Abstract
During the vaccination phase of the Rotavirus Efficacy and Safety Trial (REST), the period between the administration of dose 1 through 13 days after the administration of dose 3, there were more wild-type rotavirus gastroenteritis (RVGE) cases among vaccine recipients compared with placebo recipients using the protocol-specified microbiological plaque assay in the clinical-efficacy cohort, a subset of subjects where vaccine efficacy against RVGE of any severity was assessed. In this study, a rotavirus genome segment 6-based reverse transcriptase–polymerase chain reaction assay was applied post hoc to clarify the accuracy of type categorization of all these RVGE cases in vaccine recipients during the vaccination phase of REST. The assay characterized 147 (90%) of 163 re-assayed RVGE cases or rotavirus-associated health care contacts as type-determinable: either wild-type or vaccine-type rotavirus strains. In the clinical-efficacy cohort (N = 5673), 19 (18.8%) of 101 samples from RVGE cases contained wild-type rotavirus, 70 (69.3%) vaccine virus, and 12 (11.9%) were indeterminable. In the large-scale cohort (N = 68,038), 10 (34.5%) of 29 samples from RVGE-related health care contacts contained wild-type rotavirus strains, 15 (51.7%) vaccine-type rotavirus strains, and 4 (13.8%) were indeterminable. Of the 33 samples from RVGE cases in placebo recipients, all were confirmed to contain wild-type rotaviruses. Altogether, this post-hoc re-evaluation showed that the majority (75%) of type-determinable RVGE cases or health care contacts that occurred during the vaccination phase of REST in vaccine recipients were associated with vaccine-type rotavirus strains rather than wild-type rotavirus strains.
Introduction
Rotavirus is the leading etiologic agent of acute viral gastroenteritis in children under 5 y of age worldwide.Citation1-Citation3 Two rotavirus vaccines have now been licensed in many parts of the world: the live, attenuated, pentavalent rotavirus vaccine (RotaTeq®, Merck & Co, Inc.; designated RV5 by the Advisory Committee on Immunization Practices [ACIP]), and the live, attenuated human rotavirus vaccine (Rotarix®, GlaxoSmithKline Biologicals; designated RV1 by the ACIP).Citation4 Rotavirus vaccines have now been introduced into the national immunization programs of approximately 60 countries.
The efficacy of RV5 was evaluated in the pivotal Rotavirus Efficacy and Safety Trial (REST; NCT00090233).Citation5 The primary efficacy analyses were based on wild-type rotavirus gastroenteritis (RVGE) occurring 14 or more d after dose 3 of RV5 or placebo. This time frame was chosen to allow time for an immune response to develop after vaccine administration.Citation1,Citation2 REST demonstrated 98% efficacy of RV5 against naturally occurring severe RVGE and 74% efficacy against naturally occurring RVGE of any severity through the first full rotavirus season.Citation5 RV5 also reduced the rate of hospitalizations and emergency department (ED) visits for RVGE by 95% through up to 2 y of follow-upCitation5 and 94% for up to 3.1 y of follow-up.Citation6
During the vaccination phase of REST, the period beginning the day of administration of dose 1 through 13 d after the administration of dose 3, more wild-type RVGE cases occurred among the vaccine recipients compared with placebo recipients using the protocol-specified microbiological plaque assay in the clinical-efficacy (N = 5,673) trial cohort, a subset of subjects where vaccine efficacy against RVGE of any severity was assessed. In the clinical-efficacy cohort, 62% of such cases occurred among the vaccine recipients, even though the study was randomized 1:1 to receive RV5 or placebo. In this post hoc study, we clarify whether or not the rotavirus strains from the RVGE cases that occurred during the vaccination phase of REST had been accurately categorized as wild-type rotavirus strains by the original, protocol-specified microbiological plaque assay. A rotavirus gene 6-based reverse transcriptase–polymerase chain reaction [gene 6 RT-PCR] assay was developed and validated, to distinguish unequivocally vaccine-type from wild-type rotavirus strains. The gene 6 RT-PCR assay was applied to all vaccination-phase RVGE episodes categorized as wild-type by the original, protocol-specified microbiological plaque assay in vaccine recipients in the clinical-efficacy cohort and large-scale cohort, regardless of clinical severity or eventual ED visit or hospitalization of a recipient.
Results
Summary comparison of the microbiologic plaque assay and the molecular rotavirus gene 6 RT-PCR assay for characterizing strains as wild-type or vaccine-type that occurred during the vaccination phase
The rotavirus gene 6-based RT-PCR assay characterized the rotavirus type in 147 (90%) of the 163 re-tested samples among vaccine recipients (), which consisted of 101 vaccine recipients in the clinical-efficacy cohort, 29 vaccine recipients in the large-scale cohort, and 33 placebo recipients in the clinical-efficacy cohort. Of 130 rotavirus-positive samples categorized as wild-type rotavirus by the plaque assay, RT-PCR categorized 29 (22%) as wild-type, 85 (65%) as vaccine-type, and 16 (12%) as type-indeterminable (). Because RV5 was unlikely to be present in placebo recipients, only 33 of these samples were re-tested, and all tested samples were confirmed to contain wild-type rotavirus (). The majority (70%) of these re-classified RVGE cases were from Finland, where parents were very compliant with early submission of stool samples from infants with mild gastroenteritis symptoms.
