Abstract
Development of effective vaccines against emerging infectious diseases (EID) can take as much or more than a decade to progress from pathogen isolation/identification to clinical approval. As a result, conventional approaches fail to produce field-ready vaccines before the EID has spread extensively. Lassa is a prototypical emerging infectious disease endemic to West Africa for which no successful vaccine is available. We established the VaxCelerate Consortium to address the need for more rapid vaccine development by creating a platform capable of generating and pre-clinically testing a new vaccine against specific pathogen targets in less than 120 d. A self-assembling vaccine is at the core of the approach. It consists of a fusion protein composed of the immunostimulatory Mycobacterium tuberculosis heat shock protein 70 (MtbHSP70) and the biotin binding protein, avidin. Mixing the resulting protein (MAV) with biotinylated pathogen-specific immunogenic peptides yields a self-assembled vaccine (SAV). To meet the time constraint imposed on this project, we used a distributed R&D model involving experts in the fields of protein engineering and production, bioinformatics, peptide synthesis/design and GMP/GLP manufacturing and testing standards. SAV immunogenicity was first tested using H1N1 influenza specific peptides and the entire VaxCelerate process was then tested in a mock live-fire exercise targeting Lassa fever virus. We demonstrated that the Lassa fever vaccine induced significantly increased class II peptide specific interferon-γ CD4+ T cell responses in HLA-DR3 transgenic mice compared to peptide or MAV alone controls. We thereby demonstrated that our SAV in combination with a distributed development model may facilitate accelerated regulatory review by using an identical design for each vaccine and by applying safety and efficacy assessment tools that are more relevant to human vaccine responses than current animal models.
Abbreviations
| 6MDP | = | 6-muramyl dipeptide |
| BME | = | 2-Mercaptoethanol |
| CGE | = | Capillary Gel Electrophoresis |
| CLO97 | = | TLR7 ligand |
| CpG1826 | = | Synthetic Oligodeoxynucleotide containing unmethylated dinucleotide sequences (Toll-like receptor 9 agonist) |
| CTL | = | Cytotoxic T-lymphocyte |
| DARPA | = | Defense Advanced Research Projects Agency |
| EIDs | = | Emerging Infectious Diseases |
| GLP | = | Good Laboratory Practice |
| GMP | = | Good Manufacturing Practice |
| GP1 | = | Glycoprotein-1 |
| GP2 | = | Glycoprotein-2 |
| HLA | = | Human Leukocyte Antigen |
| HRP | = | Horseradish Peroxidase |
| LV | = | Lassa Fever Virus |
| MAV | = | Mycobacterium tuberculosis Heat Shock Protein 70 – Avidin |
| MtbHSP70 | = | Mycobacterium tuberculosis Heat Shock Protein 70 |
| NHP | = | Non-human Primates |
| OVA | = | Ovalbumin |
| PAGE | = | Polyacrylamide Gel Electrophoresis |
| PBMC | = | Peripheral Blood Mononuclear Cell |
| PEG | = | Polyethyleneglycol |
| RVKR | = | Furin Cleavage Site (Arginine, Valine, Lysine, Arginine) |
| SAV | = | Self-assembled vaccine |
| SAVL | = | Self-assembled vaccine formulated for Lassa Fever Virus |
Introduction
Following its failure to effectively deploy vaccines in response to the H1N1 influenza pandemic in 2009 the US federal government increased its capacity to respond to pandemic influenza threats.Citation1 The predominant focus of current emerging infectious diseases (EIDs) countermeasure development has been on broadly applicable post-exposure therapeutics rather than vaccines.Citation2 Therapeutic countermeasures have their limitations including the fact that the diversity of EID pathogens suggests that targeted approaches will still be required to provide sufficient protection and/or treatment either before or after exposure.Citation3 Broad countermeasures such as post-exposure monoclonal antibodies also face significant regulatory issues for eventual use in humans.Citation4 In contrast, prophylactic vaccines are generally thought to be the most effective means of protecting against disease and transmission and thus represent the most promising approach for preparedness.Citation2
The diversity of pathogen targets (viral, bacterial, fungal, parasitic) and host immune responses often results in significantly different vaccine strategies, which in turn produce vaccines that are quite structurally diverse. The resulting proliferation of vaccine structures complicates vaccine formulation and manufacturing requirements each new candidate adding raising new regulatory questions. Second, the potential for an EID's abrupt appearance and rapid spread requires vaccine development from pathogen identification to final product in a matter of months. In addition, the scale of deployment for such a vaccine is likely to be significant—possibly in the millions of doses required to provide broad protective coverage.
With these considerations in mind, we established the VaxCelerate Consortium in 2011 to demonstrate that rapid, scalable vaccine generation against unknown EIDs was technically possible.Citation5 VaxCelerate is based on a genome-to-vaccine approach and assumes as little as one pathogen genome would be available at the start of an outbreak. While a separate vaccine would be produced for every pathogen target, every vaccine would be made essentially the same way to reduce regulatory review using a broadly applicable vaccine platform. Excluding adjuvants would reduce regulatory and manufacturing complexities. Finally, the platform would offer a credible way to make rapid production of millions of doses of the specific vaccine possible within a matter of months.
The two key technologies leveraged to simultaneously address these requirements are in silico antigen design, and a self-assembling vaccine construct. An integrated set of computational tools is used to mine an infectious agent's genomes to identify candidate vaccine antigens with key properties such as predicted secretion and virulence. Validated immunoinformatics tools are then used to map protein sequences for HLA class I and II epitopes that are predicted to be immunogenic and broadly reactive.
The second technology, the self-assembling vaccine (SAV), enables assembly of multiple epitope peptides into a broadly applicable vaccine platform. The SAV consists of 2 portions: a fusion protein of M. tuberculosis heat shock protein 70 (MtbHSP70) and avidin (MAV) and a set of biotinylated peptides containing the class I and class II epitopes identified by the in silico screening method.
The recombinant MTBHsp70 protein's immunostimulatory functions include activation and maturation of monocytes and dendritic cells (DCs), enhancing antigen processing and presentation.Citation7 Furthermore, the mycobacterial protein stimulates production of CC-chemokines that attract macrophages, and effector T and B cells,Citation6,7 induction of the cytotoxic activity of natural killer cells,Citation8 and improved cross-priming of T cells which is dependent on DCs.Citation9 These functions and biology of HSP70 may play an important role in the function of SAV.
Here, we report efforts to generate a vaccine against Lassa fever and complete a key immunogenicity study of the vaccine in 120 d. As part of this project, early-phase good manufacturing practices were implemented for component supply and construction of the final vaccine and the mouse study of the vaccine were completed under good laboratory practices to increase the reliability and reproducibility of the data.
Lassa fever virus (LV) is an arenavirus infecting between 300,000 and 500,000 individuals per year in West Africa with a case fatality rate of 1–2% and a reported case-fatality rate climbing to 15% in hospitalized patients.Citation10 Although Lassa fever has been reported in other countries (Germany, Netherlands, United Kingdom and the United States) it is likely a product of international travels. The large morbidity/mortality toll, the possibility that LV could be developed as a biological warfare agent, the lack of timely diagnosis and limited treatment options further stress the need for the development of an effective vaccine. In spite of significant research efforts no viable vaccine clinical trial candidate is on the horizon.Citation11
The presence of LV specific IgM and IgG does not seem to affect survival outcomes and sera from convalescing patients fail to protect against infection.Citation10 In non-human primates (NHP), high titers of non-neutralizing antibodies correlate with survival providing strong evidence that cell mediated immunity is at the core of the protective immune response.Citation11,12 In NHP studies, rapid and strong cytotoxic lymphocyte (CTL) responses positively impact survival. Protective immunity to Lassa is largely a function of a T cell-mediated immune response to the virusCitation13-16 this is one of the rationales for testing the VaxCelerate platform with this virus.
The virus is composed of 2 ambisense RNA strands, L (7-kb) and S (3-kb). Its RNA encodes 5 proteins, L protein (RNA polymerase), NP (nucleocapsid), Z protein (RING, matrix), glycoprotein 1 (GP1) and glycoprotein 2 (GP2).Citation17,18 GP1 and GP2 are the products of the cleavage of the precursor protein GPC. Candidates for antigen targeting within the Lassa virus include the G protein (with the precursor molecule glycoprotein C, GPC, post-translationally cleaved into a stable signal peptide, SSP, and 2 subunits glycoproteins GP1 and GP2), the nucleoprotein (NP), the L protein and the Z protein. In a prospective study, a number of cross-reactive epitopes from various pathogenic arenaviruses were identified and shown to offer the potential for wide ranging immunity.Citation19 Recently, a DNA vaccine encoding the subunits GP1 and GP2 resulted in protection of vaccinated animals in a guinea pig challenge study,Citation20 suggesting that a subunit vaccine should confer protective immunity. We designed a subunit vaccine consisting of MtbHSP70-avidin coupled to immunogenic peptides from Lassa GP1 and GP2 proteins. To meet the constrained time requirement for this project we used a distributed R&D model that has been previously described.Citation5 The network included experts in the fields of protein engineering and expression and purification (Pfēnex), bioinformatics (EpiVax), peptide manufacture/design (21st Century Biochemicals) and Good Manufacturing Practice (GMP)/Good Laboratory Practice (GLP) and testing standards (MPI Research). The resultant vaccine candidate was tested in a transgenic HLA-DR3 mouse model to assess CD4+ T helper responses to the human HLA-specific peptides.
