Abstract
The JAK-STAT pathway represents a finely tuned orchestra capable of rapidly facilitating an exquisite symphony of responses from a complex array of extracellular signals. This review explores the evolution of the JAK-STAT pathway: the origins of the individual domains from which it is constructed, the formation of individual components from these basic building blocks, the assembly of the components into a functional pathway, and the subsequent reiteration of this basic template to fulfill a variety of roles downstream of cytokine receptors.
Introduction
The ability to respond to external signals is essential for life. At the level of the whole organism these signals are principally captured by peripheral sensory organs, which transmit them via the nervous system to the brain to mediate an appropriate response. At the cellular level, these signals are generally received by a myriad of cell-surface receptors that utilize a range of intracellular signaling pathways to communicate with the nucleus. This rapidly impacts on target gene transcription and mediates the desired cellular outcome, such as proliferation, differentiation, activation, and survival. The Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway represents a major mechanism used to transmit signals from extracellular receptors to the nucleus. The canonical pathway consists of JAK and STAT proteins activated by receptor ligation and inactivated by negative regulators, including SH2-containing protein tyrosine phosphatase (SHP) and suppressor of cytokine signaling (SOCS) proteins.
Each component of the JAK-STAT pathway consists of a collection of protein domains that collectively provide their distinct functional properties. Moreover, the pathway and its components have been replicated many times over, comprising four JAKs, seven STATs, eight SOCSs and two SHPs downstream of over 50 cytokine receptors in mammals.Citation1-Citation6 This review aims to capture in broad brush strokes the current state of knowledge on how such a complex signaling system has developed. In particular it follows the evolution of each individual component, discusses their assembly into a functional system and examines the subsequent multiple reiterations of the basic pathway.
JAK-STAT Pathway Components
To understand the evolution of the various components of the canonical JAK-STAT pathway, it is necessary to be familiar with their constituent protein domains, and how these modules are assembled into individual components ().
Figure 1. Architecture of JAK-STAT pathway components. (A) The constituent domains of JAK-STAT pathway components and their functionality. The PTK, SH2 and PTP domains contribute to the write, read and erase functionalities of the JAK-STAT pathway, as indicated, while the others have accessory roles. (B) The JAK-STAT pathway components and their domain assemblage.
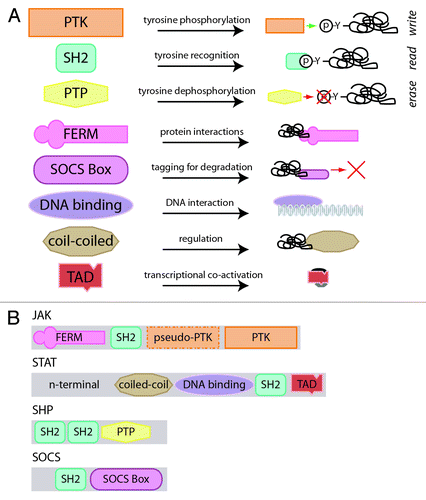
At the core of the JAK-STAT pathway is a binary code, based on specific tyrosine residues that are either unphosphorylated (off) or phosphorylated (on). Three of the protein domains utilized in the JAK-STAT pathway are specifically involved in the writing, reading and erasing of this code: the protein tyrosine kinase (PTK), Src-homology 2 (SH2) and protein tyrosine phosphatase (PTP) domains, respectively (). The PTK domain consists of ~250–300 residues, including an ATP-binding site and a catalytic region, and is able to phosphorylate tyrosine residues with a specificity determined by access to substrate and sequence context, thereby writing the code.Citation7 The SH2 domain is a ~100 residue motif that binds to phosphotyrosine residues in a manner dependent on the specific residues around the tyrosine especially those downstream,Citation8 providing a mechanism for reading the code.Citation9 Finally, the PTP domain consists of ~280 residues, with a signature CX5R motif, and is capable of dephosphorylating phosphotyrosine residues with which it is able to effectively interact, thereby erasing the code.Citation10,Citation11
JAK-STAT signaling components also consist of several other domains that provide accessory functions. The FERM domain, named after the founding proteins identified with this domain (band 4.1, ezrin, radixin and moesin), consists of three sub-domains, F1–F3, that are structurally similar to ubiquitin, acyl-CoA binding and plecstrin homology-phosphotyrosine binding domains, respectively.Citation12 The FERM domain is able to mediate protein-protein interactions, including adaptor and scaffolding interactions with membrane bound proteins.Citation13 FERM domain containing proteins can be divided into three broad groups: (1) talins and kinesins, (2) ERMS, GEF, kinases and phosphatases and (3) myosins and KIRT.Citation14 The SOCS box is a ~40 residue motif that mediates interactions with proteasomal degradation pathway components—particularly ubiquitin ligases—thereby regulating protein half-life.Citation1 The DNA-binding domain present in STAT proteins possesses an S-type immunoglobulin fold seen in members of the “p53 clan”—that includes p53 and RUNT—which mediates sequence-specific binding,Citation15-Citation17 while the coiled-coil domain is involved in protein-protein interactions. Finally, the transactivation domain (TAD)—of which there are several varieties—is important for co-factor recruitment and transcriptional responses.Citation17