Figure 1. Categorization by the gene 6-based RT-PCR assay of rotavirus-positive stool samples collected during the vaccination phase of REST among all infants who received at least 1 dose of vaccine or placebo. All specimens were characterized as wild-type by the microbiological plaque assay. (A) Categorization of rotavirus cases among participants in the clinical-efficacy cohort of REST. (B) Categorization of samples collected during rotavirus-associated health care contacts (hospitalization or ED visit) during the vaccination phase among all participants in the large-scale cohort of REST. REST, Rotavirus Efficacy and Safety Trial; RV5, pentavalent rotavirus vaccine; EIA, enzyme immunoassay. RT-PCR, reverse transcriptase–polymerase chain reaction; ED, emergency department.
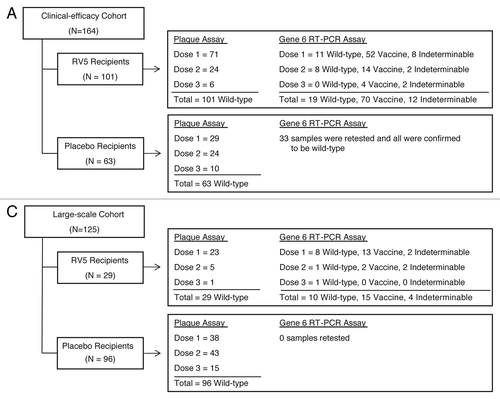
RVGE cases of any severity that occurred during the vaccination phase among infants in the clinical-efficacy cohort of REST
Among vaccine recipients in the clinical-efficacy cohort, the rotavirus gene 6 RT-PCR assay confirmed presence of wild-type rotavirus in 19 (19%) of the 101 enzyme immunoassay (EIA) rotavirus-positive RVGE cases, while 70 (69%) samples from the 101 EIA rotavirus-positive RVGE cases were shown to contain vaccine-type rotavirus (). The rotavirus type was indeterminable in 12 (12%) of the 101 samples ().
The median clinical severity scores for the RVGE cases among vaccine recipients attributed to vaccine virus were less than the clinical severity scores of RVGE cases attributed to wild-type rotavirus, as follows: for RVGE cases attributed to vaccine virus, the median clinical severity score was 4 (range, 1–11) and for RVGE cases attributed to wild-type rotavirus, the median clinical severity score was 6 (range, 1–17). The median clinical severity score of RVGE cases among placebo recipients was 10 (range, 1–19), which is indicative of moderate-to-severe RVGE. The median score reduction between the wild-type RVGE cases among placebo recipients and vaccine-type RVGE among vaccine recipients was 6 (95% confidence interval [CI]: 4, 7); whereas, the median score reduction between the wild-type RVGE cases among placebo recipients and wild-type RVGE among vaccine recipients was 4 (95% CI: –1, 7).
RVGE episodes associated with hospitalizations and ED Visits that occurred during the vaccination phase of the large-scale cohort in REST
The re-evaluation of RVGE EIA rotavirus-positive cases among vaccine recipients in the large-scale cohort also yielded a change in the distribution of vaccine-type- and wild-type-associated cases. Among vaccine recipients in the large-scale cohort, the rotavirus gene 6 RT-PCR assay confirmed presence of wild-type rotavirus in 10 (40%) of the 25 type-determinable contacts (). The rotavirus type was indeterminable in 4 (14%) of the 29 samples from this cohort. Samples from RVGE health care contacts in the placebo recipients in the large-scale cohort were not re-tested by the rotavirus gene 6 RT-PCR assay.
Distribution over time of RVGE episodes that occurred during the vaccination phase in the clinical-efficacy and large-scale cohorts of REST
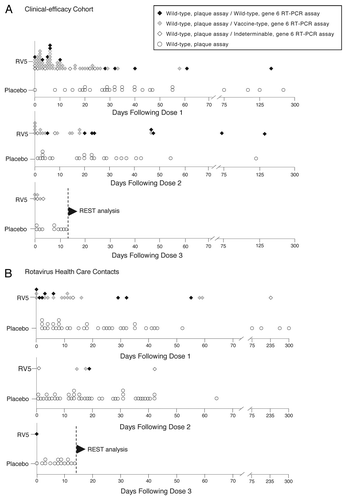
Wild-type RVGE cases were distributed differently over days in the vaccination phase between vaccine and placebo recipients, as determined by the gene 6 RT-PCR assay. For example, after dose 1, the odds of a wild-type RVGE case occurring within the first 18 d relative to after 18 d was 5.76 (95% CI: 0.97, 34.50) times higher for vaccine recipients than for placebo recipients. This phenomenon was not observed after dose 2 (). In the large-scale cohort, a similar clustering of wild-type RVGE health care contacts of ED visits or hospitalizations was not observed early after dose 1 (). The difference in the clinical-efficacy cohort after dose 1 hinges on the assumption that all 29 post-dose 1 RVGE cases during the vaccination phase in placebo recipients were by wild-type rotavirus strains, an assumption supported by the gene 6 RT-PCR testing of 33 of those 63 cases, as noted above ().