We present the successful implementation of a model vaccine development using a distributed R&D approach. First, we demonstrated in a pilot study that the self-assembled vaccine could trigger an immune response comparable to a known peptide based vaccine in the HLA-DR3 mouse model. Second, the self-assembled Lassa fever virus specific vaccine triggered a significant Th1 response in splenocytes from immunized animals when challenged with a pool of HLA class II specific peptides as compared to the peptides alone.
Results
Characterization of recombinant MtbHSP70-avidin expressed in Pseudomonas fluorescens
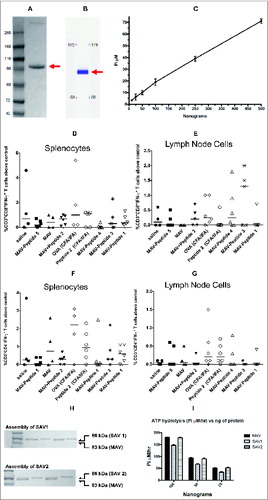
The MAV protein was produced by Pfēnex (San Diego, CA) and characterized for purity, ATPase activity and ability to bind biotin. depicts the fusion construct between MtbHSP70 and avidin. As shown in , the protein was greater than 90% pure by polyacrylamide gel electrophoresis (PAGE) and by capillary gel electrophoresis (CGE). To evaluate proper protein folding of the MtbHSP70 portion, we assessed the ability of the MAV protein to hydrolyze ATP. As shown in , the ATPase activity was linear over a wide concentration range. Finally, we assessed the ability of the avidin component to bind biotin using a biotinylated HRP assay. The protein was incubated with excess biotinylated HRP for 20 minutes. Unbound biotinylated HRP was removed with streptavidin magnetic beads. Biotinylated HRP bound to MtbHSP70-avidin remains in solution and therefore can react with tetramethylbenzidine. As shown in , the avidin portion of the fusion protein was functional as evidenced by the amount of HRP activity remaining in solution when MtbHSP70avidin was present.
Table 1. Biotin binding assay of MAV and SAV
Figure 1. Depiction of self-assembled vaccines structures. (A) Structure of MtbHSP70-avidin (MAV) and biotinylated antigens. (B) Structures of OVA peptides for SAV-OVA studies described in . (C) and (D) Structures of short (C) and long (D) Flu peptides described in for SAV1 and SAV2 used in Flu study.
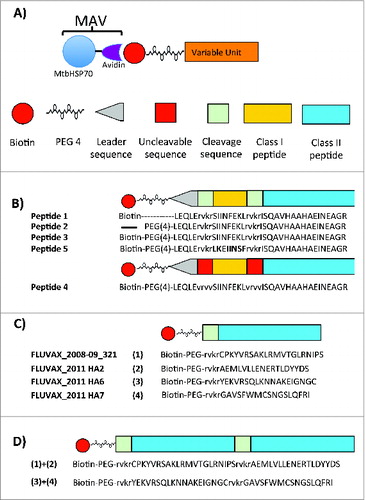
Figure 2. Characterization of Pfēnex’ MtbHSP70-avidin (MAV). (A) 500 nanograms of MAV were electrophoresed on a 4–12% Bis-Tris NuPAGE gel run in MOPS buffer at constant 200 V for 1 hr. (B) Reproduction of the capillary gel electrophoresis (CGE) profile provided by Pfēnex. 150 nl of purified MAV (976.2 ng/nl) were injected into the microfluidic channel of the LabChip GX/II using electrokinetic injection. Applying a voltage across the channel effected protein separation. Protein bands were detected via laser induced fluorescence. Arrows indicate the migration position of MtbHSP70-avidin. (C) MtbHSP70-avidin ATPase activity assessment. A series of MtbHSP70-avidin concentrations (25 – 500 ng) were incubated as described in materials and methods. (D, E, F and G) Flow cytometric analysis of splenocytes and lymph node cells from ovalbumin immunized mice as described in Materials and Methods. Splenocytes (D, F) and lymph node cells (E, G) were prepared as described in Materials and Methods and stimulated with medium alone, SIINFEKL (D, E; 10 μg/ml) or ISQAVHAAHAEINEAGR (F, G; 10 μg/ml) for 24 hours at 37°C. The percent of CD3+CD8+IFN-γ+ and CD3+CD4+IFN-γ+ cells above medium alone was determined and plotted as percent above control. (H) Gel-shift assay of SAV1 and SAV2. 400 to 800 ng of protein were denatured and subjected to electrophoresis on a Novex® 4–12% Bis-Tris NuPAGE gel run in MOPS buffer at constant 200 V for 5 hr. Arrows indicate MAV, SAV1 and SAV2. (I) SAV1 and SAV2 ATPase activity. Three different concentrations of MAV, SAV1 and SAV2 were assayed for their ability to hydrolyze ATP as described in Materials and Methods. Abbreviations: MAV - Mycobacterium tuberculosis heat shock protein 70 fused to avidin; OVA - ovalbumin; PEG - polyethylene glycol; SAV1 - MAV assembled with flu specific peptides 1–4 (); SAV2 - MAV assembled with flu specific peptides 5 and 6 ().
Peptide design optimization using an ovalbumin immunization model
In order to understand how the design of biotinylated peptides impacts antigen processing and cell mediated immunity we used the ovalbumin/C57BL/6 model to take advantage of well-characterized class I and class II epitopes.Citation21,22 We tested the effects of peptide designs incorporating furin cleavage sequences (RVKR) and a polyethylene glycol spacer on CD4+ and CD8+ T cell to SAV-OVA vaccine. Mice were immunized with self-assembled ovalbumin (OVA) peptides with and without furin cleavage sites and with and without PEG (Fig. 1B and ). All peptides successfully assembled at >95 % according to the biotinylated HRP assay (). Immunization consisted of an intradermal prime injection followed by 2 intradermal boosts. We assessed cell-mediated immune responses in isolated lymph node cells and splenocytes. After stimulating cells with the class I epitope, SIINFEKL, we analyzed the immune response by flow cytometry looking at CD3+ CD8+ cells producing interferon-γ. As shown in , we observed cell mediated immune responses in SIINFEKL-stimulated lymph node cells in the group immunized with self-assembled peptides containing biotin, PEG, a leader segment, and RVKR (peptide 3 ). Mice immunized with saline, MAV alone or the control LKEFNIIS self-assembled vaccine did not respond to SIINFEKL-stimulation.
Table 2. Epitopes and peptides sequences used for OVA, Flu and Lassa fever virus vaccines
HLA-DR3 mice respond to immunization with a self-assembled vaccine
We next tested whether the SAV platform could enhance vaccine responses in HLA-DR3 mice, the strain to be used in the Lassa vaccine study. A pilot experiment using HLA class II peptides against H1N1 influenza that induced significant class II T cell responses in a DR3 mouse model was performed.Citation23 The pilot study compared responses between a DNA/peptide vaccine previously used to deliver the H1N1 epitopes (FluVax) and an influenza SAV using the same peptides.Citation24 Sequences of the peptides used in both types of vaccines are detailed in Figure 1C and 1D and .
In light of reports that high doses of MtbHSP70 could be tolerogenic, we designed a way to reduce the amount of the core protein in SAV by increasing the immunogenic peptide amount using concatenated epitopes.Citation25,26 In this study we compared T cell mediated peptide specific immune responses to SAV assembled with both shorter (40–45 amino acids) peptides and (60–65 amino acids) concatenated peptides. The vaccine with shorter peptides (SAV1) was comprised of an equimolar mixture of 4 different protein-peptide constructs, with each construct containing one of the 4 class II epitopes. SAV2 consisted of an equimolar mixture of 2 protein-peptide constructs, with each construct containing 2 different concatenated class II epitopes. The class II peptides in SAV2 were separated by a furin cleavage site (). All the biotinylated peptides self-assembled with MtbHSP70-avidin and we characterized the product of self-assembly by gel shift assay and ATPase activity measurements (). We showed that biotinylated peptides self-assembly were essentially complete and that the ATPase activity was maintained in the process.
The immunization schedule previously established for FluVax was replicated in this study (see Method section).Citation23 SAV immunization was synchronized with this schedule by starting SAV immunizations 14 d after the initial FluVax DNA electroporation prime (). Mice were bled on days -3, 14, 28 and 35. We assessed the effectiveness of each vaccine at stimulating immune memory by isolating lymph node lymphocytes and splenocytes from each of the 4 groups, stimulating these with the individual influenza immunizing peptides and then analyzing resultant CD3+CD4+ interferon-γ expression by flow cytometry. Results of peptide stimulation are shown in . Splenocytes from Fluvax-immunized mice responded robustly when stimulated with individual influenza peptides, while splenocytes from SAV2 immunized mice showed modest responses to 3 of the influenza peptides. The robust splenocytes response from FluVax-immunized mice was appropriate to the intramuscular/intraperitoneal mode of administration and chemical adjuvanting of the FluVax peptides with 6MDP, CLO97, and CpG1826.Citation23 Lymphocytes from FluVax-, SAV1- and SAV2-immunized mice all showed measurable responses to many of the individual influenza peptides compared to medium alone. Surprisingly, lymph node lymphocytes from MAV-treated mice also responded when stimulated with influenza specific peptides. SAV2-stimulated lymphocytes also showed robust CD3+CD4+IL-4+ cell responses to peptide stimulation, which was not seen in mice immunized with FluVax, MAV or SAV1 (). Finally, analysis of sera from vaccinated and control animals revealed that only mice receiving FluVax had significant antibody titers against the peptide components of the vaccine. In contrast, mice receiving MtbHSP70-avidin or the self-assembled vaccine had measurable antibody titers against MtbHSP70 ().
