Abstract
Recent studies have suggested that a successful subunit human cytomegalovirus (CMV) vaccine requires improved formulation to generate broad-based anti-viral immunity following immunization. Here we report the development of a non-live protein-based vaccine strategy for CMV based on a polyepitope protein and CMV glycoprotein B (gB) adjuvanted with TLR4 and/or TLR9 agonists. The polyepitope protein includes contiguous multiple MHC class I-restricted epitopes with an aim to induce CD8+ T cell immunity, while gB is an important target for CD4+ T cell immunity and neutralizing antibodies. Optimal immunogenicity of this bivalent non-live protein vaccine formulation was dependent upon the co-administration of both the TLR4 and TLR9 agonist, which was associated with the activation of innate immune signatures and the influx of different DC subsets including plasmacytoid DCs and migratory CD8-DEC205+CD103-CD326- langerin-negative dermal DCs into the draining lymph nodes. Furthermore these professional antigen presenting cells also expressed IL-6, IL-12p70, TNFα, and IFNα which play a crucial role in the activation of adaptive immunity. In summary, this study provides a novel platform technology in which broad-based anti-CMV immune responses upon vaccination can be maximized by co-delivery of viral antigens and TLR4 and 9 agonists which induce activation of innate immune signatures and promote potent antigen acquisition and cross-presentation by multiple DC subsets.
Introduction
The development and widespread use of vaccines represents one of the most significant achievements of modern medicine and they have been instrumental in controlling and even eliminating life-threatening diseases including smallpox, polio, diphtheria, and measles. However, despite these successes there are a number of complex disorders (cancers, HIV, malaria, tuberculosis and other emerging infectious diseases) for which effective or preventive vaccines have not been developed. Over the past decade, several reports have specified new emerging mechanistic insights into how the innate immune system triggers protective immune responses and regulates the magnitude, quality, and persistence of vaccine-induced immunity.Citation1-Citation5 The most significant of these new insights include the identification of pathogen recognition receptors (PRRs),Citation2 the identification and transcriptional regulation of the various dendritic cells (DCs) subsets,Citation6 and the role of distinct subsets of helper T cells, in protecting against different pathogens.Citation7,Citation8 Therefore, a rational approach for directing vaccine responses toward a number of effector cell subsets of the immune system will likely improve vaccine efficacy and these mechanistic insights will potentially provide tools for designing better vaccines and improving the protective efficacy of existing vaccines.
Human Cytomegalovirus (CMV) is a widely prevalent β herpesvirus. In healthy individuals after primary infection CMV establishes a latent state with periodical reactivation and shedding from mucosal surfaces. Although primary infection is generally asymptomatic, CMV can cause significant morbidity and mortality in immunosuppressed individuals through reactivation of latent virus or reinfection with a different isolate. Congenital infection, primarily through intrauterine transmission, is a leading cause of birth defects in children.Citation9-Citation12 Based on the life-time cost to the health care system and its impact on human suffering, development of a vaccine to prevent congenital CMV infection has been assigned the highest priority by the Institute of Medicine of the National Academy of Sciences (US) and US National Vaccine Program Office. (i.e., most favorable impact—saves both money and quality-adjusted life years [QALYs]).Citation13
Recent insights in understanding the immunology, pathology, and molecular biology of CMV suggests that protection against CMV-related disease is mediated by both humoral and cellular immunity, thus, an ideal vaccine against CMV needs to induce both humoral and cellular responses.Citation14,Citation15 We have previously shown that an adenoviral vector encoding polyepitope combined with CMV encoded glycoprotein B (CMV-gB) protein to target the humoral and cellular mediated immune responses is one of the most efficient strategies for inducing CMV-gB-specific antibodies and CD4+ T cell responses and a broad spectrum of CD8+ T cell responses.Citation16 However, the issues of pre-existing immunity and safety of the use of viral vectors have impeded implementation of this vaccine in a clinical setting.Citation17 To overcome these major roadblocks, we have developed a novel platform technology to express multiple CMV CD8+ T cell epitopes as a polyepitope protein using a prokaryotic expression system and combined this with recombinant CMV glycoprotein B (gB) to generate a novel bivalent non-live protein-based vaccine formulation. In this study we have demonstrated that this bivalent vaccine formulation based on polyepitope protein and gB adjuvanted with TLR agonists can be successfully used to induce CMV-specific cellular and humoral immune responses.
Results
Rational design of the CMV polyepitope constructs, protein expression, purification, and in vitro immunogenicity studies
The CMV polyepitope was designed to encode 13 minimal CD8+ T cell epitopes without (CMVpoly) and with (CMVpoly-L) spacer sequences (). The spacer sequences were designed based upon previous observations demonstrating the enhanced liberation of T cell peptide epitopes following the inclusion of a proteasome liberation amino acid sequence and a TAP recognition motif.Citation18 The CMV polyepitope constructs were transformed into E. coli, protein expression conditions were optimised, and polyepitope proteins were purified using Ni-NTA chromatography. Results obtained from these experiments showed that both the CMVpoly and CMVpoly-L could be successfully expressed and purified to homogeneity using a bacterial expression system ().
Figure 1. Schematic design of the CMV polyepitope protein construct with and without linkers. A shows the design of CMV polyepitope protein without linkers (referred to as CMVpoly), while B shows the design of polyepitope protein with linkers (referred to as CMVpoly-L). Each of the alternate CD8+ T cell epitope sequences are italicized and underlined. HLA restriction of the every epitope is specified above epitope sequence. For CMVpoly-L each epitope sequence is separated by amino acid residue(s) which are targets for proteasomal degradation (shown in red) and a peptide transporter recognition motif (shown in blue).
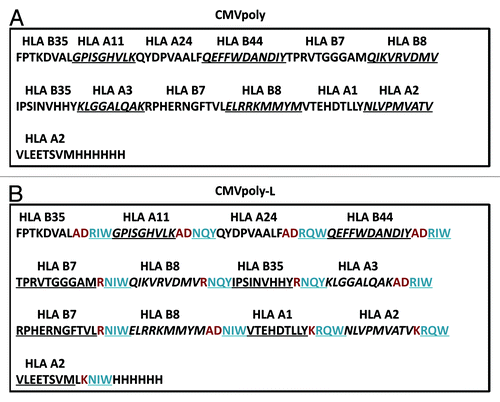
Figure 2. CMV polyepitope protein purification and in vitro assessment of processing and presentation by human cells. The DNA sequence encoding the CMV polyepitope proteins was cloned into an IPTG inducible plasmid, pJexpress 404, and transformed into E. coli for protein expression. Polyepitope protein was purified using Ni-NTA affinity chromatography. A shows SDS-PAGE analysis of purified CMVpoly and CMVpoly-L proteins. Predicted size for the CMVpoly and CMVpoly-L was 14.7Kd and 19.4Kd respectively. B shows in vitro cross-presentation of CMV polyepitope protein by human cells. EBV transformed human lymphoblastoid cell lines (LCLs) were pulsed with CMVpoly or CMVpoly-L protein (25 µg each) for two hours, washed, incubated overnight and then exposed to CMV-specific CD8+ T cells specific for HLA A2-restricted NLV (pp65), HLA A1-restricted VTE (pp50), HLA B7-restricted RPH (pp65), and HLA B7-restricted TPR (pp65) epitopes. The FACS plots shows IFN-γ expression by the CMV-specific CD8+ T cells following co-culture with CMVpoly or CMVpoly-L proteins pulsed LCLs. C shows comparative activation of CMV-specific T cells by the synthetic peptide epitopes, full-length recombinant CMV proteins (pp65 or IE-1) and CMVpoly-L protein. EBV-transformed LCLs were sensitized with synthetic peptide epitopes, full-length pp65 or IE-1 proteins or CMVpoly-L protein as indicated above and then exposed to CMV-specific CD8+ T cells. Following incubation, these CD8+ T cells were assessed for IFN-γ expression by ICS assays. The FACS plots show IFN-γ expression by CMV-specific CD8+ T cells following co-culture with antigen loaded LCLs. This data are representative of one of the three independent sets of experiments.