These domains are assembled to form the individual components of the JAK-STAT pathway ():
(1) JAKs: Possess N-terminal FERM and SH2 domains, which mediate interactions with cytokine receptors and other signaling proteins, respectively.Citation18,Citation19 A centrally located “pseudokinase” domain—so-called due to its homology to PTK domains but (now disputedCitation20) absence of catalytic function—plays a regulatory role,Citation18 while a classical PTK domain is positioned at the C-terminus.Citation18,Citation21
(2) STATs: Consist of a unique N-terminus, with roles in nuclear translocation and protein interactions,Citation18 followed by coiled-coil, DNA-binding, SH2 and variable TAD domains.Citation18 The coiled-coil domain is involved with nuclear export and regulation of tyrosine phosphorylation, the DNA-binding domain mediates recognition of sequences related to TTCN3–4GAA in the promoters of responsive genes, the SH2 domain allows for specific recognition and docking to phosphotyrosines on cytokine receptors, JAKs and other STAT molecules,Citation18 while the divergent C-terminal TAD mediates transactivation.Citation22
(3) SHPs: Possess tandem N-terminal SH2 domains that bind specifically to key substrates, including cytokine receptors, a catalytic PTP domain and a divergent C-terminal region that contains several tyrosine residues that serve as docking sites for other signaling proteins when phosphorylated.Citation11,Citation23
(4) SOCSs: Consist of a divergent N-terminal region of largely unknown function, a central SH2 domain responsible for binding to cytokine receptors, JAKs and other substrates and a C-terminal SOCS box domain that facilitates protein degradation of the cytokine receptor signaling molecules.Citation24 Specific SOCS members also possess additional motifs, such as the kinase inhibitory region (KIR) of SOCS1 and SOCS3, and the N-terminal conserved region (NTCR) of SOCS4 and SOCS5.Citation25,Citation26
The JAK-STAT Pathway
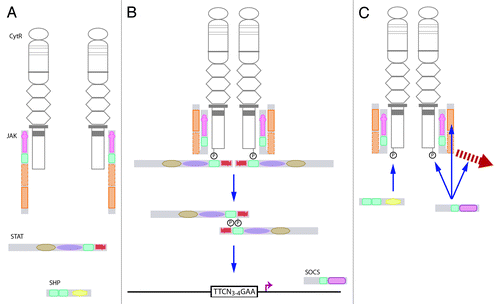
The individual components interact functionally in the canonical JAK-STAT pathway (). This pathway is activated by cytokine receptors in response to extracellular ligand binding, which leads to conformational changes in the receptor that activate the intracellularly associated JAKs. These phosphorylate the receptor complex to recruit STAT proteins, which are in turn phosphorylated, allowing dimerization and subsequent translocation to the nucleus to influence the transcription of relevant genes. The activated JAK-STAT pathway is then extinguished by negative regulators recruited to the receptor signaling complex, including latent SHPs or induced SOCSs. Additionally, the individual components are also able to signal via alternate “non-canonical” mechanisms.Citation27,Citation28
Figure 2. The canonical JAK-STAT pathway. (A) Ligand binding to a cytokine receptor results in dimerization and conformational changes that causes activation of JAKs associated with the Box 1 and Box 2 domains of the receptor intracellular domain. (B)The activated JAK is able to phosphorylate tyrosines on the receptor complex to create docking sites for signaling proteins, such as STATs. These are in turn phosphorylated, and then dimerize and translocate to the nucleus, where they bind to specific sequences in the promoters of responsive genes. (C) The pathway is extinguished by pre-existing SHPs, which serve to dephosphorylate tyrosine residues, or induced SOCSs that can act via several mechanisms, including JAK inhibition, steric interference at STAT docking sites, or mediating degradation of receptor signaling components.
Building the Building Blocks
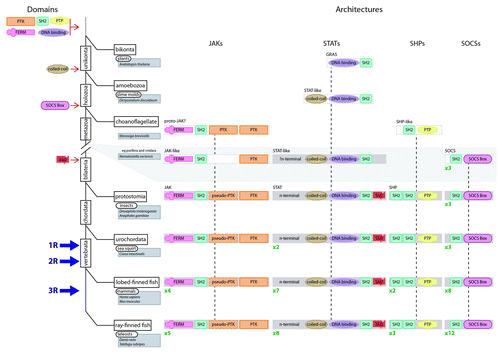
The first obligate step in the evolution of the JAK-STAT pathway was the generation of each of the individual domains that make up the various components of the pathway. Only 136 protein domains are metazoan-specific, with the majority of protein domains (> 400) emerging early in eukaryote evolution,Citation29 which is also the case for the constituent domains of the JAK-STAT pathway ().