Table 3. Immunization schemes
Table 4. Antibody responses against immunizing components of the vaccines*
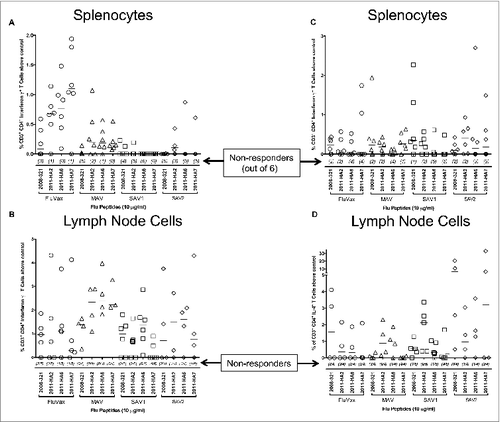
Figure 3. Flow cytometric analysis of splenocytes and lymphocytes from FluVax, SAV1 and SAV2 immunized mice. Splenocytes and lymphocytes were prepared as described in Materials and Methods and stimulated with the indicated flu peptides (10 μg/ml) or medium alone for 24 hours at 37°C. Brefeldin A was added 4 hours prior to harvest to inhibit protein transport. Cells were stained for CD3, CD4, CD8, IL-4 and interferon-γ fixed and analyzed on a BD LSR FortessaTM. The percent of CD3+CD4+IFN-γ+ cells above medium alone was determined and plotted as percent above control. (A) Splenocytes response, (B) lymph node cells response. The percent of CD3+CD4+IL-4+ cells above medium alone was determined and plotted as percent above control for (C) splenocytes response, (D) lymph node cells response. Numbers in parenthesis indicate non-responders.
Design of immunogenic Lassa fever virus peptides
The key considerations used in constructing targeting peptides for the test vaccine were as follows: 1) limit class I and class II peptides to the Lassa glycoproteins, 2) ensure peptides include class I and class II targets from both GP1 and GP2, 3) ensure coverage of all HLA super types with both with both the class I and class II peptides, 4) maximize the number of peptides delivered per unit of MtbHSP70-avidin, 5) optimize the solubility of the peptide strings, 6) combine class I and class II peptides in each peptide string, 7) include cleavage sequences to direct antigen processing and cross presentation, and 8) minimize potential junctional immunogenicity created by juxtaposition of epitopes and flanking sequences that could result in non-specific immunogenic responses.
The resultant peptide set generated using these criteria is shown in . The sequences of the final peptide constructs used in the SAVLassa (SAVL), including the location of the antigen processing cleavage sequences, are shown in . Assembly of each peptide to MtbHSP70-avidin was monitored and was reported to be >95 % complete. The ATPase activity of the self-assembled vaccine is shown in A. We observed a 22 to 50% reduction in ATPase activity when MTBHsp70-avidin was assembled with the SAV1 peptide (MAVf) or with the long Lassa fever virus specific peptides (). We surmise that this is related to the length and structure of the bound peptide(s). This is in contrast to the SAV2 vaccine, which was assembled with concatenated Flu peptides ().
Figure 4. Structure and sequences of Lassa fever virus immunogenic peptides. (A) Basic framework for the synthesis of immunogenic concatenated class I and class II peptides. (B) Biotinylated Lassa fever virus specific peptides.
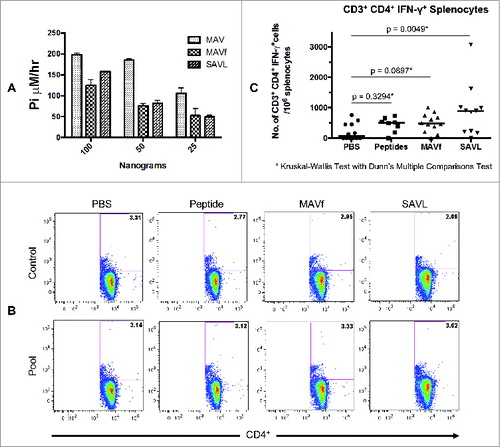
Figure 5. Characterization of the self-assembled Lassa fever virus vaccine (SAVL). A) ATPase activity of MAV, MAVf, and SAVL using 25, 50 and 100 ng. The assay was carried out as described in Materials and Methods. (B) Flow cytometric analysis of splenocytes from mice immunized with SAVL. Splenocytes from mice immunized with PBS, peptides, MAVf and SAVL were prepared as described and stimulated with medium alone or a pool of class II peptides (10 μg/ml). Total incubation time was 24 hours at 37°C. Brefeldin A was added 4 hours prior to harvest. (C) Comparison of splenocytes response to class II Lassa fever virus specific peptides between different immunization groups.
Testing the efficacy of a self-assembled vaccine for Lassa fever
The fully assembled Lassa SAV (SAVL) was formulated for injection and then tested for its ability to induce a CD4 T helper cell response in HLA-DR3 mice. Prior to immunization, the flow cytometry intracellular assays were validated for the detection of interferon-γ in CD3+CD4+ lymphocytes from spleens and lymph node cells. The validation included intra-assay precision, inter-analyst precision, and stability testing post-fixation and sample analysis with and without PMA/Ionomycin. Other endpoints such as the presence of IL-4, Granzyme B and IL-17 were optimized but were not part of the cGLP validation studies.
Immunization was performed as described in . Isolated lymph node cells and splenocytes were stimulated with a pool of class II peptides to assess immune memory. illustrates the flow cytometric analysis and gating strategy used for CD3+CD4+IFN-γ+ splenocytes. For each mouse, we calculated the resultant values from splenocytes or lymphocytes stimulated with medium alone versus peptides. These were expressed either as the number of net positive cells per 106 splenocytes or lymph node cells, or the percentage of net positive cells of the total CD4+ T cell population. As shown in , splenocytes from 10 of 11 SAVL immunized mice responded to the pool of class II Lassa fever virus specific peptides. Similarly, splenocytes from 7 of 8 mice immunized with peptides alone and splenocytes from 11 of 12 mice immunized with MAVf responded to the pool of class II peptides compared to mice injected with PBS only.
The results of the study were positive with respect to the primary end point, showing that splenocytes from mice vaccinated with SAVL generated a statistically significant increase in CD3+CD4+IFN-γ+ T cells in response to stimulation with the Lassa-specific class II peptides compared to splenocytes from mice exposed to PBS alone (; p = 0.0049; Kruskal-Wallis with Dunn's multiple comparisons test). The median values for the SAVL vaccinated mice were 891 CD3+CD4+IFN-γ+ T cells per 106 splenocytes (0.48% of the CD4+ T cells). Splenocytes from mice vaccinated with either peptide alone or with MAVf also showed an increase in these values (median values of 508 and 482 CD3+CD4+IFN-γ+ T cells per 106 splenocytes respectively), but these were not statistically significant when compared to the PBS exposed group (median value of 64 CD3+CD4+IFN-γ+ T cells per 106 splenocytes).
Results from class II peptides stimulation of splenocytes and lymph node cells were not statistically significant for CD3+CD4+IL-4+, CD3+CD4+Gzm-B+, and CD3+CD4+IL-17+ and all CD8+ T cell responses. Since HLA-DR3 mice do not express class I MHC no CD8+ T cell responses were anticipated or detected (data not shown).
Discussion
The VaxCelerate II project provides support for the concept of a rapid and scalable vaccine platform for addressing abruptly appearing EIDs of significant public health importance. We have demonstrated that within 120 d a de novo vaccine could be generated based only on genomic information using scalable manufacturing approaches and that the putative vaccine could induce potent cell mediated immune responses in a human-relevant mouse model ( for time line).
Production of the protein component of the vaccine was optimized to consistently yield a highly purified functional protein as assessed by CGE, LDS-PAGE, ATPase activity and ability to bind biotinylated horse radish peroxidase ( and ). It is interesting to note that, in contrast to a self-assembled vaccine complex consisting of Streptavidin and endogenous TLR4, we did not observe tetramer formation.Citation27 The small shift observed upon assembly of peptides and MtbHSP70-avidin made it difficult to rapidly assess completion of the process. The use of biotinylated-HRP to assess the number of remaining biotin binding sites allowed us to test many assembly conditions.
