Abstract
Cytokine signals are essential for generating a robust and specialized immune response. These signals are typically transmitted via canonical STAT homodimers. However, the number of STAT molecules utilized by cytokine signaling cascades within immune cells are limited, and so the mechanism used to deliver complex signals remains elusive. Heterodimerization of STAT proteins is one potential mechanism for signals to be modified downstream of the receptor and may play an important role in dictating the targets of specific cytokine signaling. In this review, we discuss our current understanding of the prevalence of STAT heterodimers, how they are formed and what their physiologic role may be in vivo.
Keywords: :
Introduction
While stimulation through the antigen receptor (signal 1) combined with costimulation (signal 2) is absolutely required for activation of T cells,Citation1 it is now generally understood that additional signals are required for acquisition of full effector function. These additional signals, in the form of autocrine and paracrine cytokines, are required to divert the immune response into appropriately programmed effector phenotypes (signal 3).Citation2 While signal 1 can only be delivered through the TCR and signal 2 is the “net sum” of multiple costimulatory molecule ligations, signal 3 is remarkably more complex.Citation2,Citation3 The delivery of a cytokine signal relies on several intermediaries, including (1) the expression and secretion of the cytokine on the “transmitting” cell, (2) the expression and structure of the receptor on the “receiving” cell, (3) the signaling pathways downstream of the receptor(s) and (4) additional regulation of those downstream signaling pathways [e.g., suppressor of cytokine signaling (SOCS) molecules]. Furthermore, most cytokines have complex effector functions that may contribute contrasting immunomodulatory roles in vivo.
Most cytokines predominantly transmit signals via Janus kinase (just another kinase, or JAK)-signal transducer and activator of transcription (STAT) pathways.Citation4 An inherent conundrum in cytokine biology is that while over 50 cytokines have been implicated in playing distinct roles in immune cell function, there are only seven STAT molecules with which to transmit these signals.Citation5,Citation6
In this review, we will discuss one potential mechanism by which cytokines diversify their signaling and deliver complex signals to a receiving cell. Several cytokines induce the formation of heterodimers of STAT proteins. We will discuss three important questions regarding these alternative signaling complexes. First, how common are STAT heterodimers in immunity? Second, how are STAT heterodimers formed? Third, how do STAT heterodimers mediate unique functional events?
JAK-STAT Signaling in Response to Cytokines
The initial discovery of a 91-kDa DNA binding protein that was tyrosine phosphorylated by interferon gamma (IFN-γ) treatment, referred to as STAT1, described a mechanism by which cytokines could induce transcription.Citation7,Citation8 The later discovery of JAK proteins that bound receptors and aided in tyrosine phosphorylation of these transcription factors further elucidated cytokine receptor function.Citation9 While TCR- and costimulatory-mediated signaling relies on the many rounds of signal amplification through multiple kinases,Citation1 it seemed as though cytokine-receptor interactions could directly phosphorylate the transcription factors that would program their function.Citation10 When STAT3 was found to signal via similar mechanisms, but in response to IL-6, leukemia inhibitory factor and other gp130-utilizing cytokines, it suggested that there are many STATs, each induced by different cytokines and promoting a distinct outcome.Citation11 Over the next 10 years, a growing number of cytokines would be linked to distinct STAT proteins, providing a network by which cytokines could presumably activate transcription and program function ().
Table 1. Known STATs and their activators
Common themes began to emerge as the “menu” of JAKs and STATs grew to size. Interferons promoted the activation of STAT1 and STAT2, resulting in the transcription of interferon-inducible genes.Citation7,Citation8 IL-6 and other gp130-utilizing cytokines could induce STAT3 activation, promoting the induction of inflammatory cytokines.Citation12 STAT3 was also to be critically important in the induction and development of T helper 17 (Th17) cells, a dominant inflammatory T cell subset.Citation13 IL-12 induces STAT4 activation, which, in T cells, promoted the acquisition of a T helper 1 (Th1) effector phenotype and the subsequent production of IFN-γ.Citation14 IL-2, an extremely important survival cytokine for T cells, could induce STAT5a/5b activation, resulting in the transcription of prosurvival genes, as well as Foxp3, aiding regulatory T cell development.Citation15,Citation16 STAT6 activation could be induced by IL-4, which programmed T helper 2 (Th2) differentiation, characterized by IL-4 secretion.Citation17 In addition, the mechanism by which STATs mediated transcription was elucidated; tyrosine phosphorylation of STAT proteins by receptor binding and JAK activation could induce homodimerization of STAT molecules.Citation4 This homodimerization would result in nuclear accumulation of these dimers. STAT dimers could then bind their target sequence, recruit coactivators and effect transcription.
