Abstract
With over 150 million people chronically infected worldwide and millions more infected annually, hepatitis C continues to pose a burden on the global healthcare system. The standard therapy of hepatitis C remains expensive, with severe associated side effects and inconsistent cure rates. Vaccine development against the hepatitis C virus has been hampered by practical and biological challenges posed by viral evasion mechanisms. Despite these challenges, HCV vaccine research has presented a number of candidate vaccines that progressed to phase II trials. However, those efforts focused mainly on HCV genotypes 1 and 2 as vaccine targets and barely enough attention was given to genotype 4, the variant most prevalent in the Middle East and central Africa. We describe herein the in silico identification of highly conserved and immunogenic T-cell epitopes from the HCV genotype 4 proteome, using the iVAX immunoinformatics toolkit, as targets for an epitope-driven vaccine. We also describe a fast and inexpensive approach for results validation using the empirical data on the Immune Epitope Database (IEDB) as a reference. Our analysis identified 90 HLA class I epitopes of which 20 were found to be novel and 19 more had their binding predictions retrospectively validated; empirical data for the remaining 51 epitopes was insufficient to validate their binding predictions. Our analysis also identified 14 HLA class II epitopes, of which 8 had most of their binding predictions validated. Further investigation is required regarding the efficacy of the identified epitopes as vaccine targets in populations where HCV genotype 4 is most prevalent.
Introduction
Hepatitis C is an infectious inflammatory liver disease caused by the hepatitis C virus (HCV). According to the World Health Organization, around 150 million people worldwide suffer from chronic HCV infection, with an estimated 3–4 million people newly infected each year.Citation1 There are 6 major genotypes of the virus distributed worldwide; type 1, and to a lesser extent type 2, predominate in the US and Europe.Citation2,Citation3 HCV genotype 4 (HCV-4) is the predominant type in the Middle East and Central Africa, accounting for >80% of infections in that region.Citation4 In Egypt, where the prevalence of HCV infection is the highest worldwide (>15% of the population is chronically infected), more than 90% of the infections are attributed to HCV-4. Annually, 350 000 people are estimated to die as a result of HCV-related liver diseases worldwide.Citation1
HCV is mainly transmitted via blood contact between a healthy and an infected individual.Citation1,Citation5 Following the onset of infection, the disease starts with an acute inflammatory phase that lasts for approximately 6 mo, within which 15–30% of patients are able to spontaneously clear the virus. In 70–85% of patients, however, the virus manages to evade the host’s immune response and the disease progresses to a chronic phase, which can last for decades and may eventually culminate in liver cirrhosis, hepatocellular carcinoma, and death.Citation1,Citation5 The virus itself is an enveloped single sense-stranded RNA virus; its approximately 9600 bases-long genome harbors a single open reading frame that encodes a single precursor polyprotein molecule around 3000 amino acids in length. This molecule is processed during and after its synthesis via viral and host cellular machinery into 3 structural and 7 non-structural (NS) proteins.Citation6 HCV replicates via the RNA-dependent RNA polymerase NS5B, which lacks proofreading capability, causing the virus to exhibit a high mutation rate. This allows HCV to evade the host’s adaptive immune response and genetically diversify into a wide array of variants that are classified based on the extent of their genetic variation into genotypes (30–35% variation), and further into subtypes (20–25% variation) and quasi-species.Citation3,Citation6
Following HCV infection of the liver, the host’s innate immune system responds via the activation of natural killer lymphocytes and the production of type I and III interferons.Citation7-Citation9 The adaptive humoral response involves the production of neutralizing antibodies that may block viral entry into healthy hepatocytes by targeting the viral envelope’s structural proteins E1 and E2.Citation6,Citation10 The cellular immune response occurs through the cytotoxic activity of CD8+ T cells on infected hepatocytes and the immuno-modulatory and antiviral cytokine-secreting actions of the CD4+ T cells.Citation7 Although spontaneous viral clearance is possible, in the majority of cases the virus succeeds in evading the immune response by several mechanisms, leading to persistent infection. Chief among the evasion mechanisms is the highly mutable nature of the virus, which allows it to produce escape variants and evade recognition by cells of the adaptive immune system. Other mechanisms include inhibition of type I IFN production, induction of regulatory T cells, and induction of T cell anergy (for a review see refs. Citation7 and Citation8). The outcome of HCV infection is believed to be determined by the strength and nature of the host’s immune response during the acute phase of the infection. For example, a strong base of evidence demonstrates that a vigorous, sustained, and multi-specific T cell response (both CD4+ and CD8+) to infection is associated with viral clearance. On the other hand, a weak, narrow, and short-lasting T cell response is associated with viral persistence.Citation8,Citation10
Despite the need for one, no HCV vaccine has been developed to date. HCV vaccine development has been hampered by the obstacle of overcoming the virus’ immune escape mechanisms and by other technical obstacles. Due to the fastidious nature of the virus, HCV could not initially be produced in tissue culture systems.Citation11,Citation12 Furthermore, the sole immuno-competent animal model for investigating the pathogenesis of the disease and the interaction of the virus with the immune system is the chimpanzee. As a result of the empirical, financial, and sometimes ethical constraints associated with the use of this animal in research, only a limited number of chimpanzees could be used in HCV vaccine testing, thus compromising the power of the conclusions reached from those studies.Citation11 A statistical meta-analysis was performed by Dahari et al. in 2010 on previous HCV vaccine studies performed on chimpanzees in order to reach meaningful conclusions from them. The conclusion of the analysis suggested that the inclusion of HCV structural proteins in an HCV vaccine may elicit T-cell responses that mitigate viral clearance.Citation13
Despite the challenges associated with HCV vaccine design, several vaccine studies have been conducted on animal models in the past decade, a small number of which have actually progressed to human phase II clinical trials (for a review see refs. Citation11 and Citation12). However, the focus of most of these studies has been directed toward HCV genotypes 1 and 2 as targets, and very little attention has been given to HCV-4. A meta-analysis performed on all HCV-related epitopes on the Immune Epitope Database (IEDB) revealed that out of 3444 unique reported epitopes that tested positive in T cell and B cell assays, only 13 were reported for HCV genotypes 4, 5, and 6 combined.Citation14 HCV-4 is highly prevalent in the Middle East and Central Africa, and is increasingly spreading to Europe and North America with travel and immigration.Citation15
To address the unmet need for an HCV-4 vaccine, and to fill the wide gap in HCV-4 epitope analysis, we describe herein the identification of a set of immunogenic and conserved T-cell epitopes from 46 HCV-4 full genomic sequences using the validated immunoinformatics toolkit iVAX (EpiVax Inc.). The toolkit is a suite of immunoinformatics tools that can be used for the in silico design of epitope-driven vaccines derived from protein sequences of interest.Citation16 To carry out this analysis, we collected HCV-4 sequences and uploaded them to the iVAX website.Citation17 Following epitope prediction and analysis, we describe the comparison of our results to the curated HCV epitope data on the Immune Epitope Database (IEDB) as a method of retrospective validation of our results.Citation18 Whereas this approach is not possible for novel pathogens, this method provides a cheap and fast alternative to the empirical validation of in silico predictions, thus accelerating the development of epitope-based immunotherapeutics for HCV-4. Furthermore, we performed an in vitro binding assay on 4 of the in silico-predicted HLA class II epitopes as a pilot evaluation of the reliability of our retrospective validation approach.