Figure 2. Summary of the re-categorization by the gene 6 RT-PCR assay of EIA rotavirus-positive stool samples collected during the vaccination phase of REST from infants who received at least 1 dose of RV5 or placebo. (A) RVGE cases among participants in the clinical-efficacy cohort of REST. (B) RVGE-attributable health care contacts (hospitalization or ED visit) among all participants in the large-scale cohort of REST. The trends observed in the clinical-efficacy cohort were observed in the large-scale cohort, but severity of illness was greater in participants hospitalized or taken to EDs than in the clinical-efficacy cohort and health care contacts were fewer in number than in the clinical-efficacy cohort. REST, Rotavirus Efficacy and Safety Trial; RV5, pentavalent rotavirus vaccine; EIA, enzyme immunoassay. RT-PCR, reverse transcriptase–polymerase chain reaction; ED, emergency department.
Discussion
A molecular RT-PCR assay based on the rotavirus gene 6 encoding viral protein (VP) 6 was developed, validated, and applied post hoc to discriminate unequivocally between wild-type and vaccine-type virus strains in stool samples collected during the vaccination phase of REST. The molecular RT-PCR assay detects the genome of the virus, whether associated with intact or degraded particles. Overall, the rotavirus gene 6 RT-PCR successfully determined the type of 147 (90%) of the 163 rotavirus strains in these archived samples. Still, 16 (10%) of all the samples re-tested (n = 163), encompassing both vaccine and placebo recipients, could not be categorized as either vaccine-type or wild-type. Review of the optical density readings from the EIA of 15 of the 16 samples, whose rotavirus strain identity could not be determined, revealed that the optical density readings were close to the cut-off value of the assay (data not shown), which indicates that the amount of rotavirus antigen was low in these samples. Coupled with the years of storage and different freeze and thaw cycles that these samples may have incurred, it is plausible that the amount of rotavirus genome was below the limit of detection (LOD) of the assay or was from degraded samples. In recent large (6,674 infants) phase III efficacy trials of RV5 in sub-Saharan Africa (Ghana, Kenya, Mali)Citation7 and Southeast Asia (Bangladesh, Vietnam),Citation8 the percent of rotavirus gene 6 non-typeable strains was 1.1% (9 out 803 rotavirus-positive samples), confirming the applicability and performance of the assay in large efficacy clinical trials.
Overall, this re-evaluation clarified the potential reason for more wild-type RVGE cases among vaccine recipients compared with placebo recipients using the protocol-specified microbiological plaque assay during the vaccination phase in the clinical-efficacy cohort of REST. The rotavirus gene 6 RT-PCR assay is the only assay that the RV5 program has used since 2008 to categorize stool samples as either vaccine-type or wild-type. Although there are other molecular assays, such as the reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay,Citation9,Citation10 which may be a valid option with high throughput for rapid detection of pathogens in human and animal samples as a screening test, sequencing following RT-PCR satisfies regulatory and licensing concerns as the actual sequence of the pathogen in the sample is determined. Overall, this assay is appropriate for use in this setting, but does depend upon the target gene's diversity, the ability to create high-affinity primers (ie, sequence diversity at the best primer sites), and the nature of the specimen being tested (eg, stool samples, as a whole, can be problematic).
Also noted from this re-evaluation in the clinical-efficacy cohort, that included RVGE cases of any severity, was the earlier clustering after dose 1 of wild-type RVGE cases among vaccine recipients than occurred among placebo recipients. These data may be a reflection of take of RV5 and timing of protection against RVGE starting after the first dose. This finding comes in addition to the per-protocol results from REST that showed that completion of the 3-dose series provides durable and optimal protection.Citation5,Citation6
Overall, according to the rotavirus gene 6 RT-PCR assay the rate of occurrence of vaccine-type RVGE was 2.5% in the clinical-efficacy cohort, which captured all severities of illness, and 0.04% in the large-scale cohort of REST, which captured all RVGE-related hospitalizations and ED visits. These results are consistent with previous findings, which showed that RV5 was well tolerated, with a slight (1.3%) increase in the rates of mild vomiting and mild diarrhea within the first week after the first dose.Citation1,Citation2,Citation11,Citation12 Although our data does indicate that RV5 can be associated with RVGE, the median clinical severity score for the RVGE cases among vaccine recipients attributed to vaccine-type rotavirus, as measured by a validated, 24-point clinical scoring system, was 4 (range, 1–11), which is indicative of mild RVGE.
