Abstract
Studies of yeast and mammalian prions introduced the idea that the protein aggregates can exist in multiple stable conformations that can be propagated by seeding. These conformational states (aka strains) were shown to have distinct physical (secondary structure, stability) and biological (cytotoxicity, infectivity) properties. For mammalian prions they were also tied to differences in disease pathology and incubation time. It was later shown that this phenomenon is not limited to prion proteins, and distinct conformational states of amyloid fibrils and oligomers derived from a variety of proteins can be propagated both in vitro and in vivo. Moreover, in some cases these conformations were preserved even when propagated into a protein with a different sequence. There is now an increasing body of evidence that strain phenomenon is a generic feature of protein aggregation, and characteristic features of amyloid strains can be transmitted between unrelated sequences.
Keywords: :
For decades our understanding of protein structure has been driven by Anfinsen’s dogma: a protein possesses a single native structure that is determined only by its amino acid sequence.Citation1 Discovery that proteins can be infectious,Citation2-Citation4 and that this infectivity is due to an autocatalytic conversion of a protein into a different, misfolded conformation,Citation5,Citation6 represented a significant milestone in biology. Together with the concept of intrinsic disorderCitation7,Citation8 this discovery showed that protein conformation can be plastic and malleable.
Initially autocatalytic conversion of proteins into misfolded, β-sheet rich conformation was proposed in order to explain the unusual properties of transmissible spongiform encephalophathies (TSEs) where prion protein (PrP) serves as an infectious agent.Citation2-Citation4 These diseases are caused by misfolding and aggregation of prion protein (PrPC) into fibrillar aggregates (PrPSc).Citation9 PrPSc fibrils can be transmitted between individuals and propagate themselves by seeding. Seeding process involves binding of a PrP monomer to a PrPSc fibril followed by conversion of this monomer to a conformation closely resembling that of an initial aggregate.Citation10 This mechanism of propagation is common for all amyloids. However, not all of them are infectious in vivo. Other factors such as the ability of protein aggregates to fragment and self-propagate in physiological conditions and their resistance to clearance are necessary to make the amyloid infectious.Citation11,Citation12
An important feature of TSEs is the presence of multiple disease strains. Strains have been initially defined as the isolates of infectious prions characterized by the specific incubation time of the disease, characteristic neuropathological lesions, and in some cases certain patterns in animal behavior that are faithfully recapitulated upon serial passage.Citation13,Citation14 For example, Hyper and Drowsy TSE strains lead to, respectively, increased and decreased activity of infected animals.Citation15 It has been shown that different disease strains correspond to distinct conformations of PrP fibrils as they can be differentiated by the protease cleavage pattern,Citation16 antibody reactivity,Citation17-Citation19 resistance to denaturation,Citation20 and FTIR spectra.Citation21 Stability of PrP fibrils was shown to be inversely proportional to the incubation time for the prion disease in vivo,Citation20 and replication rates of prion strains were shown to be inversely proportional to their conformational stability.Citation22,Citation23 Later the strain phenomenon was discovered for other amyloidogenic proteins, and the inverse relationship between rate of propagation and stability was found to hold for other proteins as well.Citation24,Citation25
Conformational polymorphism of amyloid fibrils is based on their common structure. The atomic architecture of most amyloid fibrils consists of 2 fundamental units: 1) a cross-β spine composed of in-register, parallel, intermolecularly stacked β-sheets running perpendicular to the fibril axis,Citation26-Citation28 and 2) a motif formed between tightly interdigitated side chains interface between β-sheets known as steric zipper.Citation26 Structural polymorphisms such as the variations in the folding patterns of the β-sheets,Citation28,Citation29 length of the amyloid core,Citation25 and interface between the β-sheetsCitation10 can be propagated from the initial fibrils onto the daughter fibrils by seeding. Since the aggregates are stabilized by intermolecular interactions of β-sheets, even relatively small differences in protein conformation between the aggregates can give rise to distinct protein conformations can become kinetically trapped in local minima on the potential energy surface.