Results
Identification and validation of HLA class I epitopes
By using the epitope prediction and conservation analysis tools of the iVAX toolkit (EpiMatrix and Conservatrix, respectively), the amino acid sequences of the precursor polyprotein genes of 46 HCV-4 genomic sequences were parsed into 32 286 nine-amino-acid-long peptide frames—from here on referred to as 9-mers—overlapping by 8 amino acid residues, and each 9-mer frame was assessed for its percent conservation across the 46 analyzed sequences and scored for binding to 6 HLA class I supertype alleles covering over 90% of 5 major human populations, namely A*01:01, A*02:01, A*03:01, A*24:02, B*07:02, and B*44:03.Citation19 Following this analysis, 151 9-mer peptide frames conserved in at least 90% of the HCV-4 sequences (42 out of 46) that had EpiMatrix binding prediction scores (Z-scores) ≥ 1.64 for at least one of the mentioned HLA class I alleles were selected for further analysis. Z-scores ≥ 1.64 for a given HLA allele indicate that the analyzed peptide is most likely a binder of that allele, falling among the top 5% of binders in a random set of peptides.Citation16 The high conservation of the selected epitopes across the analyzed sequences indicates that they play an important structural or functional role—as part of the structural and non-structural proteins of HCV—in the HCV life-cycle, and hence are difficult to mutate without compromising viral fitness.Citation20
The 151 peptides were then assessed for homology to the human genome using JanusMatrix, a newly developed tool that can predict possible T cell receptor (TCR) cross-reactivity between selected epitopes of interest and epitopes derived from the human genome, human microbiome, or human pathogens; where a cross-reactive (homologous) epitope is defined as one that possesses identical TCR-facing amino acid residues when bound to HLA molecules and may trigger tolerogenic or auto-immune responses when administered in the final vaccine product.Citation21 Such epitopes therefore should be excluded from the final vaccine. Based on the analysis, the 151 selected epitopes were filtered down to 90 that have no homologous counterparts—or ‘hits’—in the human genome. These epitopes were considered as good putative candidates for HCV-4 vaccine design. lists, as an example, 5 of those epitopes selected at random along with their percent conservation across the analyzed HCV-4 sequences and EpiMatrix Z-scores for binding to each of the analyzed HLA alleles. An analysis of the location of the 90 epitopes on the HCV polyprotein molecule showed that 23 of the epitopes were located in the Core protein region, 21 in the NS3 region, 15 in the NS5B region, 14 in the NS4B region, 5 in the NS5A region, 4 in the E2 region, 2 in the NS2 region, and 1 in the E1 region. No epitopes were found to be located in the p7 and NS4A regions. The 5 remaining epitopes were located at the junctions between the sequences of non-structural proteins.
Table 1. Five HLA class I candidate epitopes for HCV-4 vaccine design
Due to the similarity of polyprotein AA sequences across HCV genotypes, we performed a search on the Immune Epitope Database, a repository of manually curated immune epitope data from peer reviewed literature, patents, and direct submissions from companies and institutions, to find out whether our 90 predicted epitopes have been empirically tested in previous literature for HLA binding and/or the ability to trigger a CD8+ cytotoxic T cell response in the context of other HCV genotypes. This approach would allow us to retrospectively validate our in silico predictions without having to empirically validate them from scratch, saving experimentation costs and accelerating the process of HCV-4 vaccine testing. To perform the search, the IEDB was queried for previous records of in vitro HLA binding assays and CD8+ T cell assays performed on the exact sequences of the 90 epitopes or on sequences harboring those epitopes as substrings.
The IEDB search revealed that 20 out of the 90 epitopes have no previous records of empirical testing, which suggests that these epitopes are novel and have not been characterized earlier. For the remaining 70 epitopes, entries were found for HLA binding assays and CD8+ T cell assays performed on the epitopes. Based on the results of these assays and the nature of the peptides tested in previous literature, the 70 epitopes were divided into three groups: (1) a group of 20 epitopes that tested positive in HLA binding and/or T cell assays as 9-mer peptides (exact match), (2) a group of 22 epitopes that tested positive in HLA binding and/or T cell assays as substrings of longer peptides, and (3) a group of 28 epitopes that have no positive tests described.
A closer look at the results of the first group showed that, for 19 out of the 20 epitopes, the empirically determined restricting HLA alleles to which the epitopes were shown to bind in HLA binding assays or which mediated a positive T cell response in T cell assays were the same HLA alleles that EpiMatrix predicted the HCV-4 epitopes to bind to, or belonged to the same supertype group.Citation19 For only one epitope, the empirically determined HLA restriction was given a low Z-score by EpiMatrix (0.95); meaning that the EpiMatrix binding prediction for this epitope was a false negative. Overall, the results of the IEDB search retrospectively validate the results of the EpiMatrix predictions. Although entries with negative HLA binding and T cell assay results that contradicted with the EpiMatrix predictions were found for 9 out the 20 epitopes, these results were not used to draw conclusions regarding the validity of the EpiMatrix predictions, owing to the lack of standardization of experimental procedures and the variations between patient cohorts included in T cell assay studies that reported those negative results, and the fact that in chronic HCV patients the immune system frequently fails to develop a response to an epitope due to the capability of the virus to evade the immune system by other mechanisms rather than the epitope itself not being immunogenic. As an example of the validation results, lists the EpiMatrix Z-scores for the same 5 epitopes listed in and their empirically determined HLA restrictions, in addition to the Z-scores and empirically determined HLA restriction of the epitope that was given a false negative Z-score.
Table 2. Retrospective validation of 6 predicted HLA class I epitopes
For the second epitope group, the presence of positive results from T cell assays performed on larger peptides harboring the epitopes as substrings is an indicator that the positive response can be attributed to one of the component peptide frames of the larger peptides that may have undergone processing before epitope presentation, or due to the longer peptide itself if it is small enough to bind in the HLA class I binding groove. To confirm whether our predicted HCV-4 9-mers are responsible for the maximal positive response or not, truncation analysis must be performed; accordingly, these results cannot be used to validate the EpiMatrix predictions. Also, the results of HLA binding assays performed on longer peptides cannot be used for validation since they represent the binding affinity of the long peptide in itself. For the last group of peptides where only negative entries were found, no conclusions were drawn from the negative binding or T cell data for the same aforementioned reasons.