However, while the number of cytokines grew larger, the number of signaling molecules utilized by cytokines to transduce signals did not. It seemed that many cytokines with distinct (and sometimes opposing) functions would activate the same STAT protein.Citation5,Citation6 For instance, IL-6, a proinflammatory cytokine that utilizes gp130, promotes the activation of STAT3.Citation11 IL-10, however, does not utilize gp130, is a potent anti-inflammatory cytokine, but also promotes the activation of STAT3.Citation18-Citation20 This paradox continues to persist today, as new cytokines with distinct functions are discovered that activate one or more of the same seven STAT proteins.
As the mechanisms of STAT activation were identified, the picture became more complex with regard to post-translational modifications (PTMs) and their role (). Phosphotyrosine residues in STAT proteins can be recognized by STAT SH2 domains in its partner, mediating dimerization. STATs can also be phosphorylated on a serine residue in the transactivation domain,Citation21 thereby modulating gene target specificity and transcriptional activity. Phosphorylation can take place at the receptor or in response to other cellular signaling pathways.Citation22 While STAT1, -3 and -4 have homologous phosphoserine sites containing a P(M)SP motif, it is interesting that STAT5a/b and STAT6 lack these motifs.Citation22 However, it has recently been shown STAT5 and STAT6 are modulated by phosphorylation on alternate serine residues at the C terminus in non-homologous locations.Citation23-Citation26 Other modifications have also been identified. Arginine methylation has been shown to modulate interactions with epigenetic machinery.Citation27,Citation28 SUMOylation in the TA domain by protein inhibitor of activated STATs (PIAS) proteins inhibits the activation and nuclear accumulation of STAT proteins.Citation29 Other modifications have also been described (e.g., acetylation and ubiquitylation) but the physiologic relevance of these events has yet to be elucidated.
Figure 1. Anatomy of a STAT. STAT proteins in general consist of six domains. Post-translational modifications (PTM) that have been shown to have physiologic relevance are depicted. Note that some STATs (notably STAT5 and STAT6) lack a canonical phosphoserine site, but instead have several non-homologous serines that have varied functions in transcriptional regulation.
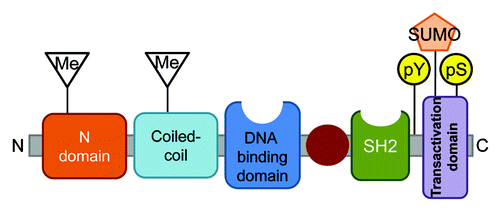
In addition, latent or unphosphorylated STAT dimers have been observed and appear to play a vital role in certain aspects of cell signaling.Citation30 These latent dimers have been identified for most STAT proteins and exhibit continuous bi-directional shuttling between the cytoplasm and nucleus.Citation31,Citation32 Importantly, these latent STAT dimers have also been shown to have distinct transcriptional targets than their phosphorylated counterpartsCitation32 These homodimeric interactions require the N-terminal domain, and are independent of the phosphotyrosyl-SH2 interactions that occur at the C terminus of the protein. N-terminal domains have been shown to be an important aspect of latent STAT homodimerization in all seven STAT proteins.Citation33 It is thought that these interactions are also important for higher order structures in the nucleus, especially in promoters that have several adjacent STAT binding sites. The N-terminal domain of STAT4 has been shown to be critical for dimerization and function, in particular IL-12-induced phosphorylation and Th1 differentiation.Citation34 These and other studies have suggested that STAT N-terminal interactions provide an alternative mode of signaling and transcription to receptor activation.Citation35
Further complicating our understanding is the fact that the majority of cytokines activate several STAT proteins (), and which STATs are activated can be heavily dependent on cell type, activation or differentiation state, the type of receptor expressed and the timing and dose of cytokine.Citation5,Citation6 Many of these problems have been addressed, at least in part, by the utilization of STAT or receptor-deficient cells. For example, IL-12 has been shown to activate STAT3 and STAT5 in addition to STAT4, but only STAT4-deficient cells seem to lack sensitivity to IL-12.Citation14,Citation36-Citation38 In other words, a cytokine, when added in excess, may activate a number of STATs, but the essential requirement of a particular STAT is determined by the impact of genetic deletion. However, when it is found that several STATs are activated (and required) by a particular cytokine, it begs the question of whether the STAT proteins are being activated in parallel, inducing separate transcriptional events, or are working together, synergizing to promote an alternative transcriptional outcome.
How Common Are STAT Heterodimers in Immunity?