To investigate the processing and presentation of the CMVpoly and CMVpoly-L proteins, we incubated human lymphoblastoid cell lines (LCLs) overnight with CMVpoly or CMVpoly-L, and then assessed the activation of a panel of CMV-specific T cells using intracellular IFN-γ analysis. Representative data presented in shows that HLA A2-restricted pp65 epitope, NLVPMVATV (referred to as NLV), HLA A1-restricted pp50 epitope, VTEHDTLLY (referred to as VTE), HLA B7-restricted pp65 epitopes RPHERNGFTVL (referred to as RPH), and TPRVTGGGAM (referred to as TPR) from CMVpoly-L were more efficiently processed and presented to CMV-specific T cells compared with LCLs pulsed with CMVpoly. To extend this analysis, we compared the processing and presentation of CMVpoly-L protein with full-length CMV proteins, including pp65 and IE-1 proteins. Data presented in shows that the HLA A2-restricted epitopes NLV and VLEETSVML (referred to as VLE) and HLA B7-restricted RPH epitope from CMVpoly-L protein were more efficiently processed and presented to CMV-specific T cells compared with the epitopes presented from the full-length pp65 or IE-1 proteins.
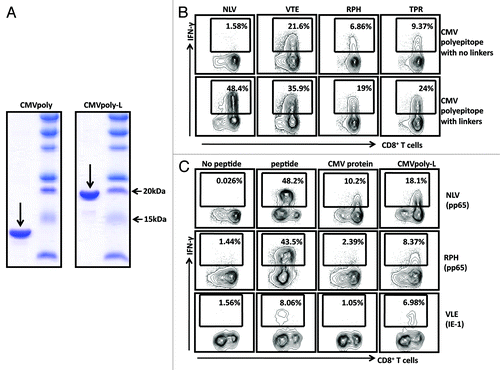
To further evaluate the immunogenicity of the CMVpoly-L protein, PBMC from 10 CMV-seropositive individuals, HLA matched for the epitopes expressed in CMVpoly-L, were incubated with the CMVpoly-L protein then cultured for 10 d in the presence of IL-2. The expansion of epitope specific T cells was then assessed by intracellular cytokine assays (ICS) assay. Representative data from one of these ICS assays is presented . The CMVpoly-L protein induced expansion of CMV specific CD8+ T cell specific in all 10 individuals and these expansions ranged from 8–8900 fold (). In the majority of the individuals expansion of T cells directed toward multiple epitopes was observed. Taken together, these data demonstrate the enhanced capacity of the CMVpoly-L protein to deliver CMV-restricted T cell epitopes for presentation to human CD8+ T cells.
Figure 3. In vitro expansions of CMV-specific T cells from healthy virus carriers following stimulation with the CMVpoly-L protein. PBMC from ten different healthy CMV-seropositive individuals were stimulated with recombinant CMVpoly-L protein and cultured for 10 d in the presence of recombinant IL-2. The percentage of expanded peptide specific CD8+ T cells producing IFN- γ was determined using an ICS assay. A shows the representative FACS plots illustrating IFN-γ expression by in vitro expanded CMV-specific CD8+ T cells following stimulation with CMVpoly-L protein. B shows the fold change in in vitro expanded CMV-specific CD8+ T cells relative to ex vivo responses from ten different CMV-seropositive donors (D1 to D10). The fold expansion of T cells for each donor was calculated using the T cell numbers before and after stimulation with CMVpoly-L protein.
CMV vaccine formulation with TLR4 and TLR9 agonists promote recruitment, activation of DC subsets and innate immune signatures in draining lymph nodes
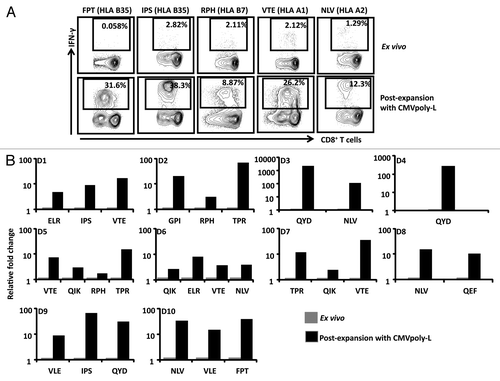
The efficient delivery of antigen in the context of the correct innate inflammatory signals is a critical component of any vaccine platform targeting the induction of adaptive immunity. Indeed a number of recent studies have successfully integrated systems biology approaches into vaccinology to identify innate immune signatures that are associated with vaccine efficacy.Citation19-Citation21 To identify the most appropriate combination of adjuvants for the CMV vaccine, we first assessed the impact of the synergistic activity of TLR4 and TLR9 adjuvanted CMV vaccine on the recruitment of professional antigen presenting cells into draining lymph nodes (DLN) which may play a crucial role in the induction of adaptive immunity. Using a panel of antibodies specific for the markers CD11c, CD8, B220, DEC205, CD103, and CD326 we identified a total of six distinct populations of CD11c+ DCs based on the differential expression of these markers similarly described in a previous study.Citation22 The gating strategy for each of these DC subpopulations is shown in . These populations included the resident CD11c+CD8+ (CD8+DCs, subset 1), CD11c+B220+(pDCs, subset 2), CD11c+CD8-DEC205+CD103-CD326-, (Langerin negative dermal DCs, subset 3), CD11c+CD8-DEC205+CD103-CD326+ (Epidermal Langerhans cells, subset 4), CD11c+CD8-DEC205+CD103+ (Langerin positive dermal DC, subset 5), and CD11c+CD8-DEC205- (resident blood derived DCs, subset 6). Data presented in shows that while we saw little impact on the number of epidermal langerhans cells and Langerin+ dermal DCs in the draining lymph nodes, immunization with the TLR4 agonist MPL lead to an increase in the frequency of Langerin- dermal DCs, pDCs, and resident blood derived DC, whereas immunization with the TLR9 agonist CpG produced a more modest increase in the frequency of Langerin-dermal DCs and a reduction in the frequency of pDCs and CD8+ DCs. Interestingly, immunization with a combination of the TLR4 and TLR9 agonists induced a similar increase in the frequency of Langerin- dermal DCs and pDCs to that seen with MPL alone. These observations suggested that the synergistic benefit provided by the TLR4 agonist MPL when delivered in combination with CpG, is through the enhanced trafficking of dendritic cells, particularly the skin derived dermal DC, into the DLN. To further address the impact the TLR agonists have upon antigen processing and presentation by professional antigen presenting cells, CD11C+ DCs were loaded with the CMVpoly-L protein, activated with the TLR agonists, then assessed for their capacity to present peptide to a T cell hybridoma specific for the HLA A2 restricted VLE epitope. Pre-treatment with the TLR9 agonist CpG alone or in combination with MPL, enhanced the cross-presentation of CMVpoly-L however, MPL failed to enhance the CMVpoly-L cross-presentation ().
Figure 4. Assessment of recruitment of distinct DC subpopulations in the DLN of HHD-I mice following vaccination with different TLR agonists. HHD-I mice were vaccinated with PBS, TLR4, TLR9, or TLR4 and TLR9. Following 48 h of vaccination CD11c+ DCs were enriched from inguinal lymph nodes. A shows representative FACS plots of different subsets of CD11c+ DCs. These cells were divided into subset 1 (CD8+ DCs), subset 2 (pDCs) based on the expression of CD8 and B220, subset 3 (Langerin negative dermal DCs) based on the expression of CD8-DEC205+CD103-CD326-, subset 4 (Epidermal Langerhans cells) based on the expression of CD8-DEC205+CD103-CD326+, subset 5 (Langerin positive dermal DC) based on the expression of CD8-DEC205+CD103+ and subset 6 (resident blood derived DCs) based on the expression of CD8-DEC205-. B shows the absolute number of different DC populations recruited in the draining lymph nodes of HHD-I mice vaccinated with different TLR ligands. Lymph nodes from 5 mice in each group were pooled for these analyses. C shows representative FACS plots of cross-presentation of CMVpoly-L protein by CD11c+ in the presence of different TLR agonists. CD11c+ DCs were pulsed with 25 µg of CMVpoly-L for two hours and then washed and resuspended in DC growth medium containing no adjuvant, TLR4, TLR9, or TLR4 and TLR9 incubated over night. CMVpoly-L pulsed CD11c+ DCs were mixed with β-gal expressing HLA A2-restricted IE-1 VLE epitope-specific murine T cell hybridoma cells and incubated for 6 h and then β-gal activity was determined using FDG substrate. Cells stimulated with adjuvants alone used as negative control and cells pulsed with VLE peptide used as a positive control. Representative data from one of the two independent experiments is presented here.