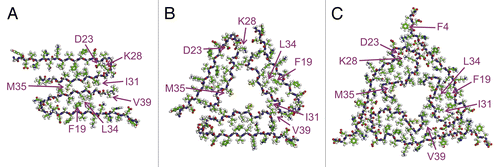
Aggregation of the Aβ40 peptide can serve as a good example of structural polymorphism of amyloids. Incubation of this peptide in buffer with shaking results in formation of fibrils consisting of 2 stacked cross-β strands with parallel in-register alignment ().Citation30 Fibrils formed by the same peptide in quiescent conditions have twisted morphology and consist of 3 cross-β strands with the same parallel alignment ().Citation28 Aβ40 fibrils isolated from the Alzheimer disease patients are also composed of 3 cross-β strands, but there are subtle structural differences between them and the fibrils prepared in vitro.Citation29 For example, fibrils isolated from an AD patient have several kinks and bends in their structure ()Citation29 that are missing in the fibrils prepared in vitro (). Moreover, similar subtle differences were observed between the fibrils isolated from different patients.Citation29 All of these fibril strains can be propagated by seeding preserving their unique structural features.
Figure 1. Molecular structures of Aβ40 fibrils. (A) Aβ40 fibrils prepared with shaking. (B) Aβ40 fibrils prepared in quiescent conditions. (C) Aβ40 fibrils isolated from the AD patient. Adapted with permission from ref. Citation29
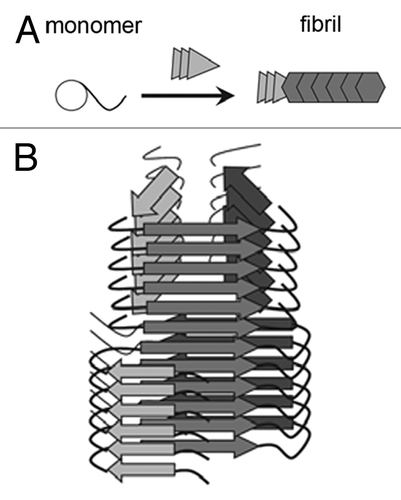
When stable strains of amyloid fibrils are propagated in the conditions different from those used in their preparation (e.g., transition from in vitro to in vivo or a change in pH), they may adapt to the altered environment by changing their structure. This phenomenon (dubbed “deformed templating”) has been especially well described for mammalian prions (),Citation31 although it has been observed for other proteins as well.Citation32 When new mammalian prion strains were created by infecting the animals with the amyloid fibrils prepared from recombinant PrP, animals often failed to show clinical signs of disease and if they did, the incubation time was very long.Citation5,Citation33 However, after multiple serial passages the incubation time shortened and the clinical signs of disease appeared.Citation33,Citation34 This process was accompanied by the decrease in conformational stability of the amyloidCitation5,Citation20 and by the dramatic change in the protein folding pattern within the fibrils.Citation35 Since a protein has to unfold before incorporation into an amyloid fibril,Citation36,Citation37 it may refold into a slightly different conformation upon incorporation into a fibril if a new conformation is more thermodynamically stable and kinetically accessible.