We used the ovalbumin immunization model to assess the optimal peptide assembly that would yield the optimal immune response.Citation22,28 Prior work with fusions of MtbHSP70 with large segments of ovalbumin containing the class I specific SIINFEKL peptide suggested that successful presentation relied on protein processing.Citation29,30 To improve class I processing of peptides in the self-assembled vaccine, we drew on the work of Del Val et al.,Citation31 showing that furin cleavage can preferentially drive class I MHC processing.Citation32-34 Lu et al., showed in a tumor model that the presence of a furin cleavage site (RVKR) and the combination of class I and class II peptides improves CTL responses to a vaccine.Citation35 To facilitate furin cleavage we included a leader segment (LEQLE) consisting of the 5 amino acid residues preceding ovalbumin's class I peptide SIINFEKL. The presence of PEG and a furin cleavage site (RVKR) resulted in optimal presentation of SIINFEKL and induction of CD8+ T cell memory. While SAV coupled to OVA peptide 3 () induced detectable class I immune responses to SIINFEKL in the OVA model, class II T cell responses were more modest compared to unvaccinated controls (). The immune response was increased in lymph node cells and was superior to whole protein immunization adjuvanted with CFA/IFA. These results demonstrated that the inclusion of a leader sequence, furin cleavage site and PEG in the peptide component of SAV improved immunogenicity. The requirement for the furin cleavage site to augment immunogenicity is consistent with prior studies showing that this component improves peptide processing and cross-presentation.Citation32-35
Before tackling the design of a Lassa fever virus specific vaccine we needed to confirm that the peptide framework would be successful when applied to already optimized immunogenic peptides. In collaboration with EpiVax we tested our approach using the Flu peptides they optimized in their FluVax vaccine using the transgenic HLA-DR3 mouse model.Citation23 To maximize the antigen dose per assembled complex, we tested the effect of concatenating 2 peptides separated by the furin cleavage site. As we had seen in the ovalbumin model, the response was more pronounced in the lymph node cells with our self-assembled vaccine although splenocytes from mice immunized with SAV2 showed some positive CD3+ CD4+ interferon-γ+ responses. Unexpectedly we observed a very strong response to Flu peptides in lymph node cells from mice immunized with MAV itself. We note that a group of investigators from Germany have published a number of studies showing that HSP70 has the capacity to bind human HLA-DR molecules and induce proliferation of CD4+ T cells.Citation36-38 This investigative team did not explore the implications of this activating HLA binding in the context of vaccines. We believe that the potentiation of the CD4+ vaccine responses by the self-assembling protein in the context of this transgenic HLA-DR3 mouse model may be reflective of this activating binding capability of MtbHSP70 and may be one mechanism of vaccine potentiation in humans. A Flu challenge study is planned in order to assess the efficacy of MAV to induce a protective immune response in transgenic mice. This immunostimulating effect seen only in transgenic HLA-DR mice or in human samples also provides a concrete example of the limitation of the wild-type mouse models for predicting human immune responses and further emphasizes the value of developing more human-like vaccine testing platforms. In order to minimize this effect in the Lassa fever vaccine study we complexed the protein with Flu peptide 4 ().
We demonstrated that the self-assembled Lassa fever vaccine is capable of inducing a significant increase in interferon-γ positive CD4+ T cells in HLA transgenic DR3 mice compared to PBS control. The vaccine group showed positive responses in splenocytes but not in lymph node cells. The CD3+CD4+IFN-γ+ T cell responses were comparable to those reported in a number of other antiviral vaccine studies in mice, non-human primates and humans, including 2 vaccines that achieved market approval in the US (). For example, when PBMCs from yellow fever vaccinees were challenged in vitro, 250 cells per millions had an interferon-γ+ response in an ELISPOT assay (). In Balb/c mice immunized against hepatitis C, 125–200 splenocytes per million showed a positive interferon-γ response (). The median number of CD4+IFN-γ+ splenocytes response in our SAVL immunized mice was 891 cells per million splenocytes thus comparing favorably with other vaccines.
Table 5. Comparison of T cell immune responses observed in this study with other EID vaccines
Unlike our results with the class II influenza peptides, the Lassa vaccine did not promote IL-4 responses. These data support the view that peptide composition plays a role in determining the type of immune response induced by the vaccine platform. It is worth noting that, while not reaching statistical significance, the peptide vaccinated group showed an increase in response to stimulation with class II peptides. The use of peptides (<55 amino acids) in absence of adjuvant do not show such responses.Citation23,39 It is possible that the use of immunogenic peptides greater than 60 amino acids and including the use of furin cleavage sites have the ability to increase immunity by themselves. Two prior studies demonstrate the immunogenicity of peptides of this length without the use of adjuvants.Citation40,41 From this perspective; the peptide-only vaccination group should not be considered to be a negative control. In addition, vaccination of mice with the MtbHSP70-avidin construct (self-assembled to a non-Lassa peptide; MAVf) showed an increase in response to stimulation with class II peptides. Again, this increase was not statistically significant. This response was similar to the potentiation of immune responses seen in the HLA-DR3 transgenic mouse influenza vaccine study. These data further support the work of Holzer et al., showing that HSP70 binding to HLA-DR molecules enhance class II-restricted peptide presentation and CD4+ T cell activation.Citation36,37
Although our vaccine included 12 class I peptides, absence of expression of human HLA class I in this mouse model precluded us from investigating the effect of the vaccine on CD8+ T cells. Potential murine/human proteosomal processing differences are likely shortfalls of testing this vaccine in double transgenic mice. A more robust approach to quantitating the immunogenicity of the designed peptides would be to assess the peptide-specific T cell immune responses of convalescent individuals who survived Lassa virus infection in endemic areas. A challenge study in non-human primates using rhesus or cynomolgus monkeys would also help assess the efficacy of our Lassa fever virus vaccine.Citation15
In the process of designing the Lassa fever vaccine, we reviewed subunit vaccine studies published over the last 15 y. These studies have shown that use of NP, GPC or the 2 G proteins or a combination of NP/GPC is protective in a variety of animal models.Citation10,14,15,42-56 Studies of convalescent sera from patients with Lassa Fever or human HLA transgenic mice identified a number of epitope targets within the NP, GP1, GP2 and GS proteins.Citation11,12,16,19,57-63 Recent work in a guinea pig model utilizing the GP1 and GP2 proteins in a DNA vaccine resulted in adequate protection from challenge virus.Citation13-16,20 Our review of Lassa vaccine development efforts did not show that the Lassa L protein has ever been used as part of the antigens in a subunit vaccine, although the L protein was shown to be protective in animal studies of other arenavirus like lymphocytic choriomeningitis virus.Citation17,18,64 In this setting, the L protein has been identified in a guinea pig model as a virulence factor for the virus.Citation19,65-67 The ML29 reassortment virus vaccine, comprising the L and Z protein from Mopeia virus—an Old World arenavirus closely related to Lassa—and the GPC and NP proteins from Lassa, have also been effective in a number of vaccine studies.Citation20,47,50,51,55 The L protein of Mopeia was chosen to serve as a replication or translational attenuation factor in the vaccine.Citation68,69 We therefore limited the peptide targets to class I and class II peptides of the Lassa glycoproteins G1 and G2. This selection also excluded targets against the Lassa nucleoprotein, which has been targeted in some subunit vaccine candidates.Citation14,21,42,44-48,50-52,55,56
Although we detected an antibody response against the MtbHSP70 component of the vaccine we failed to observe a strong antibody response against the peptide component of the vaccine. We attribute this lack of antibody response to the peptide antigen to rapid uptake and processing. While the observed strong Th1 response is promising it does not guarantee that this vaccine is protective against Lassa fever virus. In order to further qualify the chosen epitopes we plan to test PBMCs from recovering Lassa fever virus patients. This would be followed by challenge study using a suitable animal model.
In conclusion, leveraging 2 key technologies, in silico antigen design and a self-assembling vaccine construct, in combination with a distributed R&D effort and an emphasis on testing in a humanized transgenic animal model allowed us to accomplish our stated goal of generating a vaccine candidate against Lassa fever virus and performing a key immunogenicity study in 120 d. The positive CD4+ T cell responses induced by the SAV candidate further supports the feasibility of this approach to address the multiple challenges posed by EIDs. Further testing of this vaccine candidate is clearly required, including broader immunogenicity testing and pathogen challenge studies in non-human primates. Further adaptations of this vaccine strategy may contribute to improved defenses against bioterror agents, and could eventually lead to the development of personalized ‘on demand’ vaccines for civilian populations.
Materials and Methods
Mice
For the Ovalbumin study we used 6–8 weeks old female C57BL/6 mice from Jackson Laboratory. Female 6–8 weeks old HLA-DR3 mice were obtained from Taconic under license to EpiVax and were used in the Vax-Flu and Vax-Lassa studies.
Synthesis of MtbHSP70-avidin
Pfēnex Inc. used its proprietary Pseudomonas fluorescens-based platform technology to prepare 10 mg of MtbHSP70-avidin. Briefly, cells expressing the target protein were lysed, and debris was removed by centrifugation. The soluble fraction was chromatographed over a His Trap FF column (GE Healthcare Life Sciences) for initial purification. The flow through was reprocessed over a His Trap FF column to improve the yield. The pooled HisTrap elutions were further processed on a Sephacryl S-200 HR column (GE Healthcare Life Sciences) to remove co-purifying contaminants. Finally, endotoxin levels were significantly reduced by chromatography on EtoxiClearTM (ProMetic Biosciences Ltd.), as measured by the Kinetic Chromogenic LAL assay. The final endotoxin content was <50 EU/mg of protein. Protein concentration was measured on an Eppendorf Biophotometer by OD280. Correction for light scattering was performed by subtracting the absorbance at 320 nm multiplied by 1.7. The purity of the protein was assessed by capillary gel electrophoresis with detection by laser-induced florescence on a LabchipGX/II made by PerkinElmer (formerly Caliper Life Sciences, Inc.). Finally, the intact mass analysis was performed by LC-MS using an Agilent 1100 HPLC system and a Waters Q-TOF mass spectrometer with lock-mass correction. Deconvolution of the spectra from the main peak was performed with Waters MaxEnt1 software.
Gel-shift assay
Assembly of MAV with the H1N1 peptides () and Lassa fever virus peptides (; ) was assessed using a gel-shift assay. 400 to 800 ng of MAV or SAV were mixed with sample loading buffer, heated to 70°C for 10 minutes prior to addition of dithiothreitol. The proteins were subjected to electrophoresis on a 4–12% Bis-Tris NuPAGE gel run in MOPS buffer at constant 200 V for 5 hrs. Ten microliters of Novex® Sharp pre-stained protein standards were added to adjacent lanes. Protein bands were identified by staining with RAPIDstain (Geno Technologies).