We further delineated the impact of the TLR4 and/or TLR9 agonist on the molecular innate immune signatures in vivo using a customized gene expression array published by Kwisaa and colleagues.Citation6 Cells from DLN of HHD-I mice were harvested 24 h and 48 h after injection with different TLR ligands and assessed for the expression of a panel of genes associated with innate immune activation. Data presented as a gene-expression heat map in showed that 60 out of 92 genes were at least 2-fold up- or downregulated from baseline (PBS control) at 24 h post injection in TLR4 agonist alone, TLR9 agonist alone or TLR4 and TLRL9 agonist groups (n = 4). While the mice immunised with TLR4 agonist alone stimulated a weak transcriptional signature, the TLR9 agonist alone or in combination with the TLR4 agonist significantly upregulated transcription of several genes. Consistent with the data presented in , significant upregulation of expression of genes encoding inflammatory cytokines including IL12B, IL6, and IL1A was observed (). In addition, other signature genes including the type I IFN-stimulated genes (ISGs) (ISG-15, Ifit1B); chemokines involved in T cell recruitment (Cxcl-10, Cxcl-11, Ccl-13) and interferon related genes (IRF) (IRF7) were also upregulated (). These genes are known to promote T helper 1 cell development and the induction of a CD8+ T cell response. Collectively, these data indicate that TLR9 alone or a TLR4 and TLR9 combination has the ability to induce robust and sustained activation of professional antigen presenting cells and expression of genes encoding proteins involved in the initiation of inflammatory and antiviral responses in the DLN when compared with the TLR4 agonist MPL alone.
Figure 5. Pro-inflammatory innate gene transcription signature following administration of different TLR agonists. HHD-I mice (n = 4 in each group) were immunized with PBS, TLR4, TLR9, or TLR4 and TLR9 agonists. RNA was isolated after 24 h and 48 h from draining lymph nodes and then subjected to low-density array real-time PCR for a panel of 92 genes involved in innate immune responses. All genes were first normalized to the average Ct values of expression of the house keeping genes for 18S, Actb and Gusb. A shows heat map of genes upregulated and downregulated at least 2-fold relative to the PBS group following 24 h of immunization. Each column represents the experimental group (n = 4) and row represent the fold change of individual genes. B shows summary of significantly upregulated and downregulated genes relative to PBS group at the 24 and 48 h time point is shown in the bar graphs. Bar graphs represent the mean ± SEM. P values were calculated by student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001 (determined by the Student t test).
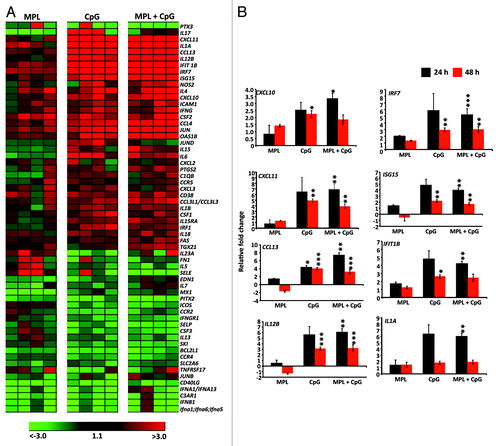
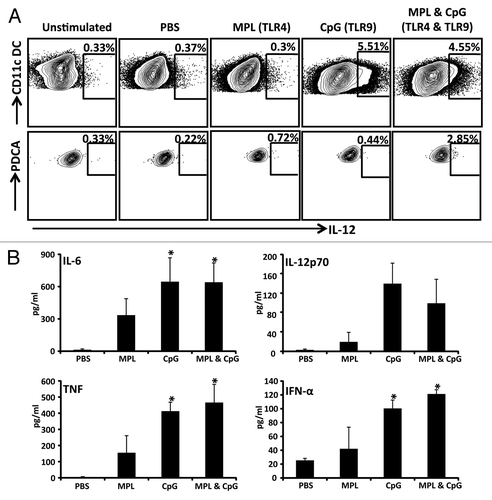
Finally we assessed the functional impact of different TLR agonist combinations have upon the activation of murine DCs. Murine CD11c+ DCs were cultured in vitro in the presence of the TLR agonists, MPL (TLR4) and/or CpG (TLR9) and the cell culture supernatants were assessed for the expression of the pro-inflammatory cytokines IL-6, IL-12p70, TNF, and IFN-α using ELISA. In addition, the intracellular expression of IL-12p70 was also assessed using an ICS assay. Data presented in shows that the TLR9 agonist alone or in combination with TLR4 agonist induced optimal expression of IL-12, IL-6, IFN-α, and TNF. More importantly, the combination of the TLR4 and TLR9 agonists showed a synergistic effect on IL-12 production by plasmacytoid DCs compared with the cells treated with TLR4 or TLR9 alone (). In summary, these data establish that CMVpoly-L protein in combination with TLR4 and TLR9 agonists induce a unique set of innate immune signatures which influence DC activation and migration into the DLN following immunization and enhance cross-presentation of antigen by CD11c+ DCs.
Figure 6. Assessment of proinflammatory cytokine production following DC stimulation with various adjuvant combinations. CD11c+ DCs were enriched using CD11c Microbeads and stimulated with PBS, TLR4 agonist alone, TLR9 agonist alone, or the TLR4 and TLR9 agonists for 20 h in a 48 well plate. A shows FACS plots of CD11c+ DCs and pDCs producing IL-12p70 following stimulation with indicated adjuvants. B shows the levels of pro-inflammatory cytokines IL-6, IL-12p70, TNF-α and IFN-α by CD11c+ DCs following stimulation with PBS, TLR4 agonist alone, TLR9 agonist alone, or the TLR4 and TLR9 agonists. These cytokines were assessed in culture supernatants using standard ELISA assays. The data represent the mean ± SEM from two independent experiments. Error bars represent the mean ± SEM *P < 0.05 (determined by the Student t test).
CMV vaccine formulated with TLR4 and 9 agonists is required for multifunctional Th1 CD4+ T cell and CD8+ T cell immunity
It is now firmly established that a successful subunit CMV vaccine requires improved formulation to generate broad-based anti-viral immunity including both cellular (CD4+ and CD8+ T cells) and humoral immunity. Data presented above indicated that a vaccine formulation adjuvanted with TLR agonists may provide an appropriate inflammatory environment to induce an effective immune response in vivo. To assess this hypothesis, we formulated a bivalent CMV vaccine which included CMVpoly-L protein (to induce CD8+ T cell responses) and the CMV-gB protein (to induce CD4+ T cell responses and anti-viral neutralizing antibody responses) in combination with the TLR4 and/or TLR9 agonists. In the first set of experiments, we assessed the ability of this vaccine formulation to induce CMV-specific cellular immunity. HHD-I mice were immunized with this vaccine formulation and then boosted with the same formulation on day 21 and sacrificed on day 50. To analyze the establishment of T cell memory, splenocytes were assessed for the presence of CMV-gB specific CD4+ T cells, and CMVpoly-L specific (HLA A2-restricted epitopes NLV and VLE) CD8+ T cells using an intracellular IFN-γ assay. CMV vaccine formulation (CMVpoly-L and CMV-gB) adjuvanted with a combination of a TLR4 and TLR9 agonist was required to induce an optimal T cell response (). While the induction of a CMV-gB-specific CD4+ T cell response could be detected ex vivo following vaccination in the presence of the TLR4 or TLR9 agonists, these responses were significantly enhanced when CMV vaccine was adjuvanted with both TLR agonists (). In contrast, CMV-specific CD8+ T cell responses were detected in mice immunized with CMV vaccine adjuvanted with CpG alone or MPL and CpG. CD8+ T cell responses were significantly lower in mice immunized with CMV vaccine adjuvanted with MPL alone. To confirm these observations, splenocytes from these immunized mice were stimulated with the HLA A2-restricted T cell synthetic peptides epitopes, NLV and VLE for 7 d, and then assessed for the frequency of IFN-γ producing CD8+ T cells. There was a dramatic expansion of CMV-specific CD8+ T cells from mice immunized with CMV vaccine adjuvanted with CpG alone or CpG and MPL ().