Figure 2. Deformed templating is a change in the β-sheet folding pattern of amyloid fibrils in the course of seeded aggregation. (A) Schematic representation of the mechanism. (B) Representation of deformed templating within an individual fibril. Adapted with permission from ref. Citation31
Similar sequences containing complementary in shape residues may form mixed steric zippers. These sequences may be derived from different parts of the same protein or from different proteins. Co-aggregation of different proteins into mixed sequence steric zippers can lead to cross-seeding of these proteins when the fibrils of one protein initiate the aggregation of another. For example, in case of curli proteins involved in bacterial biofilm formation an outer membrane protein CsgA serves as a fibril seed while the biofilm-forming fibers themselves are primarily composed from a related protein CsgB. Both CsgA and CsgB subunits can seed aggregation of CsgA proteins from other bacteria despite significant sequence differences.Citation38 Moreover, curli proteins can promote aggregation of other, unrelated proteins such as fragments of prostatic acid phosphatase.Citation39 Cross-seeding has been observed for a variety of proteins both in vitro and in vivo. In vivo cross-seeding has been observed for tau and α-synuclein,Citation40,Citation41 amyloid β and prion protein,Citation42 and for serum amyloid A and lactadhedrin.Citation43 However, the ability to cross-seed aggregation depends on both the sequences of the proteins involved and the structure of the initial aggregate. High sequence similarity is usually required for effective cross-seedingCitation44,Citation45 although there are exceptions (e.g., α-synuclein and tau or Aβ40).Citation40,Citation44,Citation46,Citation47 In some cases (e.g., Aβ40 and Aβ42) cross-seeding is unidirectional with Aβ40 fibrils able to seed the aggregation of Aβ42 but not the other way around.Citation48 Amyloid oligomers are also capable of cross-seeding and may even be more efficient at it than fibrils. For example, Aβ oligomers could seed oligomer formation from tauCitation49 and α-synuclein.Citation50
Protein conformation also plays an important role in the ability of fibrils to cross-seed their aggregation. For example, Guo et al.Citation40 have recently shown that ability of α-synuclein fibrils to seed tau aggregation in vivo depends on their conformation. While newly formed α-synuclein fibrils could seed the aggregation of endogenous α-synuclein, they could not cross-seed tau aggregation. However, when α-synuclein fibrils were propagated in vitro for several generations, their conformation was altered as indicated by their proteinase K digestion pattern and they gained the ability to seed tau aggregation.
Can the strain identity be preserved upon cross-seeding to a different sequence? Initial evidence of propagation of different fibril strains across multiple sequences came from mammalian prions. The phenomenon of “species barrier” in prion propagation occurs when a prion strain is seeded into a different host species.Citation51,Citation52 Sequence differences between prion proteins from different species are not particularly large but key amino acids in the regions important for fibril formation are often affected. Some prion strains are unable to cross the species barrier and seed aggregation of proteins with different sequences while others do it efficiently.Citation51 vCJD strains responsible for “mad cow disease” are good examples of the latter.Citation53 In some cases seeding across the species barrier will still trigger the disease but strain-specific features will be lost and the disease strain more typical for the host species will appear instead.Citation32,Citation52,Citation54 It has been proposed that the protein sequences that can adopt a wider range of conformations are capable of supporting a wider variety of prion strains.Citation55 Similar phenomena are observed for other amyloidogenic proteins. In some cases yeast prions can seed formation of each other while preserving strain identity.Citation56,Citation57 But for sequences with lower degree of similarity loss of strain identity is a more typical outcome.Citation58 The outcome can also be strain-dependent as was shown for hamster PrP fibrilsCitation59 where one strain preserved its conformation after cross-seeding into mouse PrP while another lost it.