Figure 3. Assembly of the JAK-STAT pathway components during evolution. An abbreviated tree of life for the evolution of eukaryotes (solid black lines) including major divergence points (rectangles), timing of WGD events (1R, 2R and 3R) and key organisms (rounded rectangles)/species (shaded rectangles) covered by this review. The likely time of derivation of the constituent domains is shown to the left of the tree. The proposed evolutionary steps that formed the various JAK-STAT component architectures are shown to the right of the tree, including the subsequent reiteration of individual components, the number of which is shown in green.
PTK domain
There is genomic evidence that PTK domains emerged in early eukaryotes or indeed prior to eukaryotes, as a modification of pre-existing kinase domains with alternate amino acid specificity.Citation16,Citation30-Citation32 PTK domains are certainly present in a broad range of extant non-metazoans, such as amoebozoa (such as the slime mold, Dictyostelium discoideum), basal holozoa (such as the filaseterea, Capsaspora owczarzaki) and choanoflagellates (e.g., Monosiga brevicollis), and have become extremely widespread in the metazoan lineage due to the myriad of pathways that exploit tyrosine phosphorylation.Citation33
SH2 domain
This domain, present in all JAK-STAT pathway components, can be traced to the origins of eukaryotes. Proto-SH2 domains are found in present-day plants (e.g., Arabidopsis thaliana) and slime molds, with the number of SH2 domains containing proteins expanding to > 100 in higher vertebrates.Citation8,Citation16,Citation34 Interestingly, the SH2 domains of the JAK and STAT families are closely related when compared with those of other SH2 domain-containing proteins.Citation7,Citation35
PTP domain
The PTP domain is present in prokaryotes, bacteria and archaea, and also basal eukaryotes such as malaria (e.g., Plasmodium berghei), and indeed may have also been captured by several viruses.Citation16 Like the PTK domain, this domain seems to have evolved by modification of a pre-existing enzyme to one with altered specificity for phosphotyrosines. Curiously, there are relatively few examples of PTP domains in extant representatives of plants and slime molds.Citation36-Citation38 However, the PTP domains have expanded considerably since the divergence of holozoa,Citation39 again reflecting the increasing importance of phosphotyrosine-mediated signaling along this lineage.
FERM
A number of FERM domain containing proteins are found in present-day plants and basal eukaryotes,Citation14,Citation16,Citation40 indicating a long evolutionary history. Indeed, there are six FERM domain containing proteins in present-day slime molds, although the ERMs, GEF, kinases and phosphatase group is absent,Citation14 with a total of 22 FERM domain containing proteins in extant insects, such as the fruit fly (Drosophila melanogaster).Citation13
SOCS box
Current genomic evidence suggests that the SOCS box is holozoan-specific, with a SOCS box containing protein seen in present-day choanoflagellates (e.g., Salpingoeca rosetta).Citation16 The number of SOCS box containing proteins has subsequently expanded considerably within metazoa.Citation1,Citation4
Other STAT-specific domains
The individual domains of STAT have arisen at different times during evolution. The p53 clan-type DNA binding domain has been present since early in eukaryote evolution,Citation16 with such domains described in extant plants,Citation41 early unikonta, such as amoebozoa, and holozoa, including choanoflagellates and ichthyosporea.Citation16 Although the roots are not as ancient, the coiled-coil domain arose prior to the divergence of amoebozoa, being evident in present-day slime molds and early holozoans. Finally, proto-TAD domains probably arose in the bilateria.Citation16
Component Assembly
The next important step in JAK-STAT pathway evolution was the assembly of these specific domains to generate each individual component. The assembly of domains into new protein architectures involves genomic processes such as exon-shuffling, exonization, intronization and duplication,Citation42-Citation44 while sequence divergence can modify these domains.Citation29,Citation44 These processes, rather than de novo generation of novel domains represents the major mechanism for the domain variation of proteins in eukaryotes, which is affirmed in the analysis of individual JAK-STAT pathway components.