Figure 7. Evaluation of virus-specific CD4+ and CD8+ T cell responses following immunizations with the CMV vaccine adjuvanted with TLR agonists. HHD-I mice (n = 5 in each group) were immunized with CMV vaccine formulation on day 0 and then boosted with same formulation on day 21. On Day 50, splenocytes from immunized mice were stimulated in vitro with CMV-gB protein or HLA A2-restricted NLV (pp65) and VLE (IE-1) peptide epitopes in the presence of brefeldin A, then IFN-γ production by CD4+ and CD8+ T cells was measured using an ICS assay. A shows representative FACS plots of the frequency of IFN-γ producing CMV-gB specific CD4+ T cells and CMV-specific CD8+ T cells (ex vivo and in vitro expanded). B shows comprehensive analysis of CMV-specific IFN-γ producing CD4+ and CD8+ T cell responses. C shows multifunctional potentiality of CMV-gB specific CD4+ T cells and CMV-specific CD8+ T cells. On day 50, splenocytes from immunized mice were in vitro stimulated with CMV-gB protein in the presence of brefeldin A and production of IFN-γ, TNF-α, or IL-2 was determined using multiparameter ICS. Panel D shows multifunctional potentiality of CMV-specific CD8+ T cells. On day 50, splenocytes from immunised mice were in vitro stimulated with CMV CD8+ T cell HLA A2-restricted peptide epitopes (NLV and VLE) for 10 d in the presence of IL-2 and then assessed to determine the induction IFN-γ, TNF-α, or IL-2 cytokines using multiparameter ICS. Following ICS data was analyzed using FlowJo software and percentage of antigen-specific CD4+ or CD8+ T cell inducing three cytokine, two cytokines, or one cytokine were plotted in the bar graph. Error bars represent the mean ± SEM *P < 0.05 (determined by the Student t test).
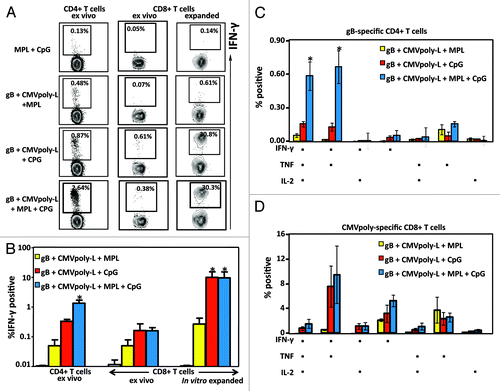
There is substantial evidence that the protective efficacy of T cell-based vaccines correlates with the frequencies of multifunctional effectors.Citation23-Citation25 To investigate the functional quality of the memory T cell response, we assessed the pattern of IL-2, TNF, and IFN-γ production using multiparametric flow cytometry either ex vivo for CMV-gB-specific CD4+ T cells or following in vitro expansion of CMVpoly-L specific CD8+ T cells (). Both CMV-gB specific CD4+ T cells and CMVpoly-L specific CD8+ T cells displayed greater polyfunctionality in mice immunized with CMV vaccine adjuvanted with MPL and CpG or CpG alone. However, frequencies of gB-specific CD4+ T cells expressing all three cytokines were significantly higher following vaccination with MPL and CpG compared with MPL alone and CpG alone. While a similar proportion of CMV-gB specific CD4+ T cells were capable of expressing all three cytokines, interestingly, a large proportion of CD4+ T cells from HHD-I mice immunized with CMV vaccine adjuvanted with MPL alone predominantly expressed TNF (). Similarly a large proportion of CMVpoly-L specific CD8+ T cells from mice immunized with CMV vaccine formulation adjuvanted with MPL expressed only TNF (). In contrast, vaccination with a vaccine formulation based on CpG or MPL and CpG predominantly generated CD8+ T cells which were IFN-γ+TNF+ or IFN-γ+ (). Taken together these analyses demonstrated that a CMV vaccine formulation based on CMVpoly-L and CMV-gB proteins adjuvanted with both TLR4 and TLR9 agonist was most effective in inducing CMV-specific CD4+ and CD8+ T cells with a multifunctional capability.
Induction of anti-viral neutralizing antibody responses are synergistically enhanced by TLR4 and 9 agonists
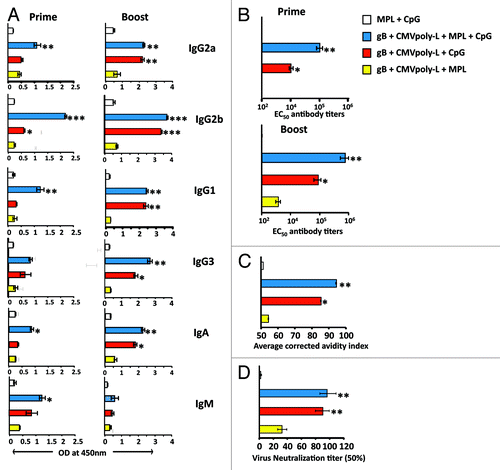
In the next set of experiments we assessed quantitative and qualitative analysis of CMV-specific humoral immune response in animals immunized with bivalent CMV vaccine adjuvanted with different TLR agonists. The combination of TLR4 and TLR9 agonist induced high titer CMV-gB-specific antibody response which included multiple isotypes, including the IgG2b, IgG3, IgG2a (Th1 like Ig isotypes), IgG1 (Th2 like Ig isotype), IgA, and IgM (). The CMV vaccine formulated with the TLR4 and TLR9 agonist induced significantly higher titers of CMV-gB specific antibody responses following the primary immunization which were sustained at high levels until at least day 50 post-immunization, when compared with the vaccine formulated with CpG or MPL alone (). To assess the quality of the CMV-gB specific antibody response generated following vaccination in the different adjuvant combinations, we assessed the functional avidity of CMV-gB specific antibody and the level of CMV-specific neutralizing antibody using a micro-neutralization assay. Antibodies generated in mice vaccinated with the TLR4 and TLR9 agonists displayed significantly higher functional avidity compared with mice immunized with MPL alone, and to a lesser extent to mice immunized with CpG (). Similarly, while antibodies from mice immunized with MPL alone failed to display a strong neutralizing response, the levels of anti-viral neutralizing responses were much greater in mice immunized with CMV vaccine adjuvanted with CpG, or MPL and CpG (). These observations provide further evidence that a CMV vaccine formulation based on CMV polyepitope and CMV-gB proteins adjuvanted with TLR4 and TLR9 agonist provides the optimal strategy to induce both cellular and humoral immune responses.
Figure 8. Assessment of CMV-specific humoral responses following vaccination with CMV vaccine adjuvanted with TLR agonists. To study the synergistic or antagonistic effect of TLR4 and TLR9, four groups (n = 5/group) of HHD-I mice were immunized subcutaneously with CMV-gB and CMVpoly-L proteins adjuvanted with TLR4 alone, TLR9 alone, or TLR4 and TLR9. On day 21 these mice were tail bled and a booster dose was given. Mice were sacrificed on day 50 to assess CMV-gB specific humoral immune responses. A shows gB-specific isotype (IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM) antibody responses in pooled serum samples from treatment groups assayed in triplicates on days 21 and 50. The data represent the mean ± SEM ***P < 0.001, **P < 0.01, and *P < 0.05 (calculated using students’s t test). B shows the CMV-gB specific 50% effective concentration (EC50) antibody titers determined by ELISA on day 21 in pooled sera and on day 50 in individual mouse serum. Error bars represent the mean ± SEM ** P < 0.01. C shows avidity maturation of CMV-gB specific antibody responses. Pooled serum samples from immunized mice at day50 were tested for their CMV-gB specific binding avidity using urea dissociation avidity assay. Bar graphs represent the average corrected avidity index. *P < 0.05, **P < 0.01. D shows 50% CMV-specific neutralizing antibody titers induced following immunization. On day 50, sera from individual groups were pooled, serially diluted, and pre-incubated with HCMV AD169 strain. MRC-5 cells were infected with serum-treated virus and virus infectivity was determined using an IE-1/IE-2 micro-neutralization assay. Neutralizing assay was performed in triplicates. **P < 0.01.