In pre-licensure clinical trials of RV5, shedding of vaccine virus strains, as measured by the microbiological plaque assay, occurred in 32 (8.9%) of 360 vaccine recipients after the first dose.Citation5 Among these recipients, 15 were identified as WI79-4 (P1), 4 as WI78-8 (G3), and 1 as BrB-9 (G4) by RNA electropherotyping. Potential reassortment among the vaccine strains was revealed to occur in the other 12 recipients, with confirmed reassortant vaccine strains between parent strains WI79-9 (G1) and P1 (n = 1) and the G3 and P1 (n = 2). The remaining 9 recipients shed other potential combinations of at least 2 vaccine strains and possibly wild-type strains. In total, only 17 recipients with stool samples that contained vaccine virus strains were collected based on a suspected AGE episode, but only 6 (35%) of these 17 stool samples met the established clinical AGE definition and the vaccine virus strains among these varied. Post-marketing surveillance has shown similar reassortment in vivo at low frequencies in several vaccinated populations,Citation13-Citation15 and these reassorted vaccine strains have occasionally caused gastroenteritis symptoms in vaccine recipients and, in some rare instances, may have transmitted the virus to close contacts.Citation16,Citation17 These findings are not unexpected as RV5 is a live viral vaccine, which replicates in the recipients’ gut and was observed in prelicensure studies.Citation5,Citation11 The benefits of vaccination outweigh any small risk of vaccine-associated gastroenteritis as RV5 has been shown to be highly effective in preventing disease with routine use of the vaccine.Citation18
In retrospect, the use of the microbiological plaque assay to determine wild-type vs. vaccine strain was not optimal for efficacy calculations. The plaque assay was utilized in REST because RV5 readily forms plaques in susceptible cell monolayers upon direct inoculation, whereas wild-type rotaviruses do not.Citation19 Hence, it was used to determine vaccine-type or wild-type rotavirus strains.Citation5 In the plaque assay, a rotavirus particle is able to form a plaque when an intact virus particle infects a cell, replicates, and then lyses that cell.Citation19 However, it is important to note that the plaque assay may not always detect fully intact, infectious rotavirus particles present in all samples; the virus in the stool may not replicate in cell culture because some stool specimens are cytotoxic and inhibitors prevent replication of fully intact, infectious rotavirus particles. With this caveat, the plaque assay was thought to only detect infectious vaccine virus but if the vaccine-virus in the stool sample was not infectious (or failed to grow in the plaque assay), then it was assumed that no vaccine-virus was present in the sample. The post hoc analyses presented here indicate that the plaque assay did not accurately type all rotavirus cases during the vaccination phase of REST. If the molecular VP6 RT-PCR assay had been originally used to determine whether the RVGE cases were due to wild-type or vaccine-type, then wild-type RVGE cases that occurred during the vaccination phase that would have been re-classified as vaccine-type would not have been excluded from the per-protocol primary efficacy analysis, which, alone, would have resulted in a higher efficacy estimate. The rotavirus gene 6 RT-PCR unequivocally categorizes the identity of the rotavirus present in the stool samples as either wild-type or vaccine-type because the RT-PCR assay detects the virus genome in the stool, whether infectious or not, and allows confirmation by sequencing.
With the recognition that the rotavirus gene 6 RT-PCR assay is suitable to determine whether the rotavirus strains present in stool samples are vaccine-type or wild-type in origin, this assay has been utilized in subsequent RV5 phase III efficacy clinical trials in Africa and Southeast Asia,Citation7,Citation8 and Japan,Citation20 as well as in an epidemiological surveillance study in Nicaragua.Citation21
Methods
Study population
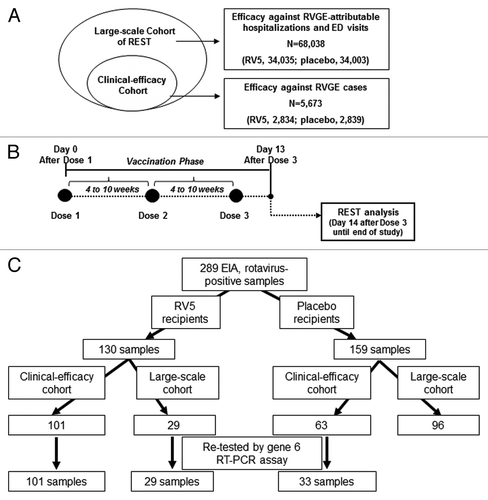
The samples re-evaluated here were from the vaccination phase of REST, a multicenter, placebo-controlled, randomized pivotal clinical trial that enrolled approximately 70 000 healthy infants.Citation5 Per protocol, infants were to receive 3 oral doses of vaccine or placebo. For this re-evaluation, all stool specimens in the intent-to-treat trial participants in the clinical-efficacy and large-scale cohorts of REST who received at least 1 dose were eligible for inclusion. Vaccine efficacy against RVGE-attributable hospitalizations and ED visits was evaluated in the large-scale cohort of 68 038 infants (vaccine, 34 035; placebo, 34 003; ). The clinical-efficacy cohort (vaccine, 2834 infants; placebo, 2839 infants; ) was a subset of the large-scale cohort in which we assessed vaccine efficacy against RVGE of any severity, based upon a severity score, whether that case resulted in a health care contact or not. In total, 20 subjects in the clinical-efficacy cohort with affected samples were also in the large-scale cohort: 6 vaccine recipients and 14 placebo recipients.
Figure 3. Overview of REST and EIA rotavirus-positive stool samples collected during the vaccination phase of the trial. (A) In REST, the clinical-efficacy cohort was nested within the large-scale cohort. (B) The vaccination phase began the day of administration of dose 1 through 13 d after the administration of dose 3. Dose 1 was administered at 6 to 12 wk of age, dose 3 was administered no later than 32 wk of age, and the interval between doses was 4 to 10 wk. The per-protocol primary monitoring period for vaccine efficacy began 14 d after dose 3, i.e., once the vaccination phase was completed. (C) Flowchart indicating the number of EIA rotavirus-positive stool samples collected from RV5 and placebo recipients during the vaccination phase of REST. The number of samples re-tested by the gene 6 RT-PCR assay is indicated at the bottom. REST, Rotavirus Efficacy and Safety Trial; EIA, enzyme immunoassay.