Given the presence of multiple aggregation-prone proteins in human proteome, the possibility of their cross-seeding or co-aggregation is very important. In general, cross-seeding by amyloid fibrils is subject to several conditions that need to be met simultaneouslyCitation44 and thus relatively rare. However, many disease-associated amyloidogenic proteins (tau, amyloid β, α-synuclein, IAPP) are capable of cross-seeding. Since these proteins tend to be foldable IDPs, disorder may allow for higher structural flexibility necessary to accommodate diverse conformations required for effective cross-seeding.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science 1973; 181:223 - 30; http://dx.doi.org/10.1126/science.181.4096.223; PMID: 4124164
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136 - 44; http://dx.doi.org/10.1126/science.6801762; PMID: 6801762
- Griffith JS. Self-replication and scrapie. Nature 1967; 215:1043 - 4; http://dx.doi.org/10.1038/2151043a0; PMID: 4964084
- Alper T, Cramp WA, Haig DA, Clarke MC. Does the agent of scrapie replicate without nucleic acid?. Nature 1967; 214:764 - 6; http://dx.doi.org/10.1038/214764a0; PMID: 4963878
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science 2004; 305:673 - 6; http://dx.doi.org/10.1126/science.1100195; PMID: 15286374
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132 - 5; http://dx.doi.org/10.1126/science.1183748; PMID: 20110469
- Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta 2013; 1834:932 - 51; http://dx.doi.org/10.1016/j.bbapap.2012.12.008; PMID: 23269364
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta 2010; 1804:1231 - 64; http://dx.doi.org/10.1016/j.bbapap.2010.01.017; PMID: 20117254
- Prusiner SB. The prion diseases. Brain Pathol 1998; 8:499 - 513; http://dx.doi.org/10.1111/j.1750-3639.1998.tb00171.x; PMID: 9669700
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell 2012; 148:1188 - 203; http://dx.doi.org/10.1016/j.cell.2012.02.022; PMID: 22424229
- Soto C. Transmissible proteins: expanding the prion heresy. Cell 2012; 149:968 - 77; http://dx.doi.org/10.1016/j.cell.2012.05.007; PMID: 22632966
- Fernández-Borges N, Eraña H, Elezgarai SR, Harrathi C, Gayosso M, Castilla J. Infectivity versus Seeding in Neurodegenerative Diseases Sharing a Prion-Like Mechanism. Int J Cell Biol 2013; 2013:583498; http://dx.doi.org/10.1155/2013/583498; PMID: 24187553
- Bruce ME. Scrapie strain variation and mutation. Br Med Bull 1993; 49:822 - 38; PMID: 8137131
- Kretzschmar H, Tatzelt J. Prion disease: a tale of folds and strains. Brain Pathol 2013; 23:321 - 32; http://dx.doi.org/10.1111/bpa.12045; PMID: 23587138
- Mulcahy ER, Bessen RA. Strain-specific kinetics of prion protein formation in vitro and in vivo. J Biol Chem 2004; 279:1643 - 9; http://dx.doi.org/10.1074/jbc.M307844200; PMID: 14573620
- Kuczius T, Groschup MH. Differences in proteinase K resistance and neuronal deposition of abnormal prion proteins characterize bovine spongiform encephalopathy (BSE) and scrapie strains. Mol Med 1999; 5:406 - 18; PMID: 10415165
- Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci 2001; 10:854 - 63; http://dx.doi.org/10.1110/ps.39201; PMID: 11274476
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157 - 65; http://dx.doi.org/10.1038/2654; PMID: 9771749
- Moroncini G, Mangieri M, Morbin M, Mazzoleni G, Ghetti B, Gabrielli A, Williamson RA, Giaccone G, Tagliavini F. Pathologic prion protein is specifically recognized in situ by a novel PrP conformational antibody. Neurobiol Dis 2006; 23:717 - 24; http://dx.doi.org/10.1016/j.nbd.2006.06.008; PMID: 16876426
- Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci U S A 2009; 106:20417 - 22; http://dx.doi.org/10.1073/pnas.0910350106; PMID: 19915150
- Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem 1998; 273:32230 - 5; http://dx.doi.org/10.1074/jbc.273.48.32230; PMID: 9822701
- Haldiman T, Kim C, Cohen Y, Chen W, Blevins J, Qing L, Cohen ML, Langeveld J, Telling GC, Kong Q, et al. Co-existence of distinct prion types enables conformational evolution of human PrPSc by competitive selection. J Biol Chem 2013; 288:29846 - 61; http://dx.doi.org/10.1074/jbc.M113.500108; PMID: 23974118
- Laferrière F, Tixador P, Moudjou M, Chapuis J, Sibille P, Herzog L, Reine F, Jaumain E, Laude H, Rezaei H, et al. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog 2013; 9:e1003702; http://dx.doi.org/10.1371/journal.ppat.1003702; PMID: 24130496
- Kalastavadi T, True HL. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. J Biol Chem 2010; 285:20748 - 55; http://dx.doi.org/10.1074/jbc.M110.115303; PMID: 20442412
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature 2006; 442:585 - 9; http://dx.doi.org/10.1038/nature04922; PMID: 16810177
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007; 447:453 - 7; http://dx.doi.org/10.1038/nature05695; PMID: 17468747
- Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys 2008; 41:265 - 97; http://dx.doi.org/10.1017/S0033583508004733; PMID: 19079806
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A 2008; 105:18349 - 54; http://dx.doi.org/10.1073/pnas.0806270105; PMID: 19015532
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 2013; 154:1257 - 68; http://dx.doi.org/10.1016/j.cell.2013.08.035; PMID: 24034249
- Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry 2006; 45:498 - 512; http://dx.doi.org/10.1021/bi051952q; PMID: 16401079
- Makarava N, Baskakov IV. Genesis of tramsmissible protein states via deformed templating. Prion 2012; 6:252 - 5; http://dx.doi.org/10.4161/pri.19930; PMID: 22561163
- Wickner RB, Edskes HK, Shewmaker F, Kryndushkin D, Nemecek J. Prion variants, species barriers, generation and propagation. J Biol 2009; 8:47; http://dx.doi.org/10.1186/jbiol148; PMID: 19519931
- Makarava N, Kovacs GG, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Stabilization of a prion strain of synthetic origin requires multiple serial passages. J Biol Chem 2012; 287:30205 - 14; http://dx.doi.org/10.1074/jbc.M112.392985; PMID: 22807452
- Ghaemmaghami S, Watts JC, Nguyen HO, Hayashi S, DeArmond SJ, Prusiner SB. Conformational transformation and selection of synthetic prion strains. J Mol Biol 2011; 413:527 - 42; http://dx.doi.org/10.1016/j.jmb.2011.07.021; PMID: 21839745
- Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci U S A 2009; 106:16990 - 5; http://dx.doi.org/10.1073/pnas.0909006106; PMID: 19805070
- Uversky VN. Hypothesis: The unfolding power of protein dielectricity. Intrinsically Disord Proteins 2013; 1:e25725; http://dx.doi.org/10.4161/idp.25725
- Ma B, Nussinov R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J Mol Biol 2012; 421:172 - 84; http://dx.doi.org/10.1016/j.jmb.2011.11.023; PMID: 22119878
- Zhou Y, Smith D, Leong BJ, Brännström K, Almqvist F, Chapman MR. Promiscuous cross-seeding between bacterial amyloids promotes interspecies biofilms. J Biol Chem 2012; 287:35092 - 103; http://dx.doi.org/10.1074/jbc.M112.383737; PMID: 22891247
- Hartman K, Brender JR, Monde K, Ono A, Evans ML, Popovych N, Chapman MR, Ramamoorthy A. Bacterial curli protein promotes the conversion of PAP248-286 into the amyloid SEVI: cross-seeding of dissimilar amyloid sequences. PeerJ 2013; 1:e5; http://dx.doi.org/10.7717/peerj.5; PMID: 23638387
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, et al. Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 2013; 154:103 - 17; http://dx.doi.org/10.1016/j.cell.2013.05.057; PMID: 23827677