Discussion
Following extensive efforts over the past 30 y, recombinant subunit CMV vaccine formulations (gB/MF59 and DNA vaccine encoding pp65 and gB) have emerged as the most promising candidates and have successfully concluded advanced clinical trials.Citation26-Citation28 In Phase II clinical trials gB/MF59 vaccine showed 50% efficacy in young women and in transplant patients these vaccine formulations reduced viral reactivation.Citation29 However, these vaccine formulations were unable to eliminate the need for antiviral therapy in transplant patients or reduce the incidence of CMV disease.Citation30,Citation31 Therefore, a vaccine with higher efficacy to prevent CMV-associated clinical disease would be highly advantageous. In immunocompetent individuals CMV infection is maintained under strict control by the immune system by a combination of humoral and cellular immune responses. Thus, the major criteria for the design of an effective CMV vaccine should be the induction of virus-specific antibodies as well as CD4+ and CD8+ T cell responses.Citation14,Citation32,Citation33
In our previous studies we investigated a vaccine formulation combining CMV-gB with a TLR9 agonist and immune stimulating complexes (AbISCO®100).Citation34 Although this CMV-gB vaccine was able to induce antibody and cellular immune responses, a single antigen is unlikely to be broadly effective due to the complexity of CMV immunity. In addition, emerging evidence suggests that CMV-specific CD8+ T cell responses in healthy CMV-seropositive individuals are directed toward multiple CMV antigens, predominantly pp65 and IE1, but also other structural, early/late antigens and immunomodulators.Citation35-Citation39 Therefore, we employed a novel platform technology that allows the activation of CMV-specific CD8+ T cell responses directed toward multiple epitopes restricted through a range of HLA class I molecules. Using a prokaryotic expression system, multiple minimal HLA class I-restricted CD8+ T cell epitopes were expressed as a polyepitope protein. Interestingly, we found that human antigen-specific CD8+ T cells were more efficiently activated by the polyepitope protein which included proteasomal/TAP targeting flanking sequences (CMVpoly-L). These observations were in contrast to our previously published data which showed that flanking sequences are not required when a polyepitope is delivered endogenously in the context of a viral vector.Citation40,Citation41 Furthermore, we also showed that the polyepitope protein-based antigen delivery was more efficient in activating human CD8+ T cells when compared with the full-length CMV proteins.
The ability of a vaccine to skew the immunological response toward a specific type is highly important for the induction of pathogen-specific immune responses.Citation22 Emerging evidence suggests that activation of resting DCs that migrate to draining lymph nodes, produce pro-inflammatory cytokines and present antigens to T cells are critical for the induction of particular correlates of protective immunity.Citation4,Citation42-Citation46 The nature of the early innate immune signatures has also recently been shown to be important in optimizing vaccine induced immunity.Citation6 TLR agonists offer the most promising approaches to regulate DC activation and early innate immune signatures, by providing defined signaling mechanisms through known receptor mediated pathways.Citation3 The TLR4 agonist MPL and the TLR9 agonist CpG are commonly used adjuvants in pre-clinical and clinical vaccine formulations for the induction of both cellular and humoral immune responses. We therefore sought to investigate their use in formulating our novel CMV vaccine which was based on CMV-poly-L and CMV-gB proteins. The CMVpoly-L protein includes 13 contiguous MHC class I-restricted CD8+ T cells from multiple CMV antigens and is designed to induce CD8+ T cell responses. The CMV-gB protein is an important source of CD4+ T cell responses and is also crucial target anti-viral neutralizing antibody responses. We believe that a bivalent vaccine based on these two proteins should provide an advantage over the previously designed subunit CMV vaccine formulations as our vaccine strategy will induce broad-based anti-viral immunity which is crucial for long-term protection.
Our initial immunization studies with the TLR4 agonist, MPL alone resulted in poor induction of innate immune gene signatures and reduced DC pro-inflammatory cytokine release. More importantly, CMV vaccine formulation with MPL alone was also inferior for the induction of anti-viral antibody, CD4+ and CD8+ T cell responses. In contrast the TLR9 agonist, CpG, induced an innate immune signature associated with the effective induction of anti-CMV CD8+ T cell immunity. This included genes known to play a pivotal role in innate cellular defense (NOS2 and OAS1b), complement activation (C1QB), proinflammatory cytokines (IL-6, IL-12b, IL-1A, IL-1B, CSF1, and CSF2) cytokines (IFNG and IL-18), chemokines and chemokines receptors (CCL3, CCL4 CXCL10, CXCL11, and CCR5), interferon regulatory factors (IRF7), intracellular adhesion molecule (ICAM and CD38), ISGlyation (ISG15), and transcription factors (JUN). While our bivalent CMV vaccine formulated with CpG alone was sufficient to induce an optimal CD8+ T cell response, the addition of MPL significantly enhanced induction of CMV-gB specific CD4+ T cell responses and anti-viral neutralizing antibody immunity. These findings were consistent with our observations that the TLR4 agonist, MPL functions synergistically with CpG to enhance IL-12 production by PDCA1+ pDCs, and activate innate immune signatures which are crucial for induction of adaptive immunity. Earlier studies have shown that pDCs play a critical role in the induction of both cellular and humoral immune responses.Citation47,Citation48 Therefore, the synergistic effect of TLR4 and TLR9 on pDCs provides one potential mechanism via which TLR4 further enhanced the immunogenicity of the bivalent CMV vaccine which includes both CMVpoly-L and CMV-gB proteins. Alternatively, the synergistic effects of TLR4 and TLR9 may be explained by other undefined mechanisms, such as changes in the recruitment of DC subsets to the DLN. Although MPL alone was unable to induce potent innate immune signatures, MPL alone or in combination with CpG triggered expansion in the numbers of pDCs and dermal DC subsets in the DLN. pDCs are uniquely able to produce large amounts of the antiviral cytokine (IFN-α/β) in response to nucleic acid sensing, which helps pDC to increase antigen presentation capacity and induction of innate and adaptive immune responses against viral antigens.Citation49,Citation50 In addition, although their contribution remains to be fully elucidated, recently dermal DCs have been shown to play a crucial role in activating the naïve CD4+ T cells.Citation22,Citation51 Based on these observations, we hypothesize that engagement of multiple DC subsets might have played a significant role in the induction of higher frequencies of CMV-gB specific CD4+ and CMVpoly-specific CD8+ T cell responses in mice immunized with bivalent CMV vaccine formulated with TLR4 and TLR9 agonists.
In summary, our findings provide a novel platform strategy for preventing CMV-associated disease/infection that maximizes humoral and cellular immunity using a protein-based vaccine with an adjuvant combination that can activate innate immune signatures, and recruit multiple resident and migratory DC subsets. More importantly, these observations may provide clues on future targeting of vaccine formulations for other persistent/chronic infections, for which broad-based immunity is essential.
Methods
Design of the expression vectors encoding CMV polyepitope protein
CMV polyepitope inserts were designed to encode multiple HLA class I restricted T-cell epitopes from three different antigens (pp65, IE-1, and pp50). The polyepitope sequences were designed in such a way that each epitope was linked to each other without a linking sequence (CMVpoly) or joined by a proteasome liberation amino acid sequence (AD or K or R) and a TAP transport motif (RIW, RQW, NIW, or NQY) (CMVpoly-L) (). In addition, a hexa-histidine tag was inserted at the c-terminus of each polyepitope protein to allow purification using a nickel-nitrilotriacetic acid (Ni-NTA) column. The amino acid sequence of each construct was translated into DNA sequence based on E. coli codon utilization and inserts were synthetically constructed (DNA2.0) and cloned into an expression plasmid (pJexpress 404) under an isopropyl–β-D-thiogalactopyraniside (IPTG) inducible promoter. These synthetically designed polyepitope constructs were transformed into chemically competent E. coli DH5α (Invitrogen) and plasmids were purified using a QIAGEN maxi prep kit (QIAGEN).
Protein expression and purification
The expression and purification of the polyepitope protein was performed as previously described.Citation52,Citation53 Briefly, following transformation of the CMV polyepitope expression vectors (CMVpoly or CMVpoly-L) into E. coli BL21 (DE3) pLysS (Invitrogen, Grand Island, NY), the expression of CMV polyepitope protein was induced for 4 h with 1 mM/mL of IPTG. The E. coli cultures were harvested by centrifugation, the cell pellet was resuspended in 80 mL of lysis buffer (25 mM Tris pH 7.4, 0.5% TritonX100, 150 mM NaCl, 0.5 mg/mL lysozyme) supplemented with a protease inhibitor cocktail (Roche) and the CMV polyepitope proteins were purified using a 5 mL of Ni-NTA (QIAGEN) metal-affinity chromatography matrix. Following analysis of the eluted fractions using SDS-PAGE, positive fractions were pooled and protein was dialysed against 25 mM MES (2-[N-morpholino] ethanesulfonic acid) buffer pH 5.6. Protein concentration was determined using a Bradford assay kit (Bio-Rad).