JAKs
Each of the constituent domains of the JAKs, with the exception of the pseudo-kinase domain, was present throughout eukaryote evolution. So, how did the unique JAK architecture arise? The FERM domain of JAK is most closely related to that of focal adhesion kinase (FAK), which consists of an N-terminal FERM domain, SH2-binding domain (not actually a SH2 domain), PTK domain and a C-terminal focal adhesion targeting (FAT) domain,Citation14 which is evident in basal holozoans, such as present-day filasterea (e.g., C. owczarzaki).Citation30 Interestingly, the JAK SH2 domain shows highest homology with the N-terminal SH2 domains of two other kinases, SYK and ZAP70, which possess dual SH2 domains and an adjacent PTK domain.Citation45 The earliest SYK-like protein is exemplified by that in present-day sponge (e.g., Amphimedon queenslandica).Citation30 It remains unclear whether the JAK architecture was generated by the addition of an SH2 domain to a FAK-like protein that subsequently lost the FAT domain, or the addition of the FERM domain to a SYK-like protein, or some fusion of a FAK and SYK protein, prior to the divergence of eumetazoa and porifera. Curiously, M. brevicollis contains a protein, UTK05mod, which consists of FERM, SH2 and dual kinase domains, that is reminiscent of the metazoan JAK proteins, although the kinase domain forms a clade with the SYK kinase domain and not that the JAK kinase domain.Citation30 However, the similarity of the architecture between UTK05mod and JAK may be due to coincidence as there exist a wide variety of kinase domain containing architectures that are specific to M. brevicollis.Citation30 The first bona fide JAK-like protein is represented in the basal metazoan porifera lineage, typified by the sequence BAA81718.3 found in the present-day sponge, Ephydatia fluviatilis, which consists of FERM and SH2 domains as well as a single PTK domain related to the JAK PTK, although lacking the pseudo-PTK domain.Citation46 The canonical JAK domain architecture inclusive of a pseudo-kinase domain appears to be unique within bilateria, with this unique topology typified by present-day fruit fly Hopscotch.Citation8 It is likely that that the pseudo-kinase domain a result of local duplication and divergence of the PTK domain within a JAK-like sequence, although there exists the possibility that it was by modification of the first PTK domain of a UTK05mod-related sequence.
STATs
The STAT family has more ancient roots, with a range of STAT-like proteins present in extant eukaryotes. The most divergent are the GRAS proteins of the plant kingdom (named after the founding members, GAI, RGA and SCR), which possess an SH2-like domain that is remarkably similar to the metazoan STAT SH2 domain and also a putative DNA binding domain.Citation41 By the time of divergence of the amoebozoa, a family of proteins can be identified that resemble metazoan STATs, with coiled-coil, DNA-binding and SH2 domains, although lacking transactivation domains and the N-terminal sequence, as exemplified by those in present-day slime molds.Citation47,Citation48 Extant members of the cnidara, the sea anemone (Nematostella vectensis) and nematode (Caenorhabditis elegans), both contain at least one STAT with architectures similar to that seen in slime mold. Furthermore, the nematode STAT paralogs also contain a TAD,Citation49-Citation51 while the sea anemone STAT contains an N-terminal sequence, but not a TAD, although it possesses a long C-terminus containing multiple tyrosine and serine residues. The canonical STAT, replete with both TAD domain and N-terminal sequence, first appeared in the bilateria, as exemplified by present-day fruit fly Marelle (STAT92E).Citation3,Citation51
SHPs
The constituent domains of the SHPs were also present throughout eukaryotes evolution, but there is no evidence of these domains coming together in the bikonta or amoebozoa. However, a hypothetical SHP-like protein, is found in present-day choanoflagellates (e.g., XP_001747983 in M. brevicollis), consisting of a single SH2 domain followed by a PTP domain,Citation37-Citation39 which suggests that this architecture had evolved by the time of holozoa. The canonical SHP was finalised early in metazoan evolution, with the extant nematode ortholog, ptp-2, and fruit fly Corkscrew displaying the unique dual N-terminal SH2 domain architecture.Citation52 This most likely arose via local duplication of the adjacent SH2 domain in the precursor as the N-terminal and C-terminal SH2 domains of SHP are more similar to each other than the SH2 domains of other proteins.Citation35
SOCSs