Immunogenic epitopes identification
Immunoinformatics
Epitope identification. The Lassa virus Josiah strain complete sequence was downloaded from the National Center for Biotechnology Information (Accessions HQ688674 and HQ688672).Citation70 Antigen sequences were downloaded in GenPept format, where the accession number and corresponding amino acid sequence of each of the 4 antigens were exported and then uploaded to an in-house database. Per antigen, the amino acid sequence was parsed into 9-mer peptides overlapping by 8 amino acids and analyzed using EpiMatrix 1.2 to identify potential T cell epitopes.Citation71 Peptides scoring ≥1 .64 on the EpiMatrix “Z” scale (typically the top 5% of any given sample), are likely to be human leukocyte antigen (HLA) ligands and are considered “hits." Class II epitopes were identified for 8 archetypal alleles that cover >90 % of the human population (DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, DRB1*1301 and DRB1*1501).Citation72 Class I epitopes were identified from among 9-mer and 10-mer peptides for 6 “supertype” alleles: A*0101, A*0201, A*0301, A*2402, B*0702, B*4401.Citation73 Lassa antigens were screened for regions of high Class II epitope density using the ClustiMer algorithm for construction of broadly reactive vaccine immunogens 15–25 amino acids long.Citation74 Class I sequences do not cluster like Class II hits and did not undergo ClustiMer analysis.
Homology analysis. The JanusMatrix algorithm was used to analyze epitopes for human or commensal homologies.Citation75 JanusMatrix screens HLA ligands for T cell receptor-facing sequence patterns shared between pathogen and human or between pathogen and commensal, which may activate pre-existing cross-reactive (inducible) regulatory T cells. Epitopes containing high cross-reactive potential were discarded and Lassa virus-specific epitopes were retained for vaccine formulations. Epitopes were also screened for homology to other Lassa strains and other arenaviruses using JanusMatrix. Finally, epitopes were cross-referenced against the Immune Epitope Database (IEDB) for Lassa virus sequences that were reported to bind HLA and/or elicit a T cell response.
Synthesis of immunogenic peptides
Five mg of each biotinylated immunogenic peptide selected as defined above were prepared by Fmoc solid phase synthesis (21st Century Biochemicals) and HPLC purified. Purity was assessed by HPLC (peak area) and all peptides were >90% purity. The mass and peptide sequence were confirmed by ESI-MS and tandem MS, respectively, utilizing a QSTAR XL Pro Qo-TOF MS (ABI-SCIEX). Three peptide designs were used in a process of optimization for immunogenicity.
For the Ovalbumin study we tested the effect of Polyethyleneglycol (PEG) and the furin cleavage substrate RVKR, on the immunogenicity of the peptide. Therefore, peptides were designed with and without PEG and with and without RVKR. These are listed in . Flu peptides were composed of PEG and a series of 4 epitopes similar to those present in FluVax. These are listed in . Finally; the Lassa fever virus specific peptides are listed in .
Vaccine self-assembly and assessment of reaction
Each peptide was assembled separately by simply mixing a 10-fold molar excess of biotinylated peptide with MAV in PBS. The reaction was allowed to proceed at room temperature for 12 hours by mixing end over end. Assembly was assessed as described below and found to have reached greater than 95% completion.
Assembly was assessed by measuring the amount of biotinylated-HRP that could still bind to MAV. Briefly, the assay consisted of taking a reaction aliquot (10 μl) and mixing it with biotinylated HRP for 15 minutes in a final reaction volume of 100 μl. Excess biotinylated HRP was removed by addition of 10 μl streptavidin magnetic beads (Pierce) and incubation for 5 minutes. 50 μl of supernatants were transferred to an ELISA plate followed by addition of 100 μl of TMB and incubation in the dark for 10 minutes. The reaction was stopped by addition of 2N H2SO4 and read at 450 nm subtracting background at 650 nm. A low read out is taken as indicative that all avidin-binding sites are occupied with peptides while a high read out means that sites remain available. These readouts are always compared to MtbHSP70-avidin unexposed to peptides.
ATPase assay
The ATPase assay was performed according to the manufacturer's recommendations (Innova Biosciences, ATPase Assay Kit cat# 601–0121). Briefly, 25 to 500 ng of assembled or unassembled MAV protein were incubated in 50 mM Tris pH 7.5 supplemented with 2.5 mM MgCl2 and 0.5 mM ATP. The assay was carried out at 37°C for 20 minutes and stopped by addition of an acidic proprietary form of malachite green. Negative controls consisted of adding the protein mix after addition of the stop solution. Color was allowed to develop for 30 minutes at room temperature. ATP hydrolysis was assessed by determining the amount of Pi released based on the intensity of the OD at 650 nm.
Immunization protocols
Three immunization protocols were performed.
Ovalbumin immunization study
A total of 9 groups comprised of 6 animals per group were used in this study. A series of peptides, described in , were tested in order to determine the optimum configuration for self-assembly. Groups consisted of Saline, Ovalbumin (CFA/IFA), peptide 2 (unbiotinylated) (CFA/IFA), MAV, MAV + peptide 2 (unbiotinylated), MAV-Peptide 5 (no SIINFEKL), MAV-Peptide 4 (no furin cleavage), MAV-Peptide 3 (furin cleavage), MAV-Peptide 1 (no PEG). All animals received a prime intradermal injection on day 0, followed by intradermal boosts on days 14 and 28. Mice received the equivalent of 12 μg of MtbHSP70-avidin per injections. Mice were bled on day -3, 14, 28 and 35.
Influenza vaccine study
Twenty-four HLA-DR3 mice were divided into 4 groups (6 mice per group) immunized with FluVax2011, MtbHSP70-avidin, SAV1, SAV2. FluVax2011 immunization consisted of a prime and boost intramuscular DNA electroporation on days 0 and 14 followed by subcutaneous peptide immunization on days 28 and 42. For peptides immunization a mixture of CLO97, CpG182 and 6MDP adjuvants were used. MtbHSP70-avidin, SAV1 and SAV2 were given intradermally (doses representing 12 μg of MtbHSP70) on days 0, 14 and 28. Bleeds were taken on day -3, 14 and 38.
Lassa fever vaccine study
Four groups of mice, (12 mice per group), were immunized intradermally with PBS, peptide alone, MAVf or SAVL. MAVf consists of assembly of the fusion protein with Flu peptide number 4 described in . The immunization schedule is shown in and consisted of a priming dose on day 0 followed by boosting doses on days 14 and 28. Mice were bled on days -3, 14, 28 and 35. Animals were euthanized on days 35 and 37. Each immunization consisted of 2 50 μl intradermal injections. Immunizations with peptide alone consisted of 2 μg doses while MAVf and SAVL immunizations consisted of 12 μg of MAV equivalent per dose.
Cell mediated immunity assay - flow cytometric analysis
Briefly, splenocytes were harvested from control and vaccinated mice. Red blood cells were lysed using M-Lyse (R&D systems). The remaining lymphocytes were suspended in RPMI–10% fetal bovine serum–1% penicillin/streptomycin–1% l-glutamine–0.1% BME to a concentration of 20 × 106 cells/ml. Single cell splenocytes/lymphocytes suspensions were transferred at 1.0 to 2.0 × 106 cells/well to round bottom plates. Individual and pooled peptides were evaluated at 10 μg/ml. Controls consisted of wells incubated with medium alone or PMA/Ionomycin. Plates were incubated for 20 hours at 37°C, 5% CO2 prior to addition of Brefeldin A and returned to the incubator for another 4 hours. At the end of the incubation period cells were harvested and stained for flow cytometry.
At 24 hours, cells were spun down, and washed by addition of FACS staining buffer (5% FBS in PBS). Antibodies against CD3, CD4, CD8, CD45, were added and cells were incubated on ice in the dark for 30 minutes. After washing with FACS staining buffer, cells were fixed and permeabilized for 20 minutes on ice. After washing with perm/wash buffer, cells were stained for interferon-γ, IL-4, granzyme B (Ovalbumin and Flu), interferon-γ, granzyme B, IL-4, and IL-17a (Lassa), for 30 minutes on ice. Cells were washed with perm/wash buffer and rinsed with FACS staining buffer.
Flow cytometry data acquisition was performed on a BD LSR FortessaTM using FACsDIVA Software v6.2. A total of 100,000 events were collected. Flow data were analyzed on FlowJo version 8.8.7.
Antibody response
A standard ELISA was used to determine antibody responses against MtbHSP70, Flu and Lassa Fever virus specific peptides as described below:
Anti-MtbHSP70
Wells of a 96-wells plate were coated with 100 μl of MtbHSP70 (2.5 μg/ml) in PBS overnight at 4°C. Unbound MtbHSP70 was removed with 2 PBS washes. Wells were blocked at room temperature for 2 hours with 2% BSA, 2% sucrose, and 1% normal goat serum in PBS. After removal of block solution, 100 μl of sera diluted in block solution were added to each well and incubated at room temperature for 1 hour. Plates were washed 4 times (0.05% Tween 20 in PBS) followed by addition of 100 μl of a 20,000 fold dilution of HRP-conjugated goat anti-mouse (Pierce) in blocking buffer. After incubation at room temperature for 1 hour, plates were washed 4 times as before, and rinsed once with water. 100 μl of TMB (warmed to room temperature) were added to each well and the plates incubated in the dark for 20 minutes at room temperature. The reaction as stopped by addition of 50 μl of 2N H2SO4. The optical density at 450 nm minus background at 650 nm was obtained using an EMAX plate reader from Molecular Device.