Presentation of CD8+ T cell epitopes from CMV polyepitope protein by human cells
Epstein-Barr virus (EBV) transformed lymphoblastoid cell lines (LCLs) were pulsed with 25 µg/ml of CMVpoly, CMVpoly-L, pp65, or IE-1 proteins for two hours at 37 °C, washed twice with RPMI 1640 medium, then resuspended in growth medium (RPMI 1640 with 10% FCS). After overnight incubation, LCLs were exposed to CMV-specific T cells at a responder to stimulator ratio of 4:1 for four hours at 37 °C; T cells were assessed for IFN-γ expression using an intracellular cytokine assay (ICS) as described previously.Citation54,Citation55
Expansion of CMV specific T-cells from healthy donors PBMC
Peripheral blood mononuclear cells from healthy virus carriers were incubated with 25 μg/ml of purified CMVpoly-L protein at 37 °C, 6.5% CO2 for 2 h. CMV-poly-L pulsed PBMC were then mixed with autologous PBMC at a 1:1 ratio, and cultured in a 24 well plate for 10 d at 37 °C Cultures were supplemented with recombinant IL-2 on days 3 and 6. T cell specificity was then assessed using an ICS assay.
Immunizations
HLA A2 transgenic mice (referred to as HHD-I) were bred and maintained under specific pathogen-free conditions at the QIMR. All protocols were followed in compliance with the QIMR animal ethics committee. In each group at least 5, six-to-eight week old mice were immunized subcutaneously (s.c.) at the base of the tail with the CMV-gB (5 μg/mouse) and CMVpoly-L (20 μg) vaccine formulated with the TLR4 agonist (Monophosphoryl Lipid A (MPL; 25 µg), alone, the TLR9 agonist CpG ODN1826; (CpG; 50 µg) alone or with both the TLR4 and TLR9 agonists. The TLR agonists alone were used as a negative control. Mice were bled on day 21 prior to a booster dose. Mice were sacrificed on day 50 and assessed for both humoral and cellular immune responses. The TLR agonists were purchased from InvivoGen.
Intracellular cytokine staining to assess IFN-γ or multiple cytokine response
Following vaccination, isolated PBMC or splenocytes were cultured for 2 h with 10 μg/ml of CMV-gB protein or 0.2 μg of CMV peptides. Cells were then incubated overnight in the presence of GolgiPlug (BD PharMingen), washed twice, then incubated with APC-conjugated anti-CD3, FITC-conjugated anti-CD4 and PerCP conjugated anti-CD8. Cells were fixed and permeabilised using a BD Cytofix/Cytoperm kit, then incubated with PE conjugated anti-IFN-γ. To assess the expression of multiple cytokines, cells were stained intracellularly with PE-conjugated anti-IFN-γ, PE-Cy7 conjugated anti-TNF and APC conjugated anti-IL2. Cells were acquired on a BD FACSCanto II and data was analyzed using FlowJo software. For this analysis, lymphocytes were first gated for expression of CD3 and then assessed for the expression of either CD4 or CD8. CD3+CD4+CD8- or CD3+CD8+CD4- T cell populations were then gated for the expression of IFN-γ, TNF, or IL-2. Polyfunctional cytokine expression was then determined using Boolean gate analysis.
Assessment of CMV-gB-specific antibody titers and avidity
Anti-CMV-gB antibody titers were determined using ELISA as previously described.Citation56 The isotypes of anti-CMV-gB antibodies in vaccinated mice serum samples were determined by ELISA using the mouse monoclonal antibody isotyping kit (Sigma). CMV-gB specific avidity was evaluated as previously described.Citation57
Micro-neutralization assay
The assay procedure was followed as described previously.Citation58 The percentage of neutralization of viral infectivity was calculated using the following formula: ([number of IE1+ nuclei of CMV infected cells—number of IE1+ nuclei of serum treated CMV infected cells/number of IE1+ nuclei of CMV infected cells] × 100]).
RNA isolation and innate gene expression analysis
Inguinal lymph nodes were collected from HHD-I mice 24 or 48 h after vaccination with PBS, TLR4 alone, TLR9 alone, or TLR4 and TLR9, and single cell suspension was made using enzymatic digestion mix. Total RNA was purified using an RNeasy mini kit (QIAGEN GmbH) and the quality of RNA was determined using a ND-1000 spectrophotometer (Nanodrop Technologies). RNA was reverse transcribed using a High Capacity RNA-to-cDNA kit (Applied Biosystems). The Quantitative Real-Time PCR Gene Expression Assays for 92 genes were custom designed by Applied Biosystems. To further enrich gene specific targets, cDNA was preamplified using a pool of TaqMan Gene Expression Assays as a source of primers and TaqMan PreAmp kit (Applied Biosystems). Preamplified cDNA samples were loaded into Taqman Array 384 well Micro Fluidic Cards (Applied Biosystemes) and sequences were amplified using the Applied Biosystems Viia 7 Real-Time PCR system. Raw data were obtained using SDS software, initially cycling threshold (Ct) values were normalized relative to the geometric mean of 3 housekeeping genes, Actb (β-actin), Gusb (β-glucuronidase), and 18s rRNA. The quality control of all assays, relative quantification delta-delta Ct analysis; fold difference between the samples were determined using Ingenuity systems IPA software.
Murine dendritic cell subset analysis and impact of TLR agonists
Dendritic cells (DCs) were isolated from the spleens or inguinal lymph nodes of HHD-I mice by positive selection using CD11c MicroBeads (Miltenyi Biotech GmbH) and cultured in mouse DC growth medium (RPMI 1640 with 10% FCS, penicillin, streptomycin, non-essential amino acids, sodium pyruvate, L-glutamine, and β-mercaptoethanol) as described earlier.Citation59,Citation60 For subset analysis, purified DCs were stained at 4 °C for 30mins with different staining panels, including CD11c (AF700) (eBioscience), CD8 (PerCP) (BD Biosciences, New Jersey, MD), B220 (PE) (BD Biosciences), CD103 (FITC) (eBioscience), CD326 (PE-Cy7) (eBioscience), and DEC205 (APC) (eBioscience). Cells were washed and acquired on FACS Canto II to determine exact cell numbers.
To determine the inflammatory cytokine response, CD11c+DCs were incubated with MPL and/or CpG for 20 h. Supernatants were the collected and levels of IL-6, IL-12p70, TNF, and IFNα were assessed using an ELISA assay (eBioscience). Expression of IL-12p70 in CD11c+ DCs was also analyzed by intracellular staining. Briefly, these cells were incubated with AF700-conjugated anti-CD11c and FITC-conjugated anti-PDCA-1 monoclonal antibodies (eBioscience), then fixed and permeabilised, and stained with PE-conjugated anti-IL-12p70 (BD Biosciences). Cells were acquired on a FACSCanto II and analyzed using FlowJo software.
Generation of VLE-specific T cell hybridoma
Spleen cells were collected from HHD-I mice immunized with an adenoviral vector encoding CMV epitopesCitation16 were stimulated with irradiated, HLA A2-restricted IE-1 epitope VLEETSVML (referred to as VLE)-pulsed spleen cells in the presence of rIL-2. Fresh stimulator cells and rIL-2 were added every 7 d for 3 wk to generate VLE-specific murine CD8+ T cells. These VLE-specific CD8+ T cells were then fused with BWZ.36 cells and selected as previously described.Citation61,Citation62 BWZ.36 cells are stably transfected with NFAT-lacZ construct which is specifically induced in response to T-cell receptor interaction with peptide-MHC complexes on antigen presenting cells.Citation61,Citation62 The T cell hybridoma were then sub-cloned by limited dilution and CD3+ clones were selected to check VLE-specificity by evaluating T cell hybridoma activation after stimulation with VLE-pulsed MRC-5 cells as previously described.Citation63 Clone with CD3 expression and VLE-specificity were VLE-specific T cell hybridoma.