Evolution of the SOCS proteins has followed a slightly more unique path. The most ancient SOCS box-containing protein architecture consisted of three N-terminal ankyrin repeats and a C-terminal SOCS box, similar to that of metazoan ankyrin repeat and SOCS box (ASB) proteins.Citation16 The SOCS SH2 domain is most closely related to that of dual adaptor of phosphotyrosine and 3-phosphoinositides (DAPP1), which together are part of a much larger clade that includes the SHP SH2 domain.Citation35 Presumably domain accretion to fuse a SH2 domain with a SOCS box generated the archetypical SOCS. In contrast to other components, this prototype underwent several gene duplication events that established three SOCS members by the divergence of the cnidara. This is exemplified in extant cnidara (e.g., N. vectensis), which possess three proteins with the signature SOCS protein architecture.Citation16 These are related to each of the three distinct SOCS clades seen in higher vertebrates: SOCS1/2/3/CISH, SOCS4/5 and SOCS6/7.Citation4
When JAK Met STAT and SOCS and SHP: The Canonical JAK-STAT Pathway
At the time of the emergence of bilateria, archetypal members of each of the JAK-STAT pathway components had been generated, including a single JAK, STAT and SHP, and three SOCS proteins. These came together to form the canonical JAK-STAT signaling pathway prior to the divergence of protostomia, as exemplified by the present day fruit fly Hopscotch-Marelle-Corkscrew/Socs36E pathway downstream of Domeless receptors. Several of the key innovations that were required to bring the various components together were, first, the association of the prototypal JAK with a membrane-bound receptor that lacked intracellular tyrosine kinase activity, second, appropriate tyrosines with which to recruit STATs, SHPs and SOCSs, and third, the incorporation of a negative feedback loop via the SOCS family. These innovations provided the ability to both recruit and activate the writer, the “paper” on which to write and a vehicle to inducibly block the writer or reader and/or act as a “paper shredder” to extinguish the signal, thereby complementing the erasers. Surprisingly little was required to establish these innovations. A few mutations to generate a cluster of intracellular prolines would have been sufficient for a cytokine receptor to be able to interact with the FERM domain of a prototypal JAK.Citation56,Citation57 A similar process could generate tyrosines in an appropriate sequence context, and a small number of mutations in the promoter region of a pre-existing SOCS gene would have been enough to generate an appropriate STAT binding site (TTCN3–4GAA) to enable their recruitment into an inducible feedback loop. Of course, each of the components would have necessarily participated in other processes prior to their recruitment into the canonical JAK-STAT pathway. Indeed, the legacy of this is evident in some of the non-canonical roles played by the pathway components in alternate signaling paradigms, such as downstream of receptor tyrosine kinases.Citation28,Citation31,Citation52,Citation54,Citation55
Making the Most of a Good Thing
Since the establishment of the canonical JAK-STAT pathway, it has subsequently been replicated multiple times to establish the diversity seen within extant higher vertebrates. However, the pathway itself has not been replicated per se, but rather the individual components have undergone replication, creating an opportunity for the generation of new pathway combinations.
Duplication events, either local or global, have largely facilitated the expansion of gene families in vertebrates. At the global level, three whole genome duplications (WGDs) have molded vertebrate evolution: the so-called 1R and 2R, which occurred before the divergence of lobe-finned and ray-finned fishes,Citation58 and 3R, which was limited to teleost fish ().Citation59 While rapid loss of many duplicated genes occurred following WGD, some have been retained because of either division of function between the duplicated genes or the generation of new functions for one of the duplicates.Citation58,Citation60,Citation61 We and others have shown that WGDs supplemented by local duplications have generated the diversity of JAK-STAT pathway components seen in higher vertebrates.Citation3,Citation4
Following the establishment of the archetypal JAK (typified by fruit fly Hopscotch) prior to the divergence of protostomes and deuterostomes, there has been no increase in the number of JAK homologs until the emergence of vertebrates, with existing urochordates such as sea squirt (Ciona intestinalis) retaining a single JAK.Citation62 The four JAKs in present-day tetrapods (e.g., Homo sapiens)—JAK1, JAK2, JAK3 and TYK2—were generated from 1R and 2R via JAK1/TYK2 and JAK2/JAK3 intermediates, while 3R yielded the additional teleost-specific JAK2 paralogs, named jak2a and jak2b,Citation4 such as in zebrafish (Danio rerio).