Identification and validation of HLA class II epitopes
For HLA class II epitope prediction, the same 32 286 9-mer frames parsed and assessed for conservation by Conservatrix across the 46 analyzed sequences were scored by EpiMatrix for binding to 8 HLA-DRB1 alleles covering over 90% of the human population; namely DRB1*01:01, DRB1*03:01, DRB1*04:01, DRB1*07:01, DRB1*08:01, DRB1*11:01, DRB1*13:01, and DRB1*15:01.Citation51 The results of the Conservatrix and EpiMatrix analyses were then used by another tool, EpiAssembler, to construct extended immunogenic consensus sequence (ICS) peptides which represent the HLA class II epitopes to be included in the final vaccine product owing to the extended nature of HLA class II epitopes in comparison to those of class I. The EpiAssembler tool works by selecting the most conserved and promiscuously immunogenic 9-mer peptide frames analyzed and scored by Conservatrix and EpiMatrix, and extending those peptide frames at their N- and C-termini using the most highly immunogenic and conserved forms of their overlapping 9-mer peptide frames until a maximum user-defined length is reached, or until no more overlapping sequences matching the user-defined criteria exist in the analyzed set of peptides.Citation16 From the 32,286 analyzed 9-mer frames, EpiAssembler used 156 frames to construct 14 ICS peptides, harboring within their ‘core’ sequence (excluding the 3 AA residues at their N- and C-termini) overlapping 9-mer frames that are collectively conserved in > 90% of the analyzed sequences, and which have a minimum EpiMatrix cluster immunogenicity score (an aggregate score of the EpiMatrix Z-scores of the 9-mer peptides contained within the core of the ICS) of 10. Each of the 14 ICS peptides harbor at least one promiscuous 9-mer frame in its core sequence that is predicted to bind at least 4 of the 8 analyzed HLA-DRB1 alleles, referred to as an EpiBar.Citation16 As an example of the EpiAssembler results, 4 randomly chosen ICS peptides are listed in .
Table 3. Four EpiAssembler-constructed Immunogenic Consensus Sequence (ICS) peptides
JanusMatrix analysis of the ICS peptides revealed that all 14 have some degree of homology with the human genome as evident by their JanusMatrix cluster scores, which are equal to their EpiMatrix cluster immunogenicity scores minus penalty deductions for each cross-reactive hit with the human genome for each of the component 9-mer peptides of the core ICS sequence. The bigger the difference between the EpiMatrix and JanusMatrix cluster scores, the higher the homology with the human genome. Accordingly, ICS peptides with a smaller difference between both scores are more likely to be successfully included into the final vaccine; however, further empirical testing is required to assess the safety and efficacy of each peptide before deciding whether or not to include it in the final product. lists the sequences of 4 ICS peptides, their conservation, EpiMatrix and JanusMatrix cluster scores, and their locations along the AA sequence of the HCV polyprotein gene. lists, as an example, the EpiMatrix Z-scores and percent conservation of the 9-mer overlapping peptides that were used to construct one of the listed ICS peptides. Analysis of the locations of the ICS peptides revealed that 8 are located in the NS4B region, 3 in the NS5B region, 1 in the Core region, 1 in the NS3 region, and 1 at the junction between the NS2 and NS3 protein sequences.
Table 4. Percent Conservation and EpiMatrix Z-scores of the component 9-mer frames of ICS peptide NS3 (1246–1265)
In order to retrospectively validate our EpiMatrix HLA class II binding predictions, a search was also performed on IEDB to find out whether the 14 constructed ICS peptides have been empirically tested in previous literature for HLA binding and/or the ability to trigger a CD4+ T-helper cell response in the context of other HCV genotypes. The search revealed that none of the 14 peptides has been tested earlier in the form of the exact same sequence, however, all 14 peptides have been tested as substrings of larger peptides, as 70–99% similar peptides (determined by performing a blast 70–90% search; see Materials and Methods section), and/or as smaller fragments of the peptides themselves. To validate our predictions, we only considered the IEDB entries where the sequence of the peptide tested in previous literature contained within its sequence at least one complete 9-mer frame from within the core sequence of our ICS peptides.
The IEDB search revealed that for 8 out of the 14 ICS peptides, entries with positive results were found for HLA binding and T cell assays that validated most of the binding predictions. More precisely, out of 51 EpiMatrix binding predictions performed for the 8 peptides collectively and for which corresponding empirical data was found on IEDB, 39 predictions have been validated as true positive (TP) or true negative (TN). Twelve of those predictions, however, were contradicted by the IEDB results. Before labeling these predictions as false positives (FP), it is suggested that further empirical testing be performed to confirm or refute the EpiMatrix predictions; especially considering that for most of the 12 predictions the contradicting results come from a single corresponding source. As an example of how the validations were performed, lists the validation results for the ICS peptide NS31246–1265 (SQGYKVLVLNPSVAATLGFG). For each HLA binding or T cell assay entry found on IEDB for the ICS peptide, lists the sequence of the peptide that was actually tested or to which the response was attributed, the references that mentioned testing that peptide, and whether the HLA restrictions that tested positive or negative for peptide binding confirm or refute the EpiMatrix predictions for each of the 8 analyzed HLA class II alleles.
Table 5. Retrospective validation of the ICS peptide NS3 (1246–1265)
It is important to note that in all the IEDB results for the 8 ICS peptides whose EpiMatrix results were mostly validated, the sequences of the peptides that were actually tested or to which the response was attributed were composed mainly of the overlapping 9-mer frames of the core sequences of the ICS peptides; which makes our retrospective validations more reliable. It is also important to note that, where the IEDB search returned results in which the test peptides were 70–99% similar to the ICS peptides, the point mutations to which the dissimilarity was attributed mainly resided in the 3 N- and C-terminal AA residues of the ICS peptides, which are not directly involved in HLA class II binding. This is true for 7 out of the 8 ICS peptides; for the remaining peptide, a point mutation was present in the empirically tested sequences that resided in the core sequence of the ICS (data not shown). For this ICS in particular, 5 out of 7 EpiMatrix binding predictions for which corresponding empirical data were found were contradicted by the IEDB results. This suggests that the mutation affected the binding specificity of the test peptides, and further demonstrates the necessity of performing further empirical analysis on the ICS sequence itself before determining the validity of the EpiMatrix predictions.
Apart from the aforementioned results of the 8 ICS peptides, the results of the IEDB search for the remaining 6 peptides returned only results for CD4+ T cell assays where the HLA restriction was not determined. However, for 5 of the 6 peptides, the T cell assay results where positive, which encourages performing assays in the future to determine the HLA restrictions responsible for the positive response.