Cross-presentation analysis of CMV polyepitope protein by purified DCs
Enriched CD11c+ DCs from spleens were pulsed with or without 25 μg/ml of CMVpoly-L protein, incubated for 2 h at 37 °C, then washed and resuspended in mouse DC growth medium containing TLR agonists. After overnight incubation, cells were exposed to a HLA A2-restricted VLE-specific murine T cell hybridoma cells for six hours and then cells were assessed for β-galactosidase (B-gal) expression using Fluorescein Di-β-D-Galactopyranoside (FDG) substrate.Citation61,Citation62
| Abbreviations: | ||
| TLR | = | Toll-like receptors |
| CMV | = | cytomegalovirus, HLA, human leukocyte antigen |
| DC | = | dendritic cells |
| CMVpoly | = | CMV polyepitope |
| CMV-gB | = | CMV glycoprotein B |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NH&MRC) and Queensland Cancer Fund. R.K. is supported by NH&MRC Senior Principal Research Fellowship.
References
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science 2010; 327:291 - 5; http://dx.doi.org/10.1126/science.1183021; PMID: 20075244
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010; 11:373 - 84; http://dx.doi.org/10.1038/ni.1863; PMID: 20404851
- Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell 2006; 124:849 - 63; http://dx.doi.org/10.1016/j.cell.2006.02.019; PMID: 16497593
- Steinman RM. Dendritic cells in vivo: a key target for a new vaccine science. Immunity 2008; 29:319 - 24; http://dx.doi.org/10.1016/j.immuni.2008.08.001; PMID: 18799140
- Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol 2011; 12:509 - 17; http://dx.doi.org/10.1038/ni.2039; PMID: 21739679
- Kwissa M, Nakaya HI, Oluoch H, Pulendran B. Distinct TLR adjuvants differentially stimulate systemic and local innate immune responses in nonhuman primates. Blood 2012; 119:2044 - 55; http://dx.doi.org/10.1182/blood-2011-10-388579; PMID: 22246032
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 2010; 28:445 - 89; http://dx.doi.org/10.1146/annurev-immunol-030409-101212; PMID: 20192806
- Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011; 29:621 - 63; http://dx.doi.org/10.1146/annurev-immunol-031210-101400; PMID: 21314428
- Ahlfors K, Ivarsson SA, Harris S. Report on a long-term study of maternal and congenital cytomegalovirus infection in Sweden. Review of prospective studies available in the literature. Scand J Infect Dis 1999; 31:443 - 57; http://dx.doi.org/10.1080/00365549950163969; PMID: 10576123
- Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med 2001; 344:1366 - 71; http://dx.doi.org/10.1056/NEJM200105033441804; PMID: 11333993
- Novak Z, Ross SA, Patro RK, Pati SK, Kumbla RA, Brice S, Boppana SB. Cytomegalovirus strain diversity in seropositive women. J Clin Microbiol 2008; 46:882 - 6; http://dx.doi.org/10.1128/JCM.01079-07; PMID: 18216215
- Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Snydman DR, Allen U, Humar A, Transplantation Society International CMV Consensus Group. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation 2010; 89:779 - 95; http://dx.doi.org/10.1097/TP.0b013e3181cee42f; PMID: 20224515
- Arvin AM, Fast P, Myers M, Plotkin S, Rabinovich R, National Vaccine Advisory Committee. Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clin Infect Dis 2004; 39:233 - 9; http://dx.doi.org/10.1086/421999; PMID: 15307033
- Burrows SR, Moss DJ, Khanna R. Understanding human T-cell-mediated immunoregulation through herpesviruses. Immunol Cell Biol 2011; 89:352 - 8; http://dx.doi.org/10.1038/icb.2010.136; PMID: 21301481
- Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev 2009; 22:76 - 98; http://dx.doi.org/10.1128/CMR.00034-08; PMID: 19136435
- Zhong J, Rist M, Cooper L, Smith C, Khanna R. Induction of pluripotent protective immunity following immunisation with a chimeric vaccine against human cytomegalovirus. PLoS One 2008; 3:e3256; http://dx.doi.org/10.1371/journal.pone.0003256; PMID: 18806877
- Saxena M, Van TT, Baird FJ, Coloe PJ, Smooker PM. Pre-existing immunity against vaccine vectors--friend or foe?. Microbiology 2013; 159:1 - 11; http://dx.doi.org/10.1099/mic.0.049601-0; PMID: 23175507
- Bazhan SI, Karpenko LI, Ilyicheva TN, Belavin PA, Seregin SV, Danilyuk NK, Antonets DV, Ilyichev AA. Rational design based synthetic polyepitope DNA vaccine for eliciting HIV-specific CD8+ T cell responses. Mol Immunol 2010; 47:1507 - 15; http://dx.doi.org/10.1016/j.molimm.2010.01.020; PMID: 20189249
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 2009; 10:116 - 25; http://dx.doi.org/10.1038/ni.1688; PMID: 19029902
- Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity 2010; 33:516 - 29; http://dx.doi.org/10.1016/j.immuni.2010.10.006; PMID: 21029962
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 2011; 12:786 - 95; http://dx.doi.org/10.1038/ni.2067; PMID: 21743478
- Kastenmüller K, Wille-Reece U, Lindsay RW, Trager LR, Darrah PA, Flynn BJ, Becker MR, Udey MC, Clausen BE, Igyarto BZ, et al. Protective T cell immunity in mice following protein-TLR7/8 agonist-conjugate immunization requires aggregation, type I IFN, and multiple DC subsets. J Clin Invest 2011; 121:1782 - 96; http://dx.doi.org/10.1172/JCI45416; PMID: 21540549
- Gottschalk RA, Hathorn MM, Beuneu H, Corse E, Dustin ML, Altan-Bonnet G, Allison JP. Distinct influences of peptide-MHC quality and quantity on in vivo T-cell responses. Proc Natl Acad Sci U S A 2012; 109:881 - 6; http://dx.doi.org/10.1073/pnas.1119763109; PMID: 22223661
- Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med 2007; 13:843 - 50; http://dx.doi.org/10.1038/nm1592; PMID: 17558415
- Wille-Reece U, Flynn BJ, Loré K, Koup RA, Kedl RM, Mattapallil JJ, Weiss WR, Roederer M, Seder RA. HIV Gag protein conjugated to a Toll-like receptor 7/8 agonist improves the magnitude and quality of Th1 and CD8+ T cell responses in nonhuman primates. Proc Natl Acad Sci U S A 2005; 102:15190 - 4; http://dx.doi.org/10.1073/pnas.0507484102; PMID: 16219698
- Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang ML, Corey L, Hill J, Davis E, Flanigan C, et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med 2009; 360:1191 - 9; http://dx.doi.org/10.1056/NEJMoa0804749; PMID: 19297572
- Griffiths PD. Burden of disease associated with human cytomegalovirus and prospects for elimination by universal immunisation. Lancet Infect Dis 2012; 12:790 - 8; http://dx.doi.org/10.1016/S1473-3099(12)70197-4; PMID: 23017365
- Kharfan-Dabaja MA, Boeckh M, Wilck MB, Langston AA, Chu AH, Wloch MK, Guterwill DF, Smith LR, Rolland AP, Kenney RT. A novel therapeutic cytomegalovirus DNA vaccine in allogeneic haemopoietic stem-cell transplantation: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Infect Dis 2012; 12:290 - 9; http://dx.doi.org/10.1016/S1473-3099(11)70344-9; PMID: 22237175
- Pass RF. Development and evidence for efficacy of CMV glycoprotein B vaccine with MF59 adjuvant. J Clin Virol 2009; 46:Suppl 4 S73 - 6; http://dx.doi.org/10.1016/j.jcv.2009.07.002; PMID: 19647480
- Krause PR, Bialek SR, Boppana SB, Griffiths PD, Laughlin CA, Ljungman P, Mocarski ES, Pass RF, Read JS, Schleiss MR, et al. Priorities for CMV vaccine development. Vaccine 2013; 32:4 - 10; http://dx.doi.org/10.1016/j.vaccine.2013.09.042; PMID: 24129123