As STAT1 was originally found as part of a nuclear multiprotein complex, ISGF3,Citation7 it was known that it had several binding partners. However, whether or not other STATs were involved was largely unknown. Later it was shown that activated STAT1 and STAT2 were found in ISGF3 in response to type I interferons, together forming a DNA-binding complex.Citation39 However, the physiologic relevance of these heterodimers was determined later, showing that STAT1:STAT2 heterodimers bound distinct consensus sequences in a subset of interferon stimulated genes.Citation40
STAT heterodimers were also shown to be important in GM- and M-CSF signaling. While M-CSF was known to activate both STAT3 and STAT5, it was unclear if a heterodimeric complex formed. Interestingly, STAT5 homodimers as well as STAT3:STAT5 heterodimers could be formed upon M-CSF stimulation, but only STAT3:STAT5 heterodimers could bind particular consensus sequences.Citation41 Further using Stat5a–/– mice, it was shown that GM-CSF requires use of STAT5a:STAT5b heterodimers to have full function in myeloid cells, whereas STAT5a homodimers were dispensable.Citation42 Later, despite having high homology to one another, it was found that a single amino acid difference between STAT5a and STAT5b resulted in surprisingly different DNA binding specificities, highlighting the potential importance of heterodimerization to expand target gene selection.Citation43,Citation44
The common gamma chain (γc) cytokines IL-2, -4, -7, -9, -15, -21 and TSLP have each been shown to activate a broad variety of STAT proteins, with STAT5 being in common.Citation45 While each of these cytokines utilizes γc to mediate signaling, the heterodimeric/trimeric nature of these receptor complexes can promote a number of distinct patterns of STAT activation. For instance, IL-2, IL-7 and IL-21 each can activate STAT1, STAT3 and STAT5, but IL-2 promotes much stronger STAT5 activation, IL-7 induces persistent STAT5 activation, while IL-21 produces far more activated STAT3.Citation46 Further, heterodimerization of STAT5 isoforms 5a and 5b has been shown in response to IL-2 as well as IL-7, showing that while the kinetics of heterodimerization were the same, the strength of signal was remarkably different between these two cytokines.Citation47
The IL-6 superfamily of cytokines, which is characterized by complex interactions like soluble receptors and receptor-like subunits,Citation48 also utilizes STAT heterodimerization. The discovery of STAT3 showed that in response to IL-6, it can utilize STAT1:STAT3 heterodimers, especially in late signaling.Citation12,Citation49
The IL-12 family, a subunit of the IL-6 superfamily, is characterized by heterodimeric cytokines that share subunits, and has multiple immunomodulatory roles.Citation50 IL-12, made of p35 and p40, promotes a strong Th1 response by inducing T-bet expression, which programs Th1 function and IFN-γ secretion.Citation51 IL-12 works mainly through inducing mostly STAT4 homodimers.Citation52 However, the other members of the IL-12 family, including IL-23, IL-27 and IL-35, have been shown to induce heterodimers.
IL-23, important during Th17 differentiation, works to stabilize the inflammatory phenotype of Th17 cells. Early differentiated Th17 cells induce expression of the IL-23R,Citation53 which pairs with IL-12Rβ1 to confer sensitivity to IL-23.Citation54 IL-23 was shown to activate STAT3 and STAT4 downstream of these receptors.Citation54 Later studies suggested that the majority of STAT3 and STAT4 induced by IL-23 to effect transcription was present in heterodimers.Citation51,Citation54 Indeed IL-27, which uses the IL-6 receptor subunit gp130 and WSX-1 to induce STAT1 and STAT3 signaling, was also shown to form STAT1:STAT3 heterodimers, although the molecular consequences of that heterodimer are still unknown.Citation55
Our group has recently identified the receptor and signaling pathway for IL-35, the newest addition to the IL-12 family.Citation56,Citation57 IL-35 has two functions in conventional naive T cells, suppression of proliferation and conversion to an induced regulatory T cell population that suppresses via IL-35.Citation57,Citation58 IL-35 utilizes three receptors: gp130 homodimers, IL12Rβ2 homodimers and a heterodimer of gp130:IL12Rβ2. IL-35-induced IL12Rβ2 and gp130 homodimers induce STAT4 and STAT1 phosphorylation, respectively, and can do so in the absence of the other receptor. These homodimeric receptors are sufficient for suppression of proliferation, but cannot induce conversion to IL-35 production. Rather, the gp130:IL-12Rβ2 heterodimeric receptor is essential for induced expression of IL-35, which is initiated by a STAT1:STAT4 heterodimer.Citation56
Collectively, these observations suggest that within every cytokine family there appears to be at least one member that promotes STAT heterodimer formation. Furthermore, STAT heterodimerization may not be an uncommon occurrence, but rather one that has not been rigorously studied.
How Are STAT Heterodimers Formed?