Case definition of RVGE and vaccine virus shedding in REST
To meet the case definition for RVGE in REST, participants met both clinical and laboratory criteria: (1) 3 or more watery or looser-than-normal stools, or 1 watery stool, within a 24-h period and/or forceful vomiting and (2) detection of rotavirus antigen by EIA in a stool sample obtained within 14 d after the onset of symptoms from infants with at least 1 clinical criterion.Citation5,Citation22,Citation23 Stool samples were to be collected based on clinical symptoms that occurred any time after the receipt of dose 1 until the end of the study. Two stool samples were to be collected as soon as possible after the onset of symptoms occurring from day 0, the day of dose 1, until the end of the study. Optimally, the first sample was to be obtained within 24 h of the onset of symptoms and the second sample approximately 24 h later.
For EIA rotavirus-positive stool samples, the neutralization antigens VP7 (G type) and VP4 (P type) genotypes were determined utilizing RT-PCR followed by amplicon sequencing,Citation24 while a microbiological plaque assay was utilized to determine whether the stool samples contained infectious vaccine virus strains.Citation25 Given that few wild-type human rotaviruses are cytopathic in cell culture unless adapted to culture by several blind passages,Citation19 failure to form plaques in the microbiological plaque assay categorized, for REST, the rotavirus present in the stool sample as wild-type rotavirus.
Outcome measures
For infants who participated in the clinical-efficacy cohort, vaccine efficacy against all cases of RVGE of any severity was determined and the episode was scored by a validated, 24-point clinical scoring system previously described (scores ≤8 were mild, those >8 and ≤16 were moderate-to-severe, and those >16 were severeCitation5). Collection of stool sample frequencies and rates from all acute gastroenteritis episodes, those involving health care resource utilization, defined as hospitalizations and ED visits, for all infants in REST (large-scale cohort), as well as those within the subset clinical-efficacy cohort, were previously described.Citation5
Rotavirus gene 6 RT-PCR assay
A RT-PCR assay targeting the rotavirus genome segment 6 (encoding the structural viral protein 6, VP6) was developed, and validated, to determine the VP6 genotype and species of origin in stool samples from participants in epidemiology studies or clinical trials. This molecular assay utilized methodology similar to the previously validated rotavirus gene 9 (encoding VP7)Citation24 and rotavirus gene 4 (encoding VP4)Citation26 RT-PCR assays, used to determine the rotavirus genotypes in stool samples trial participants. The rotavirus gene 6 RT-PCR assay was then applied to the RVGE stool samples collected during the vaccination phase of REST to identify the species of origin of the rotavirus gene 6 of the rotavirus strain present. Because all 5 of the RV5 vaccine strains possess a gene 6 derived from the bovine strain WC3 and the gene 6 from human wild-type rotavirus are distinct from RV5 at the nucleotide sequence level, the sequencing data obtained from a gene 6 RT-PCR amplicon was able to categorize the virus in the stool sample as vaccine or a human wild-type rotavirus.
The gene 6 RT-PCR assay was adapted from that described by Iturriza-Gómara, et al.,Citation27,Citation28 with minor modifications. Ribonucleic acid (RNA) was extracted from stool samples using the QIAamp™ Viral RNA Extraction Kit (QIAGEN, Inc.) or Boom’s silica extraction method.Citation29 The algorithm followed was to first extract the RNA material using the QIAamp Viral RNA Extraction kit and if RT-PCR was negative, the RNA material was extracted with the Boom’s silica extraction method. In addition, if the extracted material using the QIAamp Viral RNA Extraction kit generated an amplicon that yielded poor sequencing data, the RNA material was extracted using the Boom’s silica extraction method. The extracted RNA was denatured at 95 °C for 2 min, and then transferred immediately to ice prior to the RT-PCR. The RT reaction in the RT-PCR reaction was performed at 50 °C, instead of the traditional 42 °C, as specified in the Qiagen OneStep RT-PCR kit, following the manufacturer’s instructions. Degenerate primers (containing a mixture of nucleotide sequences) designed to amplify a 379-base pair region spanning amino acids 241 to 367 encoded by the VP6 gene were utilized. The polarity, indicated as plus- or minus-sense, and sequences (5′ to 3′) of the primers were as follows: VP6-F (plus-sense), GAY GGN GCD ACN ACA TGG T; VP6-R (minus-sense), GTC CAR TTC ATN CCT GGY GG. The RT-PCR parameters were 30 min at 50 °C (reverse transcription) and 15 min at 95 °C (inactivation of the RT enzyme and activation of polymerase) and 40 cycles each of 45 s at 94 °C (template denaturation), 45 s at 54 °C (primer annealing), 1 min at 72 °C (product extension), and a final extension of 10 min at 72 °C. Amplicon purification, direct sequencing with dye-terminator chemistry, and nucleotide and amino acid sequence analysis were performed as described previously.Citation24 Sequences were evaluated for identity to known gene 6 sequences utilizing MacVector™ (MacVector, Inc.). The final gene 6 designation (wild-type [human strain] or vaccine-type [bovine rotavirus WC3 strain] type) was determined using a reference database containing sequences of diverse gene 6 sequences from known infected species (human, bovine [other than that of strain WC3], porcine, simian, lapine, canine, feline, equine, avian, ovine, and murine), as well as the gene 6 sequence of the RV5 strains.Citation30 A gene 6 designation of “indeterminable” was made when a definitive determination was not possible, because the quality of the sequence was poor or the viral genome quantity in the sample likely was low.