Anti-Flu peptides
For the anti-Flu peptides ELISA, streptavidin plates (Pierce) were coated with 100 μl of a mixture of peptides 1 to 4 from (; 10 μg/ml) or of peptides 5 and 6 (; 10 μg/ml) diluted in PBS and incubated at 4°C overnight. Unbound peptides were removed with 2 washes with PBS and then blocked with 1% milk (KPL) in PBS for 2 hours at room temperature. After removing the block solution, 100 μl of serially diluted sera in 1% Milk diluted in PBS were added to each well and plates incubated for 1 hour at room temperature. Plates were washed 3 times (0.05% Tween 20 in PBS) followed by addition of 100 μl of HRP-conjugated goat anti-mouse diluted 10,000 fold in 1% milk and incubated for 1 hours at room temperature. Plates were then washed 3 times (0.05% Tween 20 in PBS) followed by a water rinse. 100 μl of TMB (warmed to room temperature) were added to each well and plates incubated in the dark for 20 minutes at room temperature. The reaction was stopped by addition of 50 μl of 2N H2SO4. The optical density at 450 nm minus background at 650 nm was obtained using an EMAX plate reader from Molecular Device.
Anti-Lassa peptides
For the anti-Lassa peptides ELISA, streptavidin plates (Pierce) were coated with 100 μl of peptides S1 to S6 respectively () diluted to 10 μg/ml in PBS and incubated at 4°C overnight. Unbound peptides were removed with 2 PBS washes and then blocked with 1% milk (KPL) in PBS for 2 hours at room temperature. After removing the block solution, 100 μl of serially diluted sera in 1% Milk diluted in PBS were added to each well and plates incubated for 1 hour at room temperature. Plates were washed 3 times (0.05% Tween 20 in PBS) followed by addition of 100 μl of HRP-conjugated goat anti-mouse diluted 10,000 fold in 1% milk and incubated for 1 hour at room temperature. Plates were then washed 3 times (0.05% Tween 20 in PBS) followed by a water rinse. 100 μl of TMB (warmed to room temperature) were added to each well and the plates incubated in the dark for 20 minutes at room temperature. The reaction was stopped by addition of 50 μl of 2N H2SO4. The optical density at 450 nm minus background at 650 nm was obtained using an EMAX plate reader from Molecular Device.
Endpoint titers
Endpoint titers were determined as described by Frey et al.Citation76 Briefly, 95% confidence interval cutoff curves were obtained from dilutions of non-immune sera. Endpoint titers were determined by identifying serum dilutions yielding an optical density lower than the cutoff point. Sample for which all serum dilutions yielded optical densities below the 95% confidence interval cutoff curves were recorded as having no titer against the designated target.
Statistics
Statistical analyses, Kruskal-Wallis and Dunn's multiple comparisons test, were performed using GraphPad Prism6.
Disclosure of Potential Conflicts of Interest
Anne S. De Groot and William Martin are the founders and majority shareholders at EpiVax, Inc., a privately owned vaccine design company located in Providence, Rhode Island, USA. Lenny Moise and Christine Boyle are employees of EpiVax. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company. All other authors declare not having potential conflicts of interest.
Acknowledgments
We would like to thank Dr. Satoshi Kashiwagi for reviewing the manuscript.
Funding
This material is based upon work supported by the US. Army Research Office under contract No. W911NF-13–1–0079 and the generous support of Friends of VIC. CL was supported by CAPES Foundation, Ministry of Education of Brazil, Brasilia - DF, 70040–020, Brazil.
References
- Biological Advanced Research and Development Authority. BARDA Strategic Plan 2011-2016. 2011:1-20
- Hayden EC. The price of protection. Nature 2011; 477:150-2; PMID:21900990; http://dx.doi.org/10.1038/477150a
- Roux D, Pier GB, Skurnik D. Magic bullets for the 21st century: the reemergence of immunotherapy for multi- and pan-resistant microbes. J Antimicrob Chemother 2012; 67:2785-7; PMID:22899807; http://dx.doi.org/10.1093/jac/dks335
- Gronvall GK, Rambhia KJ, Adalja A, Cicero A, Inglesby T, Kadlec R. Next-generation monoclonal antibodies: Challenges and opportunities. 2013; Available from: http://www.upmc-biosecurity.org/website/resources/publications/2013/2013-02-04-next-gen-monoclonal-antibodies.html.
- Leblanc PR, Yuan J, Brauns T, Gelfand JA, Poznansky MC. Accelerated vaccine development against emerging infectious diseases. Hum Vaccin Immunother 2012; 8:1010-2; PMID:22777091; http://dx.doi.org/10.4161/hv.20805
- Wang Y, Kelly CG, Karttunen JT, Whittall T, Lehner PJ, Duncan L, MacAry P, Younson JS, Singh M, Oehlmann W, et al. CD40 is a cellular receptor mediating mycobacterial heat shock protein 70 stimulation of CC-chemokines. Immunity 2001; 15:971-83; http://dx.doi.org/10.1016/S1074-7613(01)00242-4
- Floto RA, MacAry PA, Boname JM, Mien TS, Kampmann B, Hair JR, Huey OS, Houben ENG, Pieters J, Day C, et al. Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science [Internet] 2006; 314:454-8. Available from: https://phsexchweb.partners.org/exchange/PLEBLANC/Inbox/FW:%20needed%20PDFs.EML/science2006.pdf/C58EA28C-18C0-4a97-9AF2-036E93DDAFB3/science2006.pdf?attach=1; PMID:17053144; http://dx.doi.org/10.1126/science.1133515
- Vorobiev DS, Kiselevskii MV, Chikileva IO, Semenova IB. Effect of recombinant heat shock protein 70 of mycobacterial origin on cytotoxic activity and immunophenotype of human peripheral blood mononuclear leukocytes. Bull Exp Biol Med 2009; 148:64-7; Bull Exp Biol Med 2009; 148:64-7; PMID:19902099
- Mizukami S, Kajiwara C, Tanaka M, Kaisho T, Udono H. Differential MyD88/IRAK4 requirements for cross-priming and tumor rejection induced by heat shock protein 70-model antigen fusion protein. Cancer Sci 2012; 103:851-9; PMID:22320267; http://dx.doi.org/10.1111/j.1349-7006.2012.02233.x
- Richmond JK, Baglole DJ. Lassa fever: epidemiology, clinical features, and social consequences. BMJ 2003; 327:1271-5; PMID:14644972; http://dx.doi.org/10.1136/bmj.327.7426.1271
- Ölschläger S, Flatz L. Vaccination Strategies against Highly Pathogenic Arenaviruses: The Next Steps toward Clinical Trials. PLoS Pathog 2013; 9:e1003212; http://dx.doi.org/10.1371/journal.ppat.1003212
- Hensley LE, Smith MA, Geisbert JB, Fritz EA, Daddario-DiCaprio KM, Larsen T, Geisbert TW. Pathogenesis of Lassa fever in cynomolgus macaques. Virol J 2011; 8:205; PMID:21548931; http://dx.doi.org/10.1186/1743-422X-8-205
- Jahrling PB. Protection of Lassa virus-infected guinea pigs with Lassa-immune plasma of guinea pig, primate, and human origin. J Med Virol 1983; 12:93-102; http://dx.doi.org/10.1002/jmv.1890120203
- Morrison HG, Bauer SP, Lange JV, Esposito JJ, McCormick JB, Auperin DD. Protection of guinea pigs from Lassa fever by vaccinia virus recombinants expressing the nucleoprotein or the envelope glycoproteins of Lassa virus. Virology 1989; 171:179-88; http://dx.doi.org/10.1016/0042-6822(89)90525-4
- Fisher-Hoch SP, Hutwagner L, Brown B, McCormick JB. Effective vaccine for lassa fever. J Virol 2000; 74:6777-83; http://dx.doi.org/10.1128/JVI.74.15.6777-6783.2000
- Botten J, Alexander J, Pasquetto V, Sidney J, Barrowman P, Ting J, Peters B, Southwood S, Stewart B, Rodriguez-Carreno MP, et al. Identification of protective lassa virus epitopes that are restricted by HLA-A2. J Virol 2006; 80:8351-61; PMID:16912286; http://dx.doi.org/10.1128/JVI.00896-06
- Clegg JC, Lloyd G. Structural and cell-associated proteins of Lassa virus. J Gen Virol 1983; 64:1127-36; http://dx.doi.org/10.1099/0022-1317-64-5-1127
- Bishop DH, Auperin DD. Arenavirus gene structure and organization. Curr Top Microbiol Immunol 1987; 133:5-17; PMID:2435460
- Botten J, Whitton JL, Barrowman P, Sidney J, Whitmire JK, Alexander J, Kotturi MF, Sette A, Buchmeier MJ. A multivalent vaccination strategy for the prevention of Old World arenavirus infection in humans. J Virol 2010; 84:9947-56; PMID:20668086; http://dx.doi.org/10.1128/JVI.00672-10