The archetypal STAT (typified by fruit fly Marelle) was similarly finalized by the divergence of protostomes and deuterostomes.Citation63 However, local duplication appears to have generated distinct duplicates in chordates prior to divergence of the urochordates, as observed in present-day sea-squirt (stat.a and stat.b).Citation62 We have argued that the proximity of many existing vertebrate STAT genes suggests these duplicates originally lay adjacent as a consequence of local duplication of the original STAT gene, with this STAT cluster being subsequently duplicated en bloc via 1R and 2R. Subsequent gene loss has resulted in three discrete copies of this cluster, STAT3-STAT5, STAT2-STAT6 and STAT1-STAT4. Finally, 3R resulted in retained duplicates of both STAT1 (stat1.a and stat1.b) and STAT5 (stat5.1 and stat5.2), while local duplication has occurred along the mammalian lineage to generate adjacent STAT5 genes (STAT5A and STAT5B).Citation4
In contrast, the evolution of the SHP family appears to have been driven solely by WGD processes in concert with poor duplicate retention. The archetypal SHP (typified by fruit fly Corkscrew), established prior to the protosome and deuterosome split, was duplicated via 1R and 2R, with subsequent gene loss yielding three members (SHP1, SHP2 and SHP3) in non-mammalian tetrapods. The 3R WGD failed to generate any further expansion of SHP family members in teleosts, while SHP3 has been lost in the mammalian lineage.Citation4
The common ancestor of protostomes and deuterostomes possessed three SOCS members: SOCS1/2/3/CISH, SOCS4/5 and SOCS6/7.Citation4 A further duplication of SOCS6/7, to generate SOCS6 and SOCS7, occurred prior to the divergence of urochordates, as exemplified in present-day sea squirt. WGDs have mediated the on-going expansion of the family within vertebrates. SOCS1, SOCS2, SOCS3 and CISH were generated from the SOCS1/2/3/CISH precursor during 1R and 2R, via SOCS1/SOCS3 and SOCS2/CISH intermediates. Concurrent gene loss limited the expansion of the other genes to yield SOCS4 and SOCS5 from the SOCS4/5 precursor and no further expansion of SOCS6 and SOCS7. In contrast, the 3R event produced paralogs for SOCS3 (socs3a and socs3b), SOCS4 (socs4a and socs4b), SOCS5 (socs5a and socs5b) and CISH (cish.a and cish.b), as seen in the present-day teleost, D. rerio.Citation4
Loss of duplicated genes is attributed to the generally redundant functions of the duplicate.Citation58,Citation60 Therefore, appropriate positive selective pressure is required to maintain duplicates. We have hypothesized that the key selective force in the case of JAK-STAT pathway component retention has been the expansion of the upstream cytokine receptors. This occurred co-incidently with an increase in organism complexity, including the emergence of adaptive immunity, providing an opportunity for the duplicates to develop crucial, non-redundant functions that facilitated retention.Citation53
Conclusion
There have been four critical phases in the evolution of the complex canonical JAK-STAT pathway: (1) the generation of the individual domains with discrete functions, such as the writing, reading, and erasing of a phosphotyrosine code, or other accessory roles, most of which occurred early in eukaryotic evolution, (2) the arrangement of these domains into functional proteins, which happened gradually across many years, (3) the interaction of these proteins to form an active pathway, which occurred in bilateria, and (4) the subsequent proliferation of this pathway, which was largely driven by WGD during vertebrate evolution. This has collectively facilitated the generation of a complex JAK-STAT pathway able to orchestrate responses to appropriate extracellular signals to produce the coherent melody of transcriptional outcomes that underpins hematopoiesis, immunity, growth and other essential functions in higher vertebrates.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci 2002; 27:235 - 41; http://dx.doi.org/10.1016/S0968-0004(02)02085-6; PMID: 12076535
- Renauld JC. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat Rev Immunol 2003; 3:667 - 76; http://dx.doi.org/10.1038/nri1153; PMID: 12974481
- Wang Y, Levy DE. Comparative evolutionary genomics of the STAT family of transcription factors. JAK-STAT 2012; 1:23 - 36; http://dx.doi.org/10.4161/jkst.19418
- Liongue C, O’Sullivan LA, Trengove MC, Ward AC. Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS One 2012; 7:e32777; http://dx.doi.org/10.1371/journal.pone.0032777; PMID: 22412924
- Liongue C, Ward AC. Evolution of Class I cytokine receptors. BMC Evol Biol 2007; 7:120; http://dx.doi.org/10.1186/1471-2148-7-120; PMID: 17640376
- Boulay JL, O’Shea JJ, Paul WE. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity 2003; 19:159 - 63; http://dx.doi.org/10.1016/S1074-7613(03)00211-5; PMID: 12932349
- Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell 2010; 142:661 - 7; http://dx.doi.org/10.1016/j.cell.2010.08.023; PMID: 20813250
- Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell 2006; 22:851 - 68; http://dx.doi.org/10.1016/j.molcel.2006.06.001; PMID: 16793553
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006; 127:635 - 48; http://dx.doi.org/10.1016/j.cell.2006.09.026; PMID: 17081983
- Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J 2009; 417:401 - 9; http://dx.doi.org/10.1042/BJ20081986; PMID: 19099538
- Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol 2001; 21:7117 - 36; http://dx.doi.org/10.1128/MCB.21.21.7117-7136.2001; PMID: 11585896
- Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 2000; 101:259 - 70; http://dx.doi.org/10.1016/S0092-8674(00)80836-3; PMID: 10847681
- Tepass U. FERM proteins in animal morphogenesis. Curr Opin Genet Dev 2009; 19:357 - 67; http://dx.doi.org/10.1016/j.gde.2009.05.006; PMID: 19596566
- Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol 2010; 11:802 - 14; http://dx.doi.org/10.1038/nrm2996; PMID: 20966971
- Berardi MJ, Sun C, Zehr M, Abildgaard F, Peng J, Speck NA, et al. The Ig fold of the core binding factor alpha Runt domain is a member of a family of structurally and functionally related Ig-fold DNA-binding domains. Structure 1999; 7:1247 - 56; http://dx.doi.org/10.1016/S0969-2126(00)80058-1; PMID: 10545320
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res 2012; 40:Database Issue D290 - 301; http://dx.doi.org/10.1093/nar/gkr1065; PMID: 22127870
- Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci 2000; 25:496 - 502; http://dx.doi.org/10.1016/S0968-0004(00)01624-8; PMID: 11050435
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 2002; 285:1 - 24; http://dx.doi.org/10.1016/S0378-1119(02)00398-0; PMID: 12039028
- Stark GR, Darnell JE Jr.. The JAK-STAT pathway at twenty. Immunity 2012; 36:503 - 14; http://dx.doi.org/10.1016/j.immuni.2012.03.013; PMID: 22520844
- Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol 2011; 18:971 - 6; http://dx.doi.org/10.1038/nsmb.2099; PMID: 21841788
- Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol 1998; 16:293 - 322; http://dx.doi.org/10.1146/annurev.immunol.16.1.293; PMID: 9597132
- Park C, Lecomte MJ, Schindler C. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res 1999; 27:4191 - 9; http://dx.doi.org/10.1093/nar/27.21.4191; PMID: 10518610
- Poole AW, Jones ML. A SHPing tale: perspectives on the regulation of SHP-1 and SHP-2 tyrosine phosphatases by the C-terminal tail. Cell Signal 2005; 17:1323 - 32; http://dx.doi.org/10.1016/j.cellsig.2005.05.016; PMID: 16084691
- Yoshimura A, Nishinakamura H, Matsumura Y, Hanada T. Negative regulation of cytokine signaling and immune responses by SOCS proteins. Arthritis Res Ther 2005; 7:100 - 10; http://dx.doi.org/10.1186/ar1741; PMID: 15899058
- Feng ZP, Chandrashekaran IR, Low A, Speed TP, Nicholson SE, Norton RS. The N-terminal domains of SOCS proteins: a conserved region in the disordered N-termini of SOCS4 and 5. Proteins 2012; 80:946 - 57; http://dx.doi.org/10.1002/prot.23252; PMID: 22423360
- Piessevaux J, Lavens D, Peelman F, Tavernier J. The many faces of the SOCS box. Cytokine Growth Factor Rev 2008; 19:371 - 81; http://dx.doi.org/10.1016/j.cytogfr.2008.08.006; PMID: 18948053
- Dawson MA, Bannister AJ, Göttgens B, Foster SD, Bartke T, Green AR, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 2009; 461:819 - 22; http://dx.doi.org/10.1038/nature08448; PMID: 19783980
- Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol 2008; 18:545 - 51; http://dx.doi.org/10.1016/j.tcb.2008.08.008; PMID: 18848449
- Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science 2003; 300:1701 - 3; http://dx.doi.org/10.1126/science.1085371; PMID: 12805536
- Suga H, Dacre M, de Mendoza A, Shalchian-Tabrizi K, Manning G, Ruiz-Trillo I. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci Signal 2012; 5:ra35; http://dx.doi.org/10.1126/scisignal.2002733; PMID: 22550341
- Araki T, Kawata T, Williams JG. Identification of the kinase that activates a nonmetazoan STAT gives insights into the evolution of phosphotyrosine-SH2 domain signaling. Proc Natl Acad Sci USA 2012; 109:1931 - 7; http://dx.doi.org/10.1073/pnas.1202715109; PMID: 22308335
- Shiu SH, Li WH. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol Biol Evol 2004; 21:828 - 40; http://dx.doi.org/10.1093/molbev/msh077; PMID: 14963097
- Tan CS, Pasculescu A, Lim WA, Pawson T, Bader GD, Linding R. Positive selection of tyrosine loss in metazoan evolution. Science 2009; 325:1686 - 8; http://dx.doi.org/10.1126/science.1174301; PMID: 19589966
- Liu BA, Shah E, Jablonowski K, Stergachis A, Engelmann B, Nash PD. The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes. Sci Signal 2011; 4:ra83; http://dx.doi.org/10.1126/scisignal.2002105; PMID: 22155787
- Liu BA, Engelmann BW, Nash PD. The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett 2012; 586:2597 - 605; http://dx.doi.org/10.1016/j.febslet.2012.04.054; PMID: 22569091
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol 2002; 129:908 - 25; http://dx.doi.org/10.1104/pp.004002; PMID: 12068129
- Howard PK, Gamper M, Hunter T, Firtel RA. Regulation by protein-tyrosine phosphatase PTP2 is distinct from that by PTP1 during Dictyostelium growth and development. Mol Cell Biol 1994; 14:5154 - 64; PMID: 7518559