Validation of the rotavirus gene 6 RT-PCR assay
The validation of the rotavirus gene 6 RT-PCR assay was performed and the accuracy, precision, ruggedness, and LOD of the assay were tested. The accuracy of the assay was assessed by comparing rotavirus gene 6 determinations (vaccine [bovine strain WC3] vs. human) of 44 samples with known rotavirus strains. The acceptance criteria were 100% agreement among all gene 6 determinations for all replicates of the known VP6 genotype, where a VP6 determination was obtained. The precision of the assay was assessed by comparing determinations of the rotavirus gene 6 sequences between replicate samples. The acceptance criteria were 100% agreement for sample amplification and VP6 determinations among replicates where a VP6 determination was obtained. In addition, there had to be >85% agreement in the ability to obtain a VP6 determination compared with an undeterminable (non-typeable) result for sample replicates that had been positively amplified. The ruggedness of the assay was assessed by comparing determinations between replicate samples that were tested by multiple analysts using multiple lots of master mix kits and primers. The acceptance criteria were 100% agreement for sample amplification and VP6 determinations among replicates where a VP6 determination was obtained. In addition, there had to be >85% agreement in the ability to obtain a VP6 determination compared with an undeterminable (non-typeable) result for sample replicates that had been positively amplified, regardless of analyst, master mix lot, or primer lot. The LOD was defined as the lowest concentration of a sample that can be reproducibly amplified by the RT-PCR and subsequently sequenced. The LOD was evaluated by comparing the VP6 determinations obtained for dilution series of an individual rotavirus vaccine strain (WI78–8 [G3]) and the human rotavirus strain SC2 (G2). The LOD for each extraction method was pre-defined as the concentration at which 6 results of the 8 results obtained corresponded to a correct VP6 determination. The acceptance criteria on the LOD was ≤1 × 105 plaque-forming units (pfu)/mL for both strains using the Qiagen extraction method, and ≤1 × 104 pfu/mL for both strains using the Boom’s extraction method. In addition, the criteria had to be met regardless of analyst or master mix lot or primer lot.
All pre-defined assay acceptance criteria were met (data not shown). The LOD of the rotavirus gene 6 RT-PCR assay was determined to be 1 × 105 pfu/mL and 1 × 104 pfu/mL for human rotavirus gene 6 and vaccine rotavirus (strain WC3) gene 6, respectively, using the Qiagen extraction method. The LOD using the Boom’s extraction method was determined to be 1 × 103 pfu/mL for both the human and vaccine rotavirus gene 6. Thus, the rotavirus gene 6 RT-PCR assay was deemed suitable for its intended use: the determination of the VP6 genotype that allows to categorize the rotavirus strain as either vaccine-type or wild-type in samples from participants in clinical or epidemiological studies.
Stool samples collected during the vaccination phase of REST
During the vaccination phase of REST, the time frame beginning day 0 immediately after receipt of dose 1 through 13 d after receipt of dose 3; (), there were 289 samples positive by rotavirus EIA for RVGE cases, of which 163 were retrospectively tested (), as follows: 130 stool samples were collected from vaccine recipients in the 2 cohorts, of which 101 samples were from participants in the clinical-efficacy cohort and 29 were from participants in the large-scale cohort (), and 159 stool samples were collected from placebo recipients in the 2 cohorts, of which 63 were from participants who were part of the clinical-efficacy cohort and 96 were from the large-scale cohort (). All of the stool samples from vaccine recipients during the vaccination phase from RVGE cases in the 2 cohorts were re-evaluated in this work, in addition to a subset of 33 samples from such cases among placebo recipients in the clinical-efficacy cohort (); the clinical-efficacy cohort was chosen for testing of samples from placebo recipients because the full severity score range could be re-evaluated, whereas samples from health care contact cases would include only those cases most severe. The majority (70%) of these samples was from 3 clinical sites in Finland, and the remaining samples (30%) were from US sites.
Data analysis
All RVGE cases and health care contacts that occurred during the vaccination phase were summarized for the rotavirus type by the gene 6 RT-PCR assay: wild-type or vaccine-type, further divided by dose and by treatment group, according to the results of the microbiological plaque assay (as pre-specified in REST and the clinical development of the pentavalent rotavirus vaccine) in this post hoc analysis. Clinical severity scores between vaccine and placebo recipients who were rotavirus-positive by EIA were compared via difference in medians and associated bootstrap CIs. Distributions of cases with respect to time after administration of a dose of vaccine or placebo were compared with odds ratios and associated exact CIs.