The mechanism by which receptors promote STAT heterodimerization vs. homodimerization is still elusive. We and other have suggested at least three non-mutually exclusive different mechanisms by which this occurs (). First, post-translational modification (PTM) of STATs may either induce the formation of a heterodimer or prevent the formation of a homodimer. This is evidenced by STAT3 signaling, in which serine phosphorylation of STATs, a JAK-independent but receptor-dependent phenomenon, is dispensable for heterodimerization.Citation21 Type I interferon signaling, which utilizes STAT1:STAT2 heterodimers, also may involve alternative PTMs.Citation10,Citation39 Given that STATs can undergo many types of PTM (), it stands to reason that one or many of these mechanisms may have a role in dimerization, although whether or not these modifications can specify homo vs. heterodimerization remains to be determined.Citation5
Figure 2. Potential mechanisms of STAT heterodimer formation. (A) As STATs can be extensively post-translationally modified, PTM of STAT proteins may act to inhibit a specific kind of dimerization or promote another. (B) The specific structure of particular cytokine receptors could favor the generation of heterodimers over homodimers. (C) Proximity of subunits that generate distinct STAT molecules could influence heterodimer formation. A cytokine or other cellular event bringing these subunits together could promote the formation of STAT heterodimers.
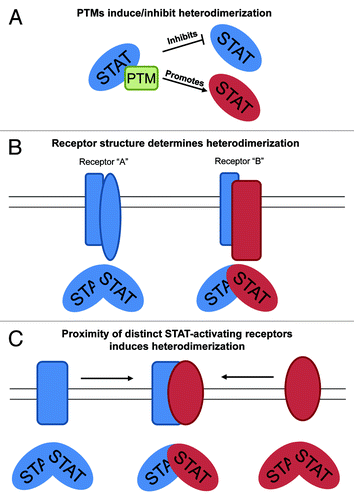
Second, it may also be that the structure of individual cytokine receptors promotes the generation of STAT heterodimers, independent of PTM or other signaling pathways. Given that expression of cytokine receptors can be regulated by cell-type, activation status and other factors, the receptor-intrinsic regulation of STAT heterodimerization provides another level of regulation that could be utilized to promote distinct function downstream of cytokine stimulation. However, testing these hypotheses presents a unique challenge. Without structural insight into what these receptors look like complexed with STAT homodimers vs. heterodimers, it would be difficult to fully ascertain how one forms over the other. However, structural changes could change the accessibility of docking motifs for binding partners, which could be assessed fairly easily. The crystal structures of several STAT homodimers have been solved, and these studies along with structure-function analyses have suggested that side groups and length of the SH2 domain might modulate a given STAT’s ability to homo or heterodimerize via phosphotyrosyl modifications.Citation59-Citation61
Third, given that most cytokine receptors are composed of two or more distinct subunits, it may also be that differing subunits can recruit and activate distinct STAT proteins, and that mere proximity can promote the formation of a heterodimer. As some receptor subunits can be spatially segregated while others are ubiquitous, this explanation, while simplistic, may prove to be important. However there has been some evidence for this in the literature. For instance, B cells stimulated with type I IFNs promote the generation of STAT2:STAT6 heterodimers, presumably due to the expression of alternative type I IFN receptors in B cells.Citation62 In addition, concomitant IL-4 and IFNα stimulation enhances IL-4 activity while promoting the generation of STAT2:STAT6 in human T cells, which the authors conclude is determined due to proximity of the IL-4R, γc and IFN-AR.Citation63 Experiments enforcing proximity of subunits paired with real-time visualization of STAT heterodimers may be vital to exploring this hypothesis.
Our understanding of the formation and role of ternary STAT structures has been further complicated by observations that STATs can homo- and hetero-tetramerize, as well as have even higher-order structures when in the nucleus.Citation64,Citation65 Recently, STAT5a/5b tetramerization was implicated, using several in vivo models, in the response to IL-2 and other γc cytokines, utilizing a mouse with a targeted mutation preventing STAT5 tetramerization while preserving dimerization.Citation66 Thus there are many potential explanations regarding how STAT heterodimers form, none being mutually exclusive. Furthermore, it is important to remember that higher-order STAT structures have been observed and are physiologically relevant.
How Do STAT Heterodimers Mediate Unique Functional Events?
The functional relevance of STAT heterodimers in vivo remains obscure. Many theories have been postulated but none have been rigorously tested, as typical molecular biology techniques can never completely rule out the generation and/or function of homodimers. Gel-shift analysis by EMSA is the “gold standard” of heterodimer identification, but these assays rely on specific probes, thereby ruling out any high-throughput, genomic analyses. ChIP-reChIP, which utilizes sequential immunoprecipitations to assay DNA bound by