- Cashman, K. A., Broderick, K. E., Wilkinson, E. R., Shaia, C. I., Bell, T. M., Shurtleff, A. C., et al. Enhanced Efficacy of a Codon-Optimized DNA Vaccine Encoding the Glycoprotein Precursor Gene of Lassa Virus in a Guinea Pig Disease Model When Delivered by Dermal Electroporation. Vaccines 2013; 1(3):262-277; http://dx.doi.org/10.3390/vaccines1030262
- Rötzschke O, Falk K, Stevanović S, Jung G, Walden P, Rammensee HG. Exact prediction of a natural T cell epitope. Eur J Immunol 1991; 21:2891-4; PMID:1718764; http://dx.doi.org/10.1002/eji.1830211136
- Lipford GB, Hoffman M, Wagner H, Heeg K. Primary in vivo responses to ovalbumin. Probing the predictive value of the Kb binding motif. J Immunol 1993; 150:1212-22; PMID:7679422
- Moise L, Tassone R, Latimer H, Terry F, Levitz L, Haran JP, Ross TM, Boyle CM, Martin WD, De Groot AS. Immunization with cross-conserved H1N1 influenza CD4+ T-cell epitopes lowers viral burden in HLA DR3 transgenic mice. Hum Vaccin Immunother 2013; 9:2060-8; PMID:24045788; http://dx.doi.org/10.4161/hv.26511
- Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine 2009; 27:6471-9; PMID:19559119; http://dx.doi.org/10.1016/j.vaccine.2009.06.018
- Spiering R, van der Zee R, Wagenaar J, Van Eden W, Broere F. Mycobacterial and mouse HSP70 have immuno-modulatory effects on dendritic cells. Cell Stress Chaperones 2013; 18:439-46; PMID:23269491; http://dx.doi.org/10.1007/s12192-012-0397-4
- Borges TJ, Wieten L, van Herwijnen MJC, Broere F, van der Zee R, Bonorino C, van Eden W. The anti-inflammatory mechanisms of Hsp70. Front Immunol 2012; 3:95; PMID:22566973; http://dx.doi.org/10.3389/fimmu.2012.00095
- Arribillaga L, Durantez M, Lozano T, Rudilla F, Rehberger F, Casares N, Villanueva L, Martinez M, Gorraiz M, Borrás-Cuesta F, et al. A fusion protein between streptavidin and the endogenous TLR4 Ligand EDA targets biotinylated antigens to dendritic cells and induces T cell responses in vivo. Biomed Res Int 2013; 2013:864720; PMID:24093105; http://dx.doi.org/10.1155/2013/864720
- Carbone FR, Bevan MJ. Induction of ovalbumin-specific cytotoxic T cells by in vivo peptide immunization. J Exp Med 1989; 169:603-12; http://dx.doi.org/10.1084/jem.169.3.603
- Huang Q, Richmond JFL, Suzue K, Eisen H, Young R. In vivo cytotoxic T lymphocyte elicitation by mycobacterial heat shock protein 70 fusion proteins maps to a discrete domain and is CD4+ T cell independent. J Exp Med 2000; 191:403-8; http://dx.doi.org/10.1084/jem.191.2.403
- Suzue K, Zhou X, Eisen HN, Young RA. Heat shock fusion proteins as vehicles for antigen delivery into the major histocompatibility complex class I presentation pathway. Proc Natl Acad Sci USA 1997; 94:13146-51; http://dx.doi.org/10.1073/pnas.94.24.13146
- Del Val M, Iborra S, Ramos M, Lázaro S. Generation of MHC class I ligands in the secretory and vesicular pathways. Cell Mol Life Sci 2011; 68:1543-52; PMID:21387141; http://dx.doi.org/10.1007/s00018-011-0661-2
- Gil-Torregrosa BC, Raúl Castaño A, Del Val M. Major histocompatibility complex class I viral antigen processing in the secretory pathway defined by the trans-Golgi network protease furin. J Exp Med 1998; 188:1105-16; http://dx.doi.org/10.1084/jem.188.6.1105
- Gil-Torregrosa BC, Castaño AR, López D, Del Val M. Generation of MHC class I peptide antigens by protein processing in the secretory route by furin. Traffic 2000; 1:641-51; http://dx.doi.org/10.1034/j.1600-0854.2000.010808.x
- Medina F, Ramos M, Iborra S, de León P, Rodríguez-Castro M, Del Val M. Furin-processed antigens targeted to the secretory route elicit functional TAP1-/-CD8+ T lymphocytes in vivo. J Immunol 2009; 183:4639-47; http://dx.doi.org/10.4049/jimmunol.0901356
- Lu J, Higashimoto Y, Appella E, Celis E. Multiepitope Trojan antigen peptide vaccines for the induction of antitumor CTL and Th immune responses. J Immunol 2004; 172:4575-82; http://dx.doi.org/10.4049/jimmunol.172.7.4575
- Haug M, Dannecker L, Schepp CP, Kwok WW, Wernet D, Buckner JH, Kalbacher H, Dannecker GE, Holzer U. The heat shock protein Hsp70 enhances antigen-specific proliferation of human CD4+ memory T cells. Eur J Immunol 2005; 35:3163-72; PMID:16245362; http://dx.doi.org/10.1002/eji.2005-35050
- Haug M, Schepp CP, Kalbacher H, Dannecker GE, Holzer U. 70-kDa heat shock proteins: specific interactions with HLA-DR molecules and their peptide fragments. Eur J Immunol 2007; 37:1053-63; PMID:17357109; http://dx.doi.org/10.1002/eji.200-636811
- Fischer N, Haug M, Kwok WW, Kalbacher H, Wernet D, Dannecker GE, Holzer U. Involvement of CD91 and scavenger receptors in Hsp70-facilitated activation of human antigen-specific CD4+ memory T cells. Eur J Immunol 2010; 40:986-97; PMID:20101615; http://dx.doi.org/10.1002/eji.200939738
- Soloway P, Fish S, Passmore H, Gefter M, Coffee R, Manser T. Regulation of the immune response to peptide antigens: differential induction of immediate-type hypersensitivity and T cell proliferation due to changes in either peptide structure or major histocompatibility complex haplotype. J Exp Med 1991; 174:847-58; http://dx.doi.org/10.1084/jem.174.4.847
- La Rosa C, Wang Z, Brewer JC, Lacey SF, Villacres MC, Sharan R, Krishnan R, Crooks M, Markel S, Maas R, et al. Preclinical development of an adjuvant-free peptide vaccine with activity against CMV pp65 in HLA transgenic mice. Blood 2002; 100:3681-9; PMID:12393676; http://dx.doi.org/10.1182/blood-2002-03-0926
- La Rosa C, Longmate J, Lacey SF, Kaltcheva T, Sharan R, Marsano D, Kwon P, Drake J, Williams B, Denison S, et al. Clinical evaluation of safety and immunogenicity of PADRE-cytomegalovirus (CMV) and tetanus-CMV fusion peptide vaccines with or without PF03512676 adjuvant. J Infect Dis 2012; 205:1294-304; PMID:22402037; http://dx.doi.org/10.1093/infdis/jis107
- Clegg JC, Lloyd G. Vaccinia recombinant expressing Lassa-virus internal nucleocapsid protein protects guineapigs against Lassa fever. Lancet 1987; 2:186-8; http://dx.doi.org/10.1016/S0140-6736(87)90767-7
- Auperin DD, Esposito JJ, Lange JV, Bauer SP, Knight J, Sasso DR, McCormick JB. Construction of a recombinant vaccinia virus expressing the Lassa virus glycoprotein gene and protection of guinea pigs from a lethal Lassa virus infection. Virus Res 1988; 9:233-48; http://dx.doi.org/10.1016/0168-1702(88)90033-0
- Morrison HG, Goldsmith CS, Regnery HL, Auperin DD. Simultaneous expression of the Lassa virus N and GPC genes from a single recombinant vaccinia virus. Virus Res 1991; 18:231-41; http://dx.doi.org/10.1016/0168-1702(91)90021-M
- Pushko P, Geisbert J, Parker M, Jahrling P, Smith J. Individual and bivalent vaccines based on alphavirus replicons protect guinea pigs against infection with Lassa and Ebola viruses. J Virol 2001; 75:11677-85; PMID:11689649; http://dx.doi.org/10.1128/JVI.75.23.11677-11685.2001
- Geisbert TW, Jones S, Fritz EA, Shurtleff AC, Geisbert JB, Liebscher R, Grolla A, Ströher U, Fernando L, Daddario KM, et al. Development of a new vaccine for the prevention of Lassa fever. PLoS Med 2005; 2:e183; PMID:15971954
- Lukashevich IS, Patterson J, Carrion R, Moshkoff D, Ticer A, Zapata J, Brasky K, Geiger R, Hubbard GB, Bryant J, et al. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol 2005; 79:13934-42; PMID:16254329; http://dx.doi.org/10.1128/JVI.79.22.13934-13942.2005
- Rodriguez-Carreno MP, Nelson MS, Botten J, Smith-Nixon K, Buchmeier MJ, Whitton JL. Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology 2005; 335:87-98; PMID:15823608; http://dx.doi.org/10.1016/j.virol.2005.01.019
- Bredenbeek PJ, Molenkamp R, Spaan WJM, Deubel V, Marianneau P, Salvato MS, Moshkoff D, Zapata J, Tikhonov I, Patterson J, et al. A recombinant yellow fever 17D vaccine expressing Lassa virus glycoproteins. Virology 2006; 345:299-304; PMID:16412488; http://dx.doi.org/10.1016/j.virol.2005.12.001