| Abbreviations: | ||
| CI | = | confidence interval |
| ED | = | emergency department |
| EIA | = | enzyme immunoassay |
| REST | = | Rotavirus Efficacy and Safety Trial |
| RNA | = | ribonucleic acid |
| RT-PRC | = | reverse transcriptase–polymerase chain reaction |
| RV | = | rotavirus |
| RV5 | = | pentavalent rotavirus vaccine |
| RVGE | = | rotavirus gastroenteritis |
| VP | = | viral protein |
Disclosure of Potential Conflicts of Interest
D.O.M. received no compensation from Merck & Co., Inc. for conduct of the research described in this report; he has been a consultant to Merck & Co., Inc., GSK, PATH, and the NIAID; a speaker for Merck & Co., Inc., and GSK; and a grant recipient from Merck & Co., Inc. T.V. has been a consultant and speaker for Merck & Co., Inc., Sanofi Pasteur-Merck Sharp & Dohme (SPMSD), MedImmune, Novartis, and GlaxoSmithKline Biologicals (GSK); any compensation received from Merck & Co., Inc., was directly related to the reasonable costs of conducting the research as specified in the research agreement from Merck & Co., Inc. P.D. has no other conflict of interest. Compensation received from Merck & Co., Inc., was directly related to the reasonable costs of conducting the research as specified in the research agreement. M.G.G. is an employee of Merck & Co., Inc., and may own stock or stock options in the company. M.C., M.J.D., and R.F.I. were employed by Merck at the time the primary study (REST) was conducted and may own stock or stock options in the company.
Acknowledgments
The authors would like to acknowledge Mark J DiNubile (Merck & Co., Inc.) and Susan Starcevic, and Tonya Goodman (Arbor Communications, Inc.) for editorial support in the writing of this article. We also thank Laura Mallette and Penny M Heaton (both formerly of Vaccines–Clinical Research, Merck & Co., Inc.) for the development and validation work of the rotavirus gene 6 RT-PCR assay and insightful discussions, respectively. This study was funded by Merck & Co., Inc.
References
- Heaton PM, Ciarlet M. Vaccines: the pentavalent rotavirus vaccine: discovery to licensure and beyond. Clin Infect Dis 2007; 45:1618 - 24; http://dx.doi.org/10.1086/522997; PMID: 18198497
- Ciarlet M, Schödel F. Development of a rotavirus vaccine: clinical safety, immunogenicity, and efficacy of the pentavalent rotavirus vaccine, RotaTeq. Vaccine 2009; 27:Suppl 6 G72 - 81; http://dx.doi.org/10.1016/j.vaccine.2009.09.107; PMID: 20006144
- Matthijnssens J, Bilcke J, Ciarlet M, Martella V, Bányai K, Rahman M, Zeller M, Beutels P, Van Damme P, Van Ranst M. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol 2009; 4:1303 - 16; http://dx.doi.org/10.2217/fmb.09.96; PMID: 19995190
- Cortese MM, Parashar UD, Centers for Disease Control and Prevention (CDC). Prevention of rotavirus gastroenteritis among infants and children: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2009; 58:RR-2 1 - 25; PMID: 19194371
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, et al, Rotavirus Efficacy and Safety Trial (REST) Study Team. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 2006; 354:23 - 33; http://dx.doi.org/10.1056/NEJMoa052664; PMID: 16394299
- Vesikari T, Karvonen A, Ferrante SA, Kuter BJ, Ciarlet M. Sustained efficacy of the pentavalent rotavirus vaccine, RV5, up to 3.1 years following the last dose of vaccine. Pediatr Infect Dis J 2010; 29:957 - 63; http://dx.doi.org/10.1097/INF.0b013e3181e28e6e; PMID: 20442684
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 2010; 376:606 - 14; http://dx.doi.org/10.1016/S0140-6736(10)60889-6; PMID: 20692030
- Zaman K, Dang DA, Victor JC, Shin S, Yunus M, Dallas MJ, Podder G, Vu DT, Le TP, Luby SP, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet 2010; 376:615 - 23; http://dx.doi.org/10.1016/S0140-6736(10)60755-6; PMID: 20692031
- Xie Z, Fan Q, Liu J, Pang Y, Deng X, Xie Z, Liji X, Khan MI. Reverse transcription loop-mediated isothermal amplification assay for rapid detection of Bovine Rotavirus. BMC Vet Res 2012; 8:133; http://dx.doi.org/10.1186/1746-6148-8-133; PMID: 22894568
- Yang BY, Liu XL, Wei YM, Wang JQ, He XQ, Jin Y, Wang ZJ. Rapid and sensitive detection of human astrovirus in water samples by loop-mediated isothermal amplification with hydroxynaphthol blue dye. BMC Microbiol 2014; 14:38; http://dx.doi.org/10.1186/1471-2180-14-38; PMID: 24524254
- Dennehy PH, Goveia MG, Dallas MJ, Heaton PM. The integrated phase III safety profile of the pentavalent human-bovine (WC3) reassortant rotavirus vaccine. Int J Infect Dis 2007; 11:Suppl 2 S36 - 42; http://dx.doi.org/10.1016/S1201-9712(07)60020-4; PMID: 18162245