- Waxman EA, Giasson BI. Induction of intracellular tau aggregation is promoted by α-synuclein seeds and provides novel insights into the hyperphosphorylation of tau. J Neurosci 2011; 31:7604 - 18; http://dx.doi.org/10.1523/JNEUROSCI.0297-11.2011; PMID: 21613474
- Morales R, Estrada LD, Diaz-Espinoza R, Morales-Scheihing D, Jara MC, Castilla J, Soto C. Molecular cross talk between misfolded proteins in animal models of Alzheimer’s and prion diseases. J Neurosci 2010; 30:4528 - 35; http://dx.doi.org/10.1523/JNEUROSCI.5924-09.2010; PMID: 20357103
- Larsson A, Malmström S, Westermark P. Signs of cross-seeding: aortic medin amyloid as a trigger for protein AA deposition. Amyloid 2011; 18:229 - 34; http://dx.doi.org/10.3109/13506129.2011.630761; PMID: 22070546
- Sarell CJ, Stockley PG, Radford SE. Assessing the causes and consequences of co-polymerization in amyloid formation. Prion 2013; 7:7; http://dx.doi.org/10.4161/pri.26415; PMID: 24025483
- Krebs MR, Morozova-Roche LA, Daniel K, Robinson CV, Dobson CM. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci 2004; 13:1933 - 8; http://dx.doi.org/10.1110/ps.04707004; PMID: 15215533
- Ono K, Takahashi R, Ikeda T, Yamada M. Cross-seeding effects of amyloid β-protein and α-synuclein. J Neurochem 2012; 122:883 - 90; http://dx.doi.org/10.1111/j.1471-4159.2012.07847.x; PMID: 22734715
- Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ, Lee VM. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science 2003; 300:636 - 40; http://dx.doi.org/10.1126/science.1082324; PMID: 12714745
- Pauwels K, Williams TL, Morris KL, Jonckheere W, Vandersteen A, Kelly G, Schymkowitz J, Rousseau F, Pastore A, Serpell LC, et al. Structural basis for increased toxicity of pathological aβ42:aβ40 ratios in Alzheimer disease. J Biol Chem 2012; 287:5650 - 60; http://dx.doi.org/10.1074/jbc.M111.264473; PMID: 22157754
- Kayed R. Anti-tau oligomers passive vaccination for the treatment of Alzheimer disease. Hum Vaccin 2010; 6:931 - 5; http://dx.doi.org/10.4161/hv.6.11.12689; PMID: 20980799
- Martin ZS, Neugebauer V, Dineley KT, Kayed R, Zhang W, Reese LC, Taglialatela G. α-Synuclein oligomers oppose long-term potentiation and impair memory through a calcineurin-dependent mechanism: relevance to human synucleopathic diseases. J Neurochem 2012; 120:440 - 52; http://dx.doi.org/10.1111/j.1471-4159.2011.07576.x; PMID: 22060133
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science 2007; 318:930 - 6; http://dx.doi.org/10.1126/science.1138718; PMID: 17991853
- Kimberlin RH, Cole S, Walker CA. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol 1987; 68:1875 - 81; http://dx.doi.org/10.1099/0022-1317-68-7-1875; PMID: 3110370
- Hill AF, Desbruslais M, Joiner S, Sidle KC, Gowland I, Collinge J, Doey LJ, Lantos P. The same prion strain causes vCJD and BSE. Nature 1997; 389:448 - 50, 526; http://dx.doi.org/10.1038/38925; PMID: 9333232
- Baskakov IV. Switching in amyloid structure within individual fibrils: implication for strain adaptation, species barrier and strain classification. FEBS Lett 2009; 583:2618 - 22; http://dx.doi.org/10.1016/j.febslet.2009.05.044; PMID: 19482025
- Cortez LM, Sim VL. Implications of prion polymorphisms. Prion 2013; 7:276 - 9; http://dx.doi.org/10.4161/pri.25566; PMID: 23807178
- Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 2005; 121:49 - 62; http://dx.doi.org/10.1016/j.cell.2005.03.008; PMID: 15820678
- Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 2009; 181:1159 - 67; http://dx.doi.org/10.1534/genetics.108.099929; PMID: 19124570
- Vishveshwara N, Liebman SW. Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol 2009; 7:26; http://dx.doi.org/10.1186/1741-7007-7-26; PMID: 19470166
- Makarava N, Ostapchenko VG, Savtchenko R, Baskakov IV. Conformational switching within individual amyloid fibrils. J Biol Chem 2009; 284:14386 - 95; http://dx.doi.org/10.1074/jbc.M900533200; PMID: 19329794