HLA class II in vitro binding assay
As a pilot assessment of the reliability of the performed retrospective validations for selecting targets for vaccine development, we performed a pilot in vitro binding assay on the 4 HLA class II ICS peptides listed in by testing their affinity for binding to 4 purified HLA class II DR molecules, namely DRB1*01:01, DRB1*04:01, DRB1*07:01, and DRB1*15:01 in competition against biotinylated standards. The results of the assay are presented in . As expected, all ICS peptides were generally found to bind with high affinity to all 4 tested HLA class II molecules, which validates the EpiMatrix binding predictions, and confirms the results of the retrospective validation. The only exception was exhibited by the ICS peptide NS4B1769–1786 (VSGIQYLAGLSTLPGNPA), which failed to bind to the allele DRB1*07:01 in contradiction to the EpiMatrix binding prediction. Interestingly, this negative result was repeated in HLA binding assays performed in three different references as demonstrated by the results of the retrospective validation.Citation57,Citation61,Citation62 Given the negative results of the previous assays and the result of this one, it is likely that the EpiMatrix binding prediction for this peptide for the DRB1*07:01 is a false positive. This demonstrates the reliability of the performed retrospective analysis for the validation of the in silico results.
Table 6. HLA class II binding assay results compared with the highest calculated Z-score per allele of the 9-mer frames of the core sequences of each ICS peptide
Discussion
In this article, we described the in silico identification of several highly immunogenic and conserved T-cell epitopes in the HCV-4 genome, using the validated iVAX immunoinformatics toolkit, that can be used for the development of an epitope-based vaccine against HCV-4 or for inclusion in an HCV vaccine that covers HCV-4 as one of its targets. We performed a retrospective validation of our results by searching the IEDB for previous records of HLA binding and T-cell assays performed on the identified epitopes, and we further performed a pilot in vitro HLA class II binding assay on 4 selected HLA class II ICS peptides to validate our in silico results and assess the reliability of our retrospective validation. Our in silico predictions lead to the identification of 90 highly conserved and immunogenic HLA class I epitopes that have no homologous counterparts in the human genome bearing identical TCR-facing AA residues, and to the construction of 14 ICS peptides (HLA class II) containing epitopes that have varying degrees of homology to epitopes derived from the human genome. Retrospective validation of the selected 90 HLA class I epitopes revealed that 20 epitopes have no record of previous empirical testing on IEDB, thus representing good targets for future investigation and testing of their role in the interaction between the immune system and HCV-4 and their efficacy as vaccine targets, and validated the results of the in silico predictions for 19 of the remaining 70 epitopes. As for the HLA class II ICS peptides, the IEDB search validated, for 8 out the 14 peptides, 39 EpiMatrix binding predictions out of 51 for which corresponding empirical data was found. The pilot in vitro HLA binding assay performed on 4 ICS peptides validated the EpiMatrix binding predictions, except for 1 peptide whose prediction for binding to the DRB1*07:01 allele was shown to be a false positive, in concurrence with the results of the retrospective validation. A flowchart summarizing the in silico epitope identification process and retrospective validation is presented in ; summarizes the results of each step of this process in selecting the HCV-4 vaccine candidates.
Figure 1. Flowchart summarizing the identification and analysis of HLA class I and II T cell epitopes from HCV-4 and the retrospective validation of the results using the IEDB.
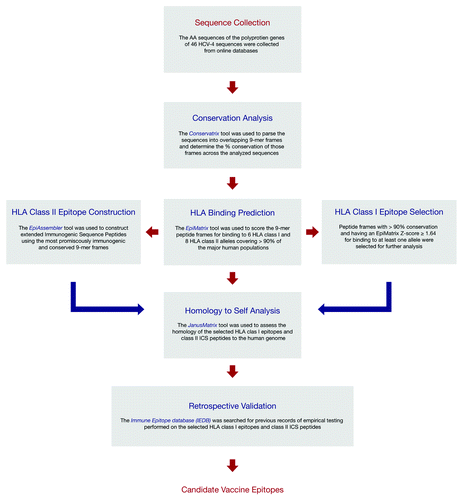
Table 7. Summary of the results of the in silico epitope identification and retrospective validation process
Analysis of the locations of the identified highly conserved HLA class I and II epitopes revealed that most of them lie in the Core, NS3, NS4B, and NS5B regions of the HCV polyprotein sequence. Twenty-three HLA class I epitopes and one ICS peptide were located in the Core protein region, the building block of the viral nucelocapsid. Twenty-one HLA class I epitopes and one ICS peptide were located in the NS3 serine protease-helicase NTPase protein region. Fourteen HLA class I epitopes and 8 HLA class II ICS peptides were found to be located in the NS4B region; which is involved in membranous web formation, an altered structure of the ER membrane that serves as a scaffold for the formation of the viral replication complex.Citation6 Finally, 15 HLA class I epitopes and 3 class II ICS peptides were found to be located in the NS5B region. The key structural and functional roles that the sequences of those proteins play in the HCV life cycle make it less likely for these sequences to mutate in the course of viral evolution.Citation20 This explains why the highest proportion of the HLA class I and II epitopes that we identified are located within those sequences. On the other hand, very few epitopes were found in the E1 and E2 envelope glycoproteins in comparison to their size; being on the surface of HCV viral particles, these proteins exhibit frequent mutations in order to evade the host’s humoral immune response.Citation63
The presence of epitopes in the final vaccine product that are homologous to epitopes derived from the human genome can potentially trigger a suppressive immune response to the vaccine, mediated by T regulatory cells, or even a detrimental autoimmune response. Accordingly, such epitopes should not be included in the final vaccine. To screen for such epitopes in our predicted data set, we used the novel JanusMatrix tool, which searches for epitopes derived from the human genome that bear identical TCR-facing AA residues while allowing the MHC-facing residues (referred to as the ‘agretope’) to vary, with the variant agretope still predicted by EpiMatrix to bind to the same HLA alleles to which the screened epitopes were predicted to bind. Homology to the human genome was recently discovered in some commensal viruses using the same tool, and was found to differ quite significantly from the level of homology to self for ‘hit-and-run’ viruses like Variola, Ebola, and Marburg.Citation64 In reality, TCRs are able to recognize their target epitopes by AA residue side-chain availability; this means that JanusMatrix cannot predict all putative cross-reactive epitopes derived from the human genome based on sequence similarity alone, however it can still help narrow down the set of epitopes that would be later tested experimentally to prove whether or not they trigger suppressive or autoimmune responses.Citation21 For our predicted HLA class I set, we stringently selected only the epitopes with zero cross-reactive hits in the human genome. For class II ICS peptides, we could not filter the epitopes based on the same stringent criteria due to their extended nature, which is required for proper HLA class II binding and the promiscuous coverage of the analyzed HLA-DR alleles using the most highly conserved forms of the 9-mer frames utilized for ICS construction. Filtering out 9-mer frames from the ICS peptides that exhibit similarity to the human genome may compromise allelic coverage, HCV-4 subtype coverage, and binding to HLA class II. A JanusMatrix analysis of a large set of empirically characterized T-effector cell epitopes has previously demonstrated that, within a certain ratio of cross-reactive hits per number of AA residues in the human genome database, a T cell epitope exhibiting cross-reactivity to the human genome may still successfully trigger a T-effector response.Citation21 Accordingly, the decision of which peptides to use for the vaccine should be taken following future empirical analysis.