- RotaTeq [prescribing information]. Whitehouse Station, NJ: Merck & Co, Inc.; 2010. Available at: https://www.merck.com/product/usa/pi_circulars/r/rotate_pi.pdf. Accessed June 9, 2014
- Hemming M, Vesikari T. Vaccine-derived human-bovine double reassortant rotavirus in infants with acute gastroenteritis. Pediatr Infect Dis J 2012; 31:992 - 4; http://dx.doi.org/10.1097/INF.0b013e31825d611e; PMID: 22581224
- Donato CM, Ch’ng LS, Boniface KF, Crawford NW, Buttery JP, Lyon M, Bishop RF, Kirkwood CD. Identification of strains of RotaTeq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J Infect Dis 2012; 206:377 - 83; http://dx.doi.org/10.1093/infdis/jis361; PMID: 22615314
- Boom JA, Sahni LC, Payne DC, Gautam R, Lyde F, Mijatovic-Rustempasic S, Bowen MD, Tate JE, Rench MA, Gentsch JR, et al. Symptomatic infection and detection of vaccine and vaccine-reassortant rotavirus strains in 5 children: a case series. J Infect Dis 2012; 206:1275 - 9; http://dx.doi.org/10.1093/infdis/jis490; PMID: 22872730
- Payne DC, Edwards KM, Bowen MD, Keckley E, Peters J, Esona MD, Teel EN, Kent D, Parashar UD, Gentsch JR. Sibling transmission of vaccine-derived rotavirus (RotaTeq) associated with rotavirus gastroenteritis. Pediatrics 2010; 125:e438 - 41; http://dx.doi.org/10.1542/peds.2009-1901; PMID: 20100758
- Hemming M, Vesikari T. Detection of rotateq vaccine-derived, double-reassortant rotavirus in a 7-year-old child with acute gastroenteritis. Pediatr Infect Dis J 2014; 33:655 - 6; http://dx.doi.org/10.1097/INF.0000000000000221; PMID: 24326415
- Rha B, Tate JE, Payne DC, Cortese MM, Lopman BA, Curns AT, Parashar UD. Effectiveness and impact of rotavirus vaccines in the United States - 2006-2012. Expert Rev Vaccines 2014; 13:365 - 76; http://dx.doi.org/10.1586/14760584.2014.877846; PMID: 24392657
- Ciarlet M, Estes MK. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol 1999; 80:943 - 8; PMID: 10211964
- Iwata S, Nakata S, Ukae S, Koizumi Y, Morita Y, Kuroki H, Tanaka Y, Shizuya T, Schödel F, Brown ML, et al. Efficacy and safety of pentavalent rotavirus vaccine in Japan: a randomized, double-blind, placebo-controlled, multicenter trial. Hum Vaccin Immunother 2013; 9:1626 - 33; http://dx.doi.org/10.4161/hv.24846; PMID: 23732903
- Khawaja S, Cardellino A, Mast TC. Hospital-based surveillance and analysis of genotype variation in nicaragua after the introduction of the pentavalent rotavirus vaccine. Pediatr Infect Dis J 2014; 33:e25 - 8; http://dx.doi.org/10.1097/INF.0000000000000074; PMID: 24042492
- Gilchrist MJ, Bretl TS, Moultney K, Knowlton DR, Ward RL. Comparison of seven kits for detection of rotavirus in fecal specimens with a sensitive, specific enzyme immunoassay. Diagn Microbiol Infect Dis 1987; 8:221 - 8; http://dx.doi.org/10.1016/0732-8893(87)90053-8; PMID: 2835201
- Ward RL, McNeal MM, Clemens JD, Sack DA, Rao M, Huda N, Green KY, Kapikian AZ, Coulson BS, Bishop RF, et al. Reactivities of serotyping monoclonal antibodies with culture-adapted human rotaviruses. J Clin Microbiol 1991; 29:449 - 56; PMID: 1709945
- DiStefano DJ, Kraiouchkine N, Mallette L, Maliga M, Kulnis G, Keller PM, Clark HF, Shaw AR. Novel rotavirus VP7 typing assay using a one-step reverse transcriptase PCR protocol and product sequencing and utility of the assay for epidemiological studies and strain characterization, including serotype subgroup analysis. J Clin Microbiol 2005; 43:5876 - 80; http://dx.doi.org/10.1128/JCM.43.12.5876-5880.2005; PMID: 16333070
- Clark HF, Furukawa T, Bell LM, Offit PA, Perrella PA, Plotkin SA. Immune response of infants and children to low-passage bovine rotavirus (strain WC3). Am J Dis Child 1986; 140:350 - 6; PMID: 3006476
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 1992; 30:1365 - 73; PMID: 1320625
- Iturriza Gómara M, Wong C, Blome S, Desselberger U, Gray J. Rotavirus subgroup characterisation by restriction endonuclease digestion of a cDNA fragment of the VP6 gene. J Virol Methods 2002; 105:99 - 103; http://dx.doi.org/10.1016/S0166-0934(02)00087-3; PMID: 12176146
- Iturriza-Gómara M, Green J, Grey J. 2000. Methods of rotavirus detection, sero- and genotyping, sequencing, and phylogenetic analysis. In: Methods in Molecular Medicine, Vol 34: Rotaviruses: Methods and Protocols. Edited by J Gray and U Desselberger, Humana Press Inc., Totowa, NJ, USA.
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 1990; 28:495 - 503; PMID: 1691208
- Matthijnssens J, Joelsson DB, Warakomski DJ, Zhou T, Mathis PK, van Maanen MH, Ranheim TS, Ciarlet M. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology 2010; 403:111 - 27; http://dx.doi.org/10.1016/j.virol.2010.04.004; PMID: 20451234