- Carrion R, Patterson JL, Johnson C, Gonzales M, Moreira CR, Ticer A, Brasky K, Hubbard GB, Moshkoff D, Zapata J, et al. A ML29 reassortant virus protects guinea pigs against a distantly related Nigerian strain of Lassa virus and can provide sterilizing immunity. Vaccine 2007; 25:4093-102; PMID:17360080; http://dx.doi.org/10.1016/j.vaccine.2007.02.038
- Lukashevich IS, Carrion R, Salvato MS, Mansfield K, Brasky K, Zapata J, Cairo C, Goicochea M, Hoosien GE, Ticer A, et al. Safety, immunogenicity, and efficacy of the ML29 reassortant vaccine for Lassa fever in small non-human primates. Vaccine 2008; 26:5246-54; PMID:18692539; http://dx.doi.org/10.1016/j.vaccine.2008.07.057
- Branco LM, Grove JN, Geske FJ, Boisen ML, Muncy IJ, Magliato SA, Henderson LA, Schoepp RJ, Cashman KA, Hensley LE, et al. Lassa virus-like particles displaying all major immunological determinants as a vaccine candidate for Lassa hemorrhagic fever. Virol J 2010; 7:279; PMID:20961433; http://dx.doi.org/10.1186/1743-422X-7-279
- Jiang X, Dalebout TJ, Bredenbeek PJ, Carrion R, Brasky K, Patterson J, Goicochea M, Bryant J, Salvato MS, Lukashevich IS. Yellow fever 17D-vectored vaccines expressing Lassa virus GP1 and GP2 glycoproteins provide protection against fatal disease in guinea pigs. Vaccine 2011; 29:1248-57; PMID:21145373; http://dx.doi.org/10.1016/j.vaccine.2010.11.079
- Grant-Klein RJ, Altamura LA, Schmaljohn CS. Progress in recombinant DNA-derived vaccines for Lassa virus and filoviruses. Virus Res 2011; 162:148-61; PMID:21925552; http://dx.doi.org/10.1016/j.virusres.2011.09.005
- Goicochea MA, Zapata JC, Bryant J, Davis H, Salvato MS, Lukashevich IS. Evaluation of Lassa virus vaccine immunogenicity in a CBA/J-ML29 mouse model. Vaccine 2012; 30:1445-52; PMID:22234266; http://dx.doi.org/10.1016/j.vaccine.2011.12.134
- Zapata JC, Poonia B, Bryant J, Davis H, Ateh E, George L, Crasta O, Zhang Y, Slezak T, Jaing C, et al. An attenuated Lassa vaccine in SIV-infected rhesus macaques does not persist or cause arenavirus disease but does elicit Lassa virus-specific immunity. Virol J 2013; 10:52; PMID:23402317; http://dx.doi.org/10.1186/1743-422X-10-52
- Meulen ter J. Lassa fever: implications of T-cell immunity for vaccine development. J Biotechnol 1999; 73:207-12; http://dx.doi.org/10.1016/S0168-1656(99)00122-4
- Meulen ter J, Badusche M, Kuhnt K, Doetze A, Satoguina J, Marti T, Loeliger C, Koulemou K, Koivogui L, Schmitz H, et al. Characterization of human CD4(+) T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J Virol 2000; 74:2186-92; http://dx.doi.org/10.1128/JVI.74.5.2186-2192.2000
- Oldstone MB, Lewicki H, Homann D, Nguyen C, Julien S, Gairin JE. Common antiviral cytotoxic t-lymphocyte epitope for diverse arenaviruses. J Virol 2001; 75:6273-8; PMID:11413293; http://dx.doi.org/10.1128/JVI.75.14.6273-6278.2001
- Meulen ter J, Sakho M, Koulemou K, Magassouba N, Bah A, Preiser W, Daffis S, Klewitz C, Bae H-G, Niedrig M, et al. Activation of the cytokine network and unfavorable outcome in patients with yellow fever. J Infect Dis 2004; 190:1821-7; PMID:15499539; http://dx.doi.org/10.1086/425016
- Boesen A, Sundar K, Coico R. Lassa fever virus peptides predicted by computational analysis induce epitope-specific cytotoxic-T-lymphocyte responses in HLA-A2.1 transgenic mice. Clin Diagn Lab Immunol 2005; 12:1223-30
- Kotturi MF, Botten J, Sidney J, Bui H-H, Giancola L, Maybeno M, Babin J, Oseroff C, Pasquetto V, Greenbaum JA, et al. A multivalent and cross-protective vaccine strategy against arenaviruses associated with human disease. PLoS Pathog 2009; 5:e1000695
- Kotturi MF, Botten J, Maybeno M, Sidney J, Glenn J, Bui H-H, Oseroff C, Crotty S, Peters B, Grey H, et al. Polyfunctional CD4+ T cell responses to a set of pathogenic arenaviruses provide broad population coverage. Immunome Res 2010; 6:4; PMID:20478058; http://dx.doi.org/10.1186/1745-7580-6-4
- Kotturi MF, Peters B, Buendia-Laysa F, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol 2007; 81:4928-40; PMID:17329346; http://dx.doi.org/10.1128/JVI.02632-06
- Riviere Y, Oldstone MB. Genetic reassortants of lymphocytic choriomeningitis virus: unexpected disease and mechanism of pathogenesis. J Virol 1986; 59:363-8
- Djavani M, Lukashevich IS, Salvato MS. Sequence comparison of the large genomic RNA segments of two strains of lymphocytic choriomeningitis virus differing in pathogenic potential for guinea pigs. Virus Genes 1998; 17:151-5; http://dx.doi.org/10.1023/A:1008016724243
- Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Oldstone MB. Genetic mapping of lymphocytic choriomeningitis virus pathogenicity: virulence in guinea pigs is associated with the L RNA segment. J Virol 1985; 55:704-9
- Lukashevich IS, Djavani M, Shapiro K, Sanchez A, Ravkov E, Nichol ST, Salvato MS. The Lassa fever virus L gene: nucleotide sequence, comparison, and precipitation of a predicted 250 kDa protein with monospecific antiserum. J Gen Virol 1997; 78( Pt 3):547-51
- Lelke M, Brunotte L, Busch C, Günther S. An N-terminal region of Lassa virus L protein plays a critical role in transcription but not replication of the virus genome. J Virol 2010; 84:1934-44; PMID:20007273; http://dx.doi.org/10.1128/JVI.01657-09
- Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, Erickson BR, Nichol ST. Efficient rescue of recombinant Lassa virus reveals the influence of S segment noncoding regions on virus replication and virulence. J Virol 2011; 85:4020-4; PMID:21307206; http://dx.doi.org/10.1128/JVI.02556-10
- De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol 2009; 131:189-201; PMID:19269256; http://dx.doi.org/10.1016/j.clim.2009.01.009
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 1998; 160:3363-73
- Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 1999; 50:201-12; http://dx.doi.org/10.1007/s002510050594
- De Groot AS. Immunomics: discovering new targets for vaccines and therapeutics. Drug Discov Today 2006; 11:203-9; http://dx.doi.org/10.1016/S1359-6446(05)03720-7
- Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Hady KMA, VerBerkmoes NC, Sztein MB, Losikoff PT, Martin WD, et al. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother 2013; 9:1577-86; PMID:23584251; http://dx.doi.org/10.4161/hv.24615
- Frey A, Di Canzio J, Zurakowski D. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods 1998; 221:35-41; http://dx.doi.org/10.1016/S0022-1759(98)00170-7
- Lindow JC, Borochoff-Porte N, Durbin AP, Whitehead SS, Fimlaid KA, Bunn JY, Kirkpatrick BD. Primary vaccination with low dose live dengue 1 virus generates a proinflammatory, multifunctional T cell response in humans. PLoS Negl Trop Dis 2012; 6:e1742; PMID:22816004; http://dx.doi.org/10.1371/journal.pntd.0001742
- Sugauchi F, Wang RYH, Qiu Q, Jin B, Alter HJ, Shih JWK. Vigorous hepatitis C virus-specific CD4+ and CD8+ T cell responses induced by protein immunization in the presence of Montanide ISA720 plus synthetic oligodeoxynucleotides containing immunostimulatory cytosine-guanine dinucleotide motifs. J Infect Dis 2006; 193:563-72; PMID:16425136; http://dx.doi.org/10.1086/499823
- Chege GK, Burgers WA, Stutz H, Meyers AE, Chapman R, Kiravu A, Bunjun R, Shephard EG, Jacobs WR, Rybicki EP, et al. Robust immunity to an auxotrophic Mycobacterium bovis BCG-VLP prime-boost HIV vaccine candidate in a nonhuman primate model. J Virol 2013; 87:5151-60; PMID:23449790; http://dx.doi.org/10.1128/JVI.03178-12
- Meseda CA, Mayer AE, Kumar A, Garcia AD, Campbell J, Listrani P, Manischewitz J, King LR, Golding H, Merchlinsky M, et al. Comparative evaluation of the immune responses and protection engendered by LC16m8 and Dryvax smallpox vaccines in a mouse model. Clin Vaccine Immunol 2009; 16:1261-71; PMID:19605597; http://dx.doi.org/10.1128/CVI.00040-09
- James EA, LaFond RE, Gates TJ, Mai DT, Malhotra U, Kwok WW. Yellow fever vaccination elicits broad functional CD4+ T cell responses that recognize structural and nonstructural proteins. J Virol 2013; 87:12794-804; PMID:24049183; http://dx.doi.org/10.1128/JVI.01160-13