In conclusion, we have demonstrated the in silico identification and selection of a set of highly conserved and immunogenic T cell epitopes from within the HCV-4 proteome as a first step toward the development of an epitope-driven vaccine against this viral genotype using one of the most accurate T cell epitope prediction tools available online.Citation65 Compared with the traditional method of empirically testing all possible overlapping peptide frames in a target antigenic sequence for immunogenicity prior to the validation of their efficacy as vaccine targets in vivo, the initial in silico prediction of immunogenic epitopes narrows down the in vivo validation process to only a selected set of epitopes, thus saving the time and costs associated with the analysis. Furthermore, we performed a retrospective validation of our results using the repository of empirical data on the IEDB; this approach provides a faster and cheaper alternative to performing empirical validations from scratch, which can accelerate vaccine development when outbreaks of a different strain of the same pathogen occurs. Using this approach, the production of population-specific or individualized vaccines against pathogens such as HCV, HIV, and Coronaviruses may be facilitated. A recent example of utilizing this approach was performed by Duvvuri et al., where an IEDB search was performed to validate the immunogenic potential of in silico identified HLA class II epitopes from the novel avian influenza H7N9 virus relative to other human influenza A viruses.Citation66 In the future, we intend to empirically characterize the 20 novel HLA class I epitopes identified by the iVAX analysis, and to further elucidate the nature of the immune response toward the identified HLA class I and II epitopes in patient populations where HCV-4 is highly prevalent, as an assessment of the efficacy of those epitopes as vaccine targets. Although the future holds promise for increasing the chances of success of HCV therapy with the continuing approval of novel direct-acting antiviral drugs to enter the market (e.g., Sofosbuvir),Citation67 these new drugs remain very expensive, making it hard for a large sector of the infected population, especially in some countries of the Middle East and Africa, to afford treatment. The development of an effective preventative or therapeutic HCV vaccine can have a strong impact on HCV prevention and therapy in such populations.
Materials and Methods
HCV-4 Polyprotein sequence collection
The amino acid sequences of 46 non-redundant HCV-4 precursor polyprotein genes were retrieved from the Los Alamos HCV sequence database (LANL)Citation68 and uploaded to the iVAX toolkit server. The accession numbers of the full genomic HCV-4 sequences harboring those genes are: DQ418785, DQ418782, DQ418783, DQ418784, DQ418787; DQ418788; DQ418789, DQ516084, DQ988073, DQ988074, DQ988075, DQ988076, DQ988077, DQ988078, DQ988079, GU814265, NC_009825, FJ025854, FJ025855, FJ025856, FJ462435, FJ462436, DQ418786, DQ516083, EU392172, FJ462437, EF589160, EF589161, EU392169, EU392170, EU392174, EU392175, FJ462432, EU392171, EU392173, FJ462438, FJ839870, FJ462433, FJ462441, FJ462440, FJ462431, FJ462434, FJ462439, FJ839869, HQ537008, and HQ537009.
Epitope prediction and analysis using the iVAX toolkit
Following sequence upload, the tools of the iVAX toolkit were consecutively used in order to identify HLA class I and II epitopes from the HCV-4 sequences and filter the results to a selected set of epitopes that can potentially be used to develop the vaccine. The Conservatrix tool was used first; it was set to parse the sequences into 9-mer peptide frames overlapping by 8 amino acids and determine the percent conservation of each frame across the 46 sequences. The matrix-based T cell epitope prediction tool EpiMatrix was then used to score the parsed 9-mer frames for binding to the 6 HLA class I supertype alleles A*01:01, A*02:01, A*03:01, A*24:02, B*07:02, and B*44:03, and the 8 HLA-DRB1 alleles DRB1*01:01, DRB1*03:01, DRB1*04:01, DRB1*07:01, DRB1*08:01, DRB1*11:01, DRB1*13:01, and DRB1*15:01. To construct the HLA class II ICS peptides, the EpiAssembler tool was set to construct peptides of a maximum length of 21 AA residues that harbor at least one 9-mer frame that is immunogenic and conserved in ≥ 90% of the analyzed HCV-4 sequences, and that possess a minimum EpiMatrix Cluster Immunogenicity score of 10; an EpiMatrix cluster score of 10 or higher indicates that the constructed ICS has a significant immunogenic potential.Citation16 The JanusMatrix tool was then used to identify possible homology between selected epitopes and ICS peptides, and epitopes derived from the human genome by searching for 9-mer epitope frames in a human protein database pre-uploaded into JanusMatrix that bear identical TCR-facing residues and were predicted by EpiMatrix to bind to the same HLA alleles as the analyzed HCV epitopes.Citation21 Identifying the locations of the identified epitopes on the HCV genome was performed using the ‘Sequence Locator’ tool of the HCV Los Alamos database.
IEDB epitope search
The Immune Epitope Database (IEDB) was queried for the selected epitopes and ICS peptides via the home page search tool (http://www.iedb.org/). The database was queried for available data on MHC binding, MHC elution, and T cell response assays performed on epitopes/peptides that exactly match the query epitopes or harbor those epitopes as substrings. For HLA class II ICS peptides, the search was expanded to include peptide sequences that are 70–90% identical to the constructed ICS peptides using the BLAST 70–90% search option. The immune recognition context parameters of the search were confined to MHC class I- or II-restricted matches (according to the queried epitope), and T cell assays were confined to those performed on blood samples from human subjects by selecting Homo sapiens (id:9606) as the host organism.
HLA class II in vitro binding assay
In four 96-well reaction plates (Fisherbrand), each non-biotinylated ICS test peptide (21st Century Biochemicals) diluted in DMSO (Sigma-Aldrich) was incubated, in triplicates and over a wide range of concentrations, with the purified HLA-DR molecules DRB1*01:01, DRB1*04:01, DRB1*07:01, and DRB1*15:01 (Benaroya Research Institute)—one molecule per plate—and standard biotinylated peptides at 25 nM (21st Century Biochemicals) in 50 μL per well of pH 5.4 citrate-phosphate buffer solution at 37 °C overnight. After the binding mixtures reached a steady equilibrium, they were transferred to 96-well ELISA coated with anti-HLA-DRA antibodies (L243, BioXCell) and were incubated for 2.5 h at 37 °C. After the incubation, the plates were thoroughly washed with PBS+0.05% Tween-20, developed by adding Europium-labeled Streptavidin (Perkin-Elmer), and analyzed using a Victor3V Microtiter Plate Reader (Perkin-Elmer, MA, USA). IC50 values were then calculated using SigmaPlot 11.1 software.
| Abbreviations: | ||
| HCV | = | hepatitis C virus |
| HCV-4 | = | hepatitis C virus genotype 4 |
| UTR | = | untranslated region |
| NS | = | non-structural |
| TCR | = | T-cell Receptor |
| IEDB | = | Immune Epitope Database |
| AA | = | amino acid |
| HIV | = | human immunodeficiency virus |
Disclosure of Potential Conflicts of Interest
Coauthors A.D.G. and F.T. are employees of EpiVax, a vaccine and therapeutic design company, and A.D.G. is a majority stockholder. These authors recognize the presence of potential conflicts of interest and affirm that the information represented in this paper is original and unbiased observations. K.A.H., H.M.E.A., A.H.G., and J.D. declare no competing interests.