- Griffiths P, Plotkin S, Mocarski E, Pass R, Schleiss M, Krause P, Bialek S. Desirability and feasibility of a vaccine against cytomegalovirus. Vaccine 2013; 31:Suppl 2 B197 - 203; http://dx.doi.org/10.1016/j.vaccine.2012.10.074; PMID: 23598482
- Dasari V, Smith C, Khanna R. Recent advances in designing an effective vaccine to prevent cytomegalovirus-associated clinical diseases. Expert Rev Vaccines 2013; 12:661 - 76; http://dx.doi.org/10.1586/erv.13.46; PMID: 23750795
- Smith C, Khanna R. Herpesvirus vaccines: challenges and future prospects. Hum Vaccin 2010; 6:1062 - 7; http://dx.doi.org/10.4161/hv.6.12.13347; PMID: 21150281
- Dasari V, Smith C, Zhong J, Scott G, Rawlinson W, Khanna R. Recombinant glycoprotein B vaccine formulation with Toll-like receptor 9 agonist and immune-stimulating complex induces specific immunity against multiple strains of cytomegalovirus. J Gen Virol 2011; 92:1021 - 31; http://dx.doi.org/10.1099/vir.0.029413-0; PMID: 21307228
- Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, Khanna R. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J Virol 2003; 77:5226 - 40; http://dx.doi.org/10.1128/JVI.77.9.5226-5240.2003; PMID: 12692225
- Elkington R, Shoukry NH, Walker S, Crough T, Fazou C, Kaur A, Walker CM, Khanna R. Cross-reactive recognition of human and primate cytomegalovirus sequences by human CD4 cytotoxic T lymphocytes specific for glycoprotein B and H. Eur J Immunol 2004; 34:3216 - 26; http://dx.doi.org/10.1002/eji.200425203; PMID: 15368271
- Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 2005; 202:673 - 85; http://dx.doi.org/10.1084/jem.20050882; PMID: 16147978
- Manley TJ, Luy L, Jones T, Boeckh M, Mutimer H, Riddell SR. Immune evasion proteins of human cytomegalovirus do not prevent a diverse CD8+ cytotoxic T-cell response in natural infection. Blood 2004; 104:1075 - 82; http://dx.doi.org/10.1182/blood-2003-06-1937; PMID: 15039282
- Khan N, Bruton R, Taylor GS, Cobbold M, Jones TR, Rickinson AB, Moss PA. Identification of cytomegalovirus-specific cytotoxic T lymphocytes in vitro is greatly enhanced by the use of recombinant virus lacking the US2 to US11 region or modified vaccinia virus Ankara expressing individual viral genes. J Virol 2005; 79:2869 - 79; http://dx.doi.org/10.1128/JVI.79.5.2869-2879.2005; PMID: 15709006
- Thomson SA, Elliott SL, Sherritt MA, Sproat KW, Coupar BE, Scalzo AA, Forbes CA, Ladhams AM, Mo XY, Tripp RA, et al. Recombinant polyepitope vaccines for the delivery of multiple CD8 cytotoxic T cell epitopes. J Immunol 1996; 157:822 - 6; PMID: 8752934
- Thomson SA, Khanna R, Gardner J, Burrows SR, Coupar B, Moss DJ, Suhrbier A. Minimal epitopes expressed in a recombinant polyepitope protein are processed and presented to CD8+ cytotoxic T cells: implications for vaccine design. Proc Natl Acad Sci U S A 1995; 92:5845 - 9; http://dx.doi.org/10.1073/pnas.92.13.5845; PMID: 7541138
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol 2000; 18:767 - 811; http://dx.doi.org/10.1146/annurev.immunol.18.1.767; PMID: 10837075
- Moser M, Murphy KM. Dendritic cell regulation of TH1-TH2 development. Nat Immunol 2000; 1:199 - 205; http://dx.doi.org/10.1038/79734; PMID: 10973276
- Kaliński P, Hilkens CM, Wierenga EA, Kapsenberg ML. T-cell priming by type-1 and type-2 polarized dendritic cells: the concept of a third signal. Immunol Today 1999; 20:561 - 7; http://dx.doi.org/10.1016/S0167-5699(99)01547-9; PMID: 10562707
- Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity 2010; 33:492 - 503; http://dx.doi.org/10.1016/j.immuni.2010.10.002; PMID: 21029960
- Jones SA. Directing transition from innate to acquired immunity: defining a role for IL-6. J Immunol 2005; 175:3463 - 8; PMID: 16148087
- Pulendran B. Modulating vaccine responses with dendritic cells and Toll-like receptors. Immunol Rev 2004; 199:227 - 50; http://dx.doi.org/10.1111/j.0105-2896.2004.00144.x; PMID: 15233738
- Takagi H, Fukaya T, Eizumi K, Sato Y, Sato K, Shibazaki A, Otsuka H, Hijikata A, Watanabe T, Ohara O, et al. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity 2011; 35:958 - 71; http://dx.doi.org/10.1016/j.immuni.2011.10.014; PMID: 22177923
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 2008; 8:594 - 606; http://dx.doi.org/10.1038/nri2358; PMID: 18641647
- Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol 2011; 29:163 - 83; http://dx.doi.org/10.1146/annurev-immunol-031210-101345; PMID: 21219184
- King IL, Kroenke MA, Segal BM. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J Exp Med 2010; 207:953 - 61; http://dx.doi.org/10.1084/jem.20091844; PMID: 20421390
- Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci 2009; 18:936 - 48; http://dx.doi.org/10.1002/pro.102; PMID: 19384993
- Crowe J, Masone BS, Ribbe J. One-step purification of recombinant proteins with the 6xHis tag and Ni-NTA resin. Mol Biotechnol 1995; 4:247 - 58; http://dx.doi.org/10.1007/BF02779018; PMID: 8680931
- Smith C, Beagley L, Khanna R. Acquisition of polyfunctionality by Epstein-Barr virus-specific CD8+ T cells correlates with increased resistance to galectin-1-mediated suppression. J Virol 2009; 83:6192 - 8; http://dx.doi.org/10.1128/JVI.00239-09; PMID: 19357166
- Smith C, Cooper L, Burgess M, Rist M, Webb N, Lambley E, Tellam J, Marlton P, Seymour JF, Gandhi M, et al. Functional reversion of antigen-specific CD8+ T cells from patients with Hodgkin lymphoma following in vitro stimulation with recombinant polyepitope. J Immunol 2006; 177:4897 - 906; PMID: 16982932
- Zhong L, Peng X, Hidalgo GE, Doherty DE, Stromberg AJ, Hirschowitz EA. Identification of circulating antibodies to tumor-associated proteins for combined use as markers of non-small cell lung cancer. Proteomics 2004; 4:1216 - 25; http://dx.doi.org/10.1002/pmic.200200679; PMID: 15049001
- Marshall BC, Adler SP. Avidity maturation following immunization with two human cytomegalovirus (CMV) vaccines: a live attenuated vaccine (Towne) and a recombinant glycoprotein vaccine (gB/MF59). Viral Immunol 2003; 16:491 - 500; http://dx.doi.org/10.1089/088282403771926319; PMID: 14733736
- Wang Z, La Rosa C, Maas R, Ly H, Brewer J, Mekhoubad S, Daftarian P, Longmate J, Britt WJ, Diamond DJ. Recombinant modified vaccinia virus Ankara expressing a soluble form of glycoprotein B causes durable immunity and neutralizing antibodies against multiple strains of human cytomegalovirus. J Virol 2004; 78:3965 - 76; http://dx.doi.org/10.1128/JVI.78.8.3965-3976.2004; PMID: 15047812
- Vremec D. The isolation of mouse dendritic cells from lymphoid tissues and the identification of dendritic cell subtypes by multiparameter flow cytometry. Methods Mol Biol 2010; 595:205 - 29; http://dx.doi.org/10.1007/978-1-60761-421-0_14; PMID: 19941115
- Vremec D, Shortman K. The isolation and identification of murine dendritic cell populations from lymphoid tissues and their production in culture. Methods Mol Biol 2008; 415:163 - 78; PMID: 18370154
- Sanderson S, Shastri N. LacZ inducible, antigen/MHC-specific T cell hybrids. Int Immunol 1994; 6:369 - 76; http://dx.doi.org/10.1093/intimm/6.3.369; PMID: 8186188
- Usherwood EJ, Hogg TL, Woodland DL. Enumeration of antigen-presenting cells in mice infected with Sendai virus. J Immunol 1999; 162:3350 - 5; PMID: 10092789
- Nolan GP, Fiering S, Nicolas JF, Herzenberg LA. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci U S A 1988; 85:2603 - 7; http://dx.doi.org/10.1073/pnas.85.8.2603; PMID: 3128790