- Howard PK, Sefton BM, Firtel RA. Analysis of a spatially regulated phosphotyrosine phosphatase identifies tyrosine phosphorylation as a key regulatory pathway in Dictyostelium. Cell 1992; 71:637 - 47; http://dx.doi.org/10.1016/0092-8674(92)90597-6; PMID: 1423620
- Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci U S A 2008; 105:9680 - 4; http://dx.doi.org/10.1073/pnas.0803161105; PMID: 18599463
- Baines AJ. A FERM-adjacent (FA) region defines a subset of the 4.1 superfamily and is a potential regulator of FERM domain function. BMC Genomics 2006; 7:85; http://dx.doi.org/10.1186/1471-2164-7-85; PMID: 16626485
- Richards DE, Peng J, Harberd NP. Plant GRAS and metazoan STATs: one family?. Bioessays 2000; 22:573 - 7; http://dx.doi.org/10.1002/(SICI)1521-1878(200006)22:6<573::AID-BIES10>3.0.CO;2-H; PMID: 10842311
- Liu M, Grigoriev A. Protein domains correlate strongly with exons in multiple eukaryotic genomes--evidence of exon shuffling?. Trends Genet 2004; 20:399 - 403; http://dx.doi.org/10.1016/j.tig.2004.06.013; PMID: 15313546
- Liu M, Walch H, Wu S, Grigoriev A. Significant expansion of exon-bordering protein domains during animal proteome evolution. Nucleic Acids Res 2005; 33:95 - 105; http://dx.doi.org/10.1093/nar/gki152; PMID: 15640447
- Schmidt EE, Davies CJ. The origins of polypeptide domains. Bioessays 2007; 29:262 - 70; http://dx.doi.org/10.1002/bies.20546; PMID: 17295290
- Filippakopoulos P, Müller S, Knapp S. SH2 domains: modulators of nonreceptor tyrosine kinase activity. Curr Opin Struct Biol 2009; 19:643 - 9; http://dx.doi.org/10.1016/j.sbi.2009.10.001; PMID: 19926274
- Suga H, Katoh K, Miyata T. Sponge homologs of vertebrate protein tyrosine kinases and frequent domain shufflings in the early evolution of animals before the parazoan-eumetazoan split. Gene 2001; 280:195 - 201; http://dx.doi.org/10.1016/S0378-1119(01)00784-3; PMID: 11738833
- Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, et al. The genome of the social amoeba Dictyostelium discoideum. Nature 2005; 435:43 - 57; http://dx.doi.org/10.1038/nature03481; PMID: 15875012
- Kawata T. STAT signaling in Dictyostelium development. Dev Growth Differ 2011; 53:548 - 57; http://dx.doi.org/10.1111/j.1440-169X.2010.01243.x; PMID: 21534947
- Dierking K, Polanowska J, Omi S, Engelmann I, Gut M, Lembo F, et al. Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe 2011; 9:425 - 35; http://dx.doi.org/10.1016/j.chom.2011.04.011; PMID: 21575913
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007; 317:86 - 94; http://dx.doi.org/10.1126/science.1139158; PMID: 17615350
- Wang Y, Levy DE. C. elegans STAT: evolution of a regulatory switch. FASEB J 2006; 20:1641 - 52; http://dx.doi.org/10.1096/fj.06-6051com; PMID: 16873887
- Gutch MJ, Flint AJ, Keller J, Tonks NK, Hengartner MO. The Caenorhabditis elegans SH2 domain-containing protein tyrosine phosphatase PTP-2 participates in signal transduction during oogenesis and vulval development. Genes Dev 1998; 12:571 - 85; http://dx.doi.org/10.1101/gad.12.4.571; PMID: 9472025
- Liongue C, John LB, Ward A. Origins of adaptive immunity. Crit Rev Immunol 2011; 31:61 - 71; http://dx.doi.org/10.1615/CritRevImmunol.v31.i1.60; PMID: 21395512
- Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet 2006; 38:1071 - 6; http://dx.doi.org/10.1038/ng1860; PMID: 16892059
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 2008; 10:489 - 96; http://dx.doi.org/10.1038/ncb1713; PMID: 18344984
- Usacheva A, Kotenko S, Witte MM, Colamonici OR. Two distinct domains within the N-terminal region of Janus kinase 1 interact with cytokine receptors. J Immunol 2002; 169:1302 - 8; PMID: 12133952
- Kouro T, Kikuchi Y, Kanazawa H, Hirokawa K, Harada N, Shiiba M, et al. Critical proline residues of the cytoplasmic domain of the IL-5 receptor alpha chain and its function in IL-5-mediated activation of JAK kinase and STAT5. Int Immunol 1996; 8:237 - 45; http://dx.doi.org/10.1093/intimm/8.2.237; PMID: 8671609
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 2005; 3:e314; http://dx.doi.org/10.1371/journal.pbio.0030314; PMID: 16128622
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004; 431:946 - 57; http://dx.doi.org/10.1038/nature03025; PMID: 15496914
- Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet 2001; 2:333 - 41; http://dx.doi.org/10.1038/35072009; PMID: 11331899
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008; 453:1064 - 71; http://dx.doi.org/10.1038/nature06967; PMID: 18563158
- Hino K, Satou Y, Yagi K, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. VI. Genes for Wnt, TGFbeta, Hedgehog and JAK/STAT signaling pathways. Dev Genes Evol 2003; 213:264 - 72; http://dx.doi.org/10.1007/s00427-003-0318-8; PMID: 12739142
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 1996; 84:411 - 9; http://dx.doi.org/10.1016/S0092-8674(00)81286-6; PMID: 8608595