Acknowledgments
This work has been funded by a grant from the American University in Cairo to Dr Hassan Azzazy and an ARRA supplement to the U19 grant to Prof. Anne De Groot. The iVAX toolkit has been made available for academic use through a unique arrangement with the University of Rhode Island. Thanks to Sheila Chandran for assisting with the in silico analysis and Lana Abdel-Hady for assisting with the artwork.
References
- World Health Organization. Hepatitis C fact sheet No164 [Internet]. [Updated 2013 Jul; Cited 2013 Nov 13]. Available from: http://www.who.int/mediacentre/factsheets/fs164/en/.
- World Health Organization. Hepatitis C [Internet]. 2002. [Cited 2013 Nov 13]. Available from: http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index2.html.
- Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 2005; 42:962 - 73; http://dx.doi.org/10.1002/hep.20819; PMID: 16149085
- Khattab MA, Ferenci P, Hadziyannis SJ, Colombo M, Manns MP, Almasio PL, Esteban R, Abdo AA, Harrison SA, Ibrahim N, et al. Management of hepatitis C virus genotype 4: recommendations of an international expert panel. J Hepatol 2011; 54:1250 - 62; http://dx.doi.org/10.1016/j.jhep.2010.11.016; PMID: 21316497
- Centers for Disease Control and Prevention. Hepatitis C Information for Health Professionals [Internet]. 2011. [Cited 2013 Nov 13]. Available from: http://www.cdc.gov/hepatitis/HCV/index.htm
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol 2007; 5:453 - 63; http://dx.doi.org/10.1038/nrmicro1645; PMID: 17487147
- Ishii S, Koziel MJ. Immune responses during acute and chronic infection with hepatitis C virus. Clin Immunol 2008; 128:133 - 47; http://dx.doi.org/10.1016/j.clim.2008.03.525; PMID: 18514579
- Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest 2009; 119:1745 - 54; http://dx.doi.org/10.1172/JCI39133; PMID: 19587449
- Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem 2010; 285:22741 - 7; http://dx.doi.org/10.1074/jbc.R109.099556; PMID: 20457596
- Zeisel MB, Cosset FL, Baumert TF. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology 2008; 48:299 - 307; http://dx.doi.org/10.1002/hep.22307; PMID: 18508291
- Halliday J, Klenerman P, Barnes E. Vaccination for hepatitis C virus: closing in on an evasive target. Expert Rev Vaccines 2011; 10:659 - 72; http://dx.doi.org/10.1586/erv.11.55; PMID: 21604986
- Yu CI, Chiang BL. A new insight into hepatitis C vaccine development. J Biomed Biotechnol 2010; 2010:548280; http://dx.doi.org/10.1155/2010/548280; PMID: 20625493
- Dahari H, Feinstone SM, Major ME. Meta-analysis of hepatitis C virus vaccine efficacy in chimpanzees indicates an importance for structural proteins. Gastroenterology 2010; 139:965 - 74; http://dx.doi.org/10.1053/j.gastro.2010.05.077; PMID: 20621699
- Kim Y, Vaughan K, Greenbaum J, Peters B, Law M, Sette A. A meta-analysis of the existing knowledge of immunoreactivity against hepatitis C virus (HCV). PLoS One 2012; 7:e38028; http://dx.doi.org/10.1371/journal.pone.0038028; PMID: 22675428
- Yahia M. Global health: a uniquely Egyptian epidemic. Nature 2011; 474:S12 - 3; http://dx.doi.org/10.1038/474S12a; PMID: 21666728
- De Groot AS. Epitope-based immunome-derived vaccines: A strategy for improved design and safety. Springer, NY; 2009.
- iCubed: Institute for Immunology and Informatics. iVAX Tool Kit [Internet]. 2012. [Cited 2013 Nov 13]. Available from: http://www.immunome.org/ivax/ivax-tool-kit/.
- Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol 2005; 3:e91; http://dx.doi.org/10.1371/journal.pbio.0030091; PMID: 15760272
- Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 1999; 50:201 - 12; http://dx.doi.org/10.1007/s002510050594; PMID: 10602880
- Petrovic D, Dempsey E, Doherty DG, Kelleher D, Long A. Hepatitis C virus--T-cell responses and viral escape mutations. Eur J Immunol 2012; 42:17 - 26; http://dx.doi.org/10.1002/eji.201141593; PMID: 22125159
- Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Abdel Hady KM, VerBerkmoes NC, Sztein MB, Losikoff PT, Martin WD, et al. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother 2013; 9:1577 - 86; http://dx.doi.org/10.4161/hv.24615; PMID: 23584251
- Neumann-Haefelin C, Killinger T, Timm J, Southwood S, McKinney D, Blum HE, Thimme R. Absence of viral escape within a frequently recognized HLA-A26-restricted CD8+ T-cell epitope targeting the functionally constrained hepatitis C virus NS5A/5B cleavage site. J Gen Virol 2007; 88:1986 - 91; http://dx.doi.org/10.1099/vir.0.82826-0; PMID: 17554032
- Neumann-Haefelin C, Timm J, Spangenberg HC, Wischniowski N, Nazarova N, Kersting N, Roggendorf M, Allen TM, Blum HE, Thimme R. Virological and immunological determinants of intrahepatic virus-specific CD8+ T-cell failure in chronic hepatitis C virus infection. Hepatology 2008; 47:1824 - 36; http://dx.doi.org/10.1002/hep.22242; PMID: 18454507
- Wentworth PA, Sette A, Celis E, Sidney J, Southwood S, Crimi C, Stitely S, Keogh E, Wong NC, Livingston B, et al. Identification of A2-restricted hepatitis C virus-specific cytotoxic T lymphocyte epitopes from conserved regions of the viral genome. Int Immunol 1996; 8:651 - 9; http://dx.doi.org/10.1093/intimm/8.5.651; PMID: 8671652
- Mashiba T, Udaka K, Hirachi Y, Hiasa Y, Miyakawa T, Satta Y, Osoda T, Kataoka S, Kohara M, Onji M. Identification of CTL epitopes in hepatitis C virus by a genome-wide computational scanning and a rational design of peptide vaccine. Immunogenetics 2007; 59:197 - 209; http://dx.doi.org/10.1007/s00251-006-0185-3; PMID: 17225159
