for electron-microscopy. J Mol Biol 1965; 13:1 - 12; http://dx.doi.org/10.1016/S0022-2836(65)80075-4; PMID: 4159383
- Orlova EV, Saibil HR. Structural analysis of macromolecular assemblies by electron microscopy. Chem Rev 2011; 111:7710 - 48; http://dx.doi.org/10.1021/cr100353t; PMID: 21919528
- Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J 1991; 60:1445 - 56; http://dx.doi.org/10.1016/S0006-3495(91)82181-6; PMID: 1663794
- Trus BL, Roden RB, Greenstone HL, Vrhel M, Schiller JT, Booy FP. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat Struct Biol 1997; 4:413 - 20; http://dx.doi.org/10.1038/nsb0597-413; PMID: 9145113
- Hagensee ME, Yaegashi N, Galloway DA. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol 1993; 67:315 - 22; PMID: 8380079
- Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 1996; 223:174 - 84; http://dx.doi.org/10.1006/viro.1996.0466; PMID: 8806551
- Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 2001; 291:324 - 34; http://dx.doi.org/10.1006/viro.2001.1220; PMID: 11878901
- Culp TD, Spatz CM, Reed CA, Christensen ND. Binding and neutralization efficiencies of monoclonal antibodies, Fab fragments, and scFv specific for L1 epitopes on the capsid of infectious HPV particles. Virology 2007; 361:435 - 46; http://dx.doi.org/10.1016/j.virol.2006.12.002; PMID: 17222883
- Wang Z, Christensen N, Schiller JT, Dillner J. A monoclonal antibody against intact human papillomavirus type 16 capsids blocks the serological reactivity of most human sera. J Gen Virol 1997; 78:2209 - 15; PMID: 9292008
- Zhao Q, Modis Y, High K, Towne V, Meng Y, Wang Y, Alexandroff J, Brown M, Carragher B, Potter CS, et al. Disassembly and reassembly of human papillomavirus virus-like particles produces more virion-like antibody reactivity. Virol J 2012; 9:52; http://dx.doi.org/10.1186/1743-422X-9-52; PMID: 22356831
- Towne V, Zhao Q, Brown M, Finnefrock AC. Pairwise antibody footprinting using surface plasmon resonance technology to characterize human papillomavirus type 16 virus-like particles with direct anti-HPV antibody immobilization. J Immunol Methods 2013; 388:1 - 7; http://dx.doi.org/10.1016/j.jim.2012.11.005; PMID: 23159495
- Shank-Retzlaff M, Wang F, Morley T, Anderson C, Hamm M, Brown M, Rowland K, Pancari G, Zorman J, Lowe R, et al. Correlation between mouse potency and in vitro relative potency for human papillomavirus Type 16 virus-like particles and Gardasil vaccine samples. Hum Vaccin 2005; 1:191 - 7; http://dx.doi.org/10.4161/hv.1.5.2126; PMID: 17012876
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A 2010; 107:6298 - 303; http://dx.doi.org/10.1073/pnas.0914604107; PMID: 20308582
- Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell 2000; 5:557 - 67; http://dx.doi.org/10.1016/S1097-2765(00)80449-9; PMID: 10882140
- Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem 2007; 282:31803 - 11; http://dx.doi.org/10.1074/jbc.M706380200; PMID: 17804402
- Booy FP, Roden RB, Greenstone HL, Schiller JT, Trus BL. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 A resolution. J Mol Biol 1998; 281:95 - 106; http://dx.doi.org/10.1006/jmbi.1998.1920; PMID: 9680478
- Shank-Retzlaff ML, Zhao Q, Anderson C, Hamm M, High K, Nguyen M, Wang F, Wang N, Wang B, Wang Y, et al. Evaluation of the thermal stability of Gardasil. Hum Vaccin 2006; 2:147 - 54; http://dx.doi.org/10.4161/hv.2.4.2989; PMID: 17012891
- Zhao Q, Allen MJ, Wang Y, Wang B, Wang N, Shi L, Sitrin RD. Disassembly and reassembly improves morphology and thermal stability of human papillomavirus type 16 virus-like particles. Nanomedicine 2012; 8:1182 - 9; PMID: 22306156
- Ruiz W, McClements WL, Jansen KU, Esser MT. Kinetics and isotype profile of antibody responses in rhesus macaques induced following vaccination with HPV 6, 11, 16 and 18 L1-virus-like particles formulated with or without Merck aluminum adjuvant. J Immune Based Ther Vaccines 2005; 3:2; http://dx.doi.org/10.1186/1476-8518-3-2; PMID: 15842730
- Murphy GE, Jensen GJ. Electron cryotomography. Biotechniques 2007; 43:413 - , 415, 417 passim; http://dx.doi.org/10.2144/000112568; PMID: 18019332
- Neeper MP, Hofmann KJ, Jansen KU. Expression of the major capsid protein of human papillomavirus type 11 in Saccharomyces cerevisae. Gene 1996; 180:1 - 6; http://dx.doi.org/10.1016/S0378-1119(96)00388-5; PMID: 8973339
- Cook JC, Joyce JG, George HA, Schultz LD, Hurni WM, Jansen KU, Hepler RW, Ip C, Lowe RS, Keller PM, et al. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr Purif 1999; 17:477 - 84; http://dx.doi.org/10.1006/prep.1999.1155; PMID: 10600468
- Wang XM, Cook JC, Lee JC, Jansen KU, Christensen ND, Ludmerer SW, McClements WL. Human papillomavirus type 6 virus-like particles present overlapping yet distinct conformational epitopes. J Gen Virol 2003; 84:1493 - 7; http://dx.doi.org/10.1099/vir.0.18872-0; PMID: 12771418
- Zhao Q, Guo HH, Wang Y, Washabaugh MW, Sitrin RD. Visualization of discrete L1 oligomers in human papillomavirus 16 virus-like particles by gel electrophoresis with Coomassie staining. J Virol Methods 2005; 127:133 - 40; http://dx.doi.org/10.1016/j.jviromet.2005.03.015; PMID: 15894387
- Mach H, Volkin DB, Troutman RD, Wang B, Luo Z, Jansen KU, Shi L. Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J Pharm Sci 2006; 95:2195 - 206; http://dx.doi.org/10.1002/jps.20696; PMID: 16871523
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol 2009; 167:11 - 8; http://dx.doi.org/10.1016/j.jsb.2009.03.019; PMID: 19361558
- Zhu Y, Carragher B, Potter CS. Contaminant detection: improving template matching based particle selection for cryo-electron microscopy. IEEE Symposium on Biomedical Imaging, Arlington, VA April 15-18, 2004. 2004:1071-74.
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy 2005; 104:8 - 29; http://dx.doi.org/10.1016/j.ultramic.2005.02.004; PMID: 15935913
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 1999; 128:82 - 97; http://dx.doi.org/10.1006/jsbi.1999.4174; PMID: 10600563
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605 - 12; http://dx.doi.org/10.1002/jcc.20084; PMID: 15264254