- Nakamoto Y, Kaneko S, Takizawa H, Kikumoto Y, Takano M, Himeda Y, Kobayashi K. Analysis of the CD8-positive T cell response in Japanese patients with chronic hepatitis C using HLA-A*2402 peptide tetramers. J Med Virol 2003; 70:51 - 61; http://dx.doi.org/10.1002/jmv.10349; PMID: 12629644
- Kaji K, Nakamoto Y, Kaneko S. Analysis of hepatitis C virus-specific CD8+ T-cells with HLA-A*24 tetramers during phlebotomy and interferon therapy for chronic hepatitis C. Oncol Rep 2007; 18:993 - 8; PMID: 17786365
- Gruener NH, Jung MC, Ulsenheimer A, Gerlach JT, Zachoval R, Diepolder HM, Baretton G, Schauer R, Pape GR, Schirren CA. Analysis of a successful HCV-specific CD8+ T cell response in patients with recurrent HCV-infection after orthotopic liver transplantation. Liver Transpl 2004; 10:1487 - 96; http://dx.doi.org/10.1002/lt.20300; PMID: 15558593
- Christie JM, Chapel H, Chapman RW, Rosenberg WM. Immune selection and genetic sequence variation in core and envelope regions of hepatitis C virus. Hepatology 1999; 30:1037 - 44; http://dx.doi.org/10.1002/hep.510300403; PMID: 10498657
- Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY, Day CL, Robbins GK, Casson DR, Reiser M, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 2004; 127:924 - 36; http://dx.doi.org/10.1053/j.gastro.2004.06.015; PMID: 15362047
- Cucchiarini M, Kammer AR, Grabscheid B, Diepolder HM, Gerlach TJ, Grüner N, Santantonio T, Reichen J, Pape GR, Cerny A. Vigorous peripheral blood cytotoxic T cell response during the acute phase of hepatitis C virus infection. Cell Immunol 2000; 203:111 - 23; http://dx.doi.org/10.1006/cimm.2000.1683; PMID: 11006009
- Grüner NH, Gerlach TJ, Jung MC, Diepolder HM, Schirren CA, Schraut WW, Hoffmann R, Zachoval R, Santantonio T, Cucchiarini M, et al. Association of hepatitis C virus-specific CD8+ T cells with viral clearance in acute hepatitis C. J Infect Dis 2000; 181:1528 - 36; http://dx.doi.org/10.1086/315450; PMID: 10823750
- Lauer GM, Ouchi K, Chung RT, Nguyen TN, Day CL, Purkis DR, Reiser M, Kim AY, Lucas M, Klenerman P, et al. Comprehensive analysis of CD8(+)-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol 2002; 76:6104 - 13; http://dx.doi.org/10.1128/JVI.76.12.6104-6113.2002; PMID: 12021343
- Grey HM, Ruppert J, Vitiello A, Sidney J, Kast WM, Kubo RT, Sette A. Class I MHC-peptide interactions: structural requirements and functional implications. Cancer Surv 1995; 22:37 - 49; PMID: 7536628
- Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell 1993; 74:929 - 37; http://dx.doi.org/10.1016/0092-8674(93)90472-3; PMID: 8104103
- Bertoni R, Sette A, Sidney J, Guidotti LG, Shapiro M, Purcell R, Chisari FV. Human class I supertypes and CTL repertoires extend to chimpanzees. J Immunol 1998; 161:4447 - 55; PMID: 9780224
- Alexander J, Del Guercio MF, Fikes JD, Chesnut RW, Chisari FV, Chang KM, Appella E, Sette A. Recognition of a novel naturally processed, A2 restricted, HCV-NS4 epitope triggers IFN-gamma release in absence of detectable cytopathicity. Hum Immunol 1998; 59:776 - 82; http://dx.doi.org/10.1016/S0198-8859(98)00080-9; PMID: 9831133
- Scognamiglio P, Accapezzato D, Casciaro MA, Cacciani A, Artini M, Bruno G, Chircu ML, Sidney J, Southwood S, Abrignani S, et al. Presence of effector CD8+ T cells in hepatitis C virus-exposed healthy seronegative donors. J Immunol 1999; 162:6681 - 9; PMID: 10352286
- Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum Immunol 2001; 62:1200 - 16; http://dx.doi.org/10.1016/S0198-8859(01)00319-6; PMID: 11704282
- Himoudi N, Abraham JD, Fournillier A, Lone YC, Joubert A, Op De Beeck A, Freida D, Lemonnier F, Kieny MP, Inchauspé G. Comparative vaccine studies in HLA-A2.1-transgenic mice reveal a clustered organization of epitopes presented in hepatitis C virus natural infection. J Virol 2002; 76:12735 - 46; http://dx.doi.org/10.1128/JVI.76.24.12735-12746.2002; PMID: 12438599
- Battegay M, Fikes J, Di Bisceglie AM, Wentworth PA, Sette A, Celis E, Ching WM, Grakoui A, Rice CM, Kurokohchi K, et al. Patients with chronic hepatitis C have circulating cytotoxic T cells which recognize hepatitis C virus-encoded peptides binding to HLA-A2.1 molecules. J Virol 1995; 69:2462 - 70; PMID: 7884894
- Ohno S, Moriya O, Yoshimoto T, Hayashi H, Akatsuka T, Matsui M. Immunogenic variation between multiple HLA-A*0201-restricted, Hepatitis C Virus-derived epitopes for cytotoxic T lymphocytes. Viral Immunol 2006; 19:458 - 67; http://dx.doi.org/10.1089/vim.2006.19.458; PMID: 16987064
- Ishizuka J, Grebe K, Shenderov E, Peters B, Chen Q, Peng Y, Wang L, Dong T, Pasquetto V, Oseroff C, et al. Quantitating T cell cross-reactivity for unrelated peptide antigens. J Immunol 2009; 183:4337 - 45; http://dx.doi.org/10.4049/jimmunol.0901607; PMID: 19734234
- Anthony DD, Valdez H, Post AB, Carlson NL, Heeger PS, Lehmann PV. Comprehensive determinant mapping of the hepatitis C-specific CD8 cell repertoire reveals unpredicted immune hierarchy. Clin Immunol 2002; 103:264 - 76; http://dx.doi.org/10.1006/clim.2001.5193; PMID: 12173301
- Fitzmaurice K, Petrovic D, Ramamurthy N, Simmons R, Merani S, Gaudieri S, Sims S, Dempsey E, Freitas E, Lea S, et al. Molecular footprints reveal the impact of the protective HLA-A*03 allele in hepatitis C virus infection. Gut 2011; 60:1563 - 71; http://dx.doi.org/10.1136/gut.2010.228403; PMID: 21551190
- Tsai SL, Sheen IS, Chien RN, Chu CM, Huang HC, Chuang YL, Lee TH, Liao SK, Lin CL, Kuo GC, et al. Activation of Th1 immunity is a common immune mechanism for the successful treatment of hepatitis B and C: tetramer assay and therapeutic implications. J Biomed Sci 2003; 10:120 - 35; http://dx.doi.org/10.1007/BF02256004; PMID: 12566993
- Tsai SL, Lee TH, Chien RN, Liao SK, Lin CL, Kuo GC, Liaw YF. A method to increase tetramer staining efficiency of CD8+ T cells with MHC-peptide complexes: therapeutic applications in monitoring cytotoxic T lymphocyte activity during hepatitis B and C treatment. J Immunol Methods 2004; 285:71 - 87; http://dx.doi.org/10.1016/j.jim.2003.11.005; PMID: 14871536
- Thammanichanond D, Moneer S, Yotnda P, Aitken C, Earnest-Silveira L, Jackson D, Hellard M, McCluskey J, Torresi J, Bharadwaj M. Fiber-modified recombinant adenoviral constructs encoding hepatitis C virus proteins induce potent HCV-specific T cell response. Clin Immunol 2008; 128:329 - 39; http://dx.doi.org/10.1016/j.clim.2008.04.002; PMID: 18524682
- Guo Z, Zhang H, Rao H, Jiang D, Cong X, Feng B, Wang J, Wei L, Chen H. DCs pulsed with novel HLA-A2-restricted CTL epitopes against hepatitis C virus induced a broadly reactive anti-HCV-specific T lymphocyte response. PLoS One 2012; 7:e38390; http://dx.doi.org/10.1371/journal.pone.0038390; PMID: 22701633
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol 2008; 9:1; http://dx.doi.org/10.1186/1471-2172-9-1; PMID: 18211710
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 1998; 160:3363 - 73; PMID: 9531296
- Diepolder HM, Gerlach JT, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, Scholz S, Santantonio T, Houghton M, Southwood S, et al. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol 1997; 71:6011 - 9; PMID: 9223492
- Pape GR, Gerlach TJ, Diepolder HM, Grüner N, Jung M, Santantonio T. Role of the specific T-cell response for clearance and control of hepatitis C virus. J Viral Hepat 1999; 6:Suppl 1 36 - 40; http://dx.doi.org/10.1046/j.1365-2893.1999.00006.x; PMID: 10760033
- Shoukry NH, Sidney J, Sette A, Walker CM. Conserved hierarchy of helper T cell responses in a chimpanzee during primary and secondary hepatitis C virus infections. J Immunol 2004; 172:483 - 92; http://dx.doi.org/10.4049/jimmunol.172.1.483; PMID: 14688358
- Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest 2003; 112:831 - 42; http://dx.doi.org/10.1172/JCI200318509; PMID: 12975468
- Castelli FA, Leleu M, Pouvelle-Moratille S, Farci S, Zarour HM, Andrieu M, Auriault C, Ménez A, Georges B, Maillere B. Differential capacity of T cell priming in naive donors of promiscuous CD4+ T cell epitopes of HCV NS3 and Core proteins. Eur J Immunol 2007; 37:1513 - 23; http://dx.doi.org/10.1002/eji.200636783; PMID: 17492804
- Lamonaca V, Missale G, Urbani S, Pilli M, Boni C, Mori C, Sette A, Massari M, Southwood S, Bertoni R, et al. Conserved hepatitis C virus sequences are highly immunogenic for CD4(+) T cells: implications for vaccine development. Hepatology 1999; 30:1088 - 98; http://dx.doi.org/10.1002/hep.510300435; PMID: 10498664
- Harcourt GC, Lucas M, Sheridan I, Barnes E, Phillips R, Klenerman P. Longitudinal mapping of protective CD4+ T cell responses against HCV: analysis of fluctuating dominant and subdominant HLA-DR11 restricted epitopes. J Viral Hepat 2004; 11:324 - 31; http://dx.doi.org/10.1111/j.1365-2893.2004.00516.x; PMID: 15230855
- Ebinuma H, Nakamoto N, Li Y, Price DA, Gostick E, Levine BL, Tobias J, Kwok WW, Chang KM. Identification and in vitro expansion of functional antigen-specific CD25+ FoxP3+ regulatory T cells in hepatitis C virus infection. J Virol 2008; 82:5043 - 53; http://dx.doi.org/10.1128/JVI.01548-07; PMID: 18337568
- Eckels DD, Bian T, Gill JC, Sønderstrup G. Epitopes of the NS3 protein of hepatitis C virus: recognition in HLA-DR4 transgenic mice. Immunol Cell Biol 2002; 80:106 - 12; http://dx.doi.org/10.1046/j.1440-1711.2002.01053.x; PMID: 11869368
- Schulze Zur Wiesch J, Sidney J, Walker B, Sette A. Direct Submission. 2006.
- Schulze zur Wiesch J, Lauer GM, Day CL, Kim AY, Ouchi K, Duncan JE, Wurcel AG, Timm J, Jones AM, Mothe B, et al. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol 2005; 175:3603 - 13; http://dx.doi.org/10.4049/jimmunol.175.6.3603; PMID: 16148104
- Wang Y, Keck ZY, Foung SK. Neutralizing antibody response to hepatitis C virus. Viruses 2011; 3:2127 - 45; http://dx.doi.org/10.3390/v3112127; PMID: 22163337
- He L, De Groot AS, Gutierrez AH, Martin WD, Moise L, Bailey-Kellogg C. Integrated assessment of predicted MHC binding and cross-conservation with self reveals patterns of viral camouflage. BMC Bioinformatics 2014; 15:S1
- De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol 2009; 131:189 - 201; http://dx.doi.org/10.1016/j.clim.2009.01.009; PMID: 19269256
- Duvvuri VR, Duvvuri B, Alice C, Wu GE, Gubbay JB, Wu J. Preexisting CD4+ T-cell immunity in human population to avian influenza H7N9 virus: whole proteome-wide immunoinformatics analyses. PLoS One 2014; 9:e91273; http://dx.doi.org/10.1371/journal.pone.0091273; PMID: 24609014
- Asselah T. Sofosbuvir for the treatment of hepatitis C virus. Expert Opin Pharmacother 2014; 15:121 - 30; http://dx.doi.org/10.1517/14656566.2014.857656; PMID: 24289735
- Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos hepatitis C sequence database. Bioinformatics 2005; 21:379 - 84; http://dx.doi.org/10.1093/bioinformatics/bth485; PMID: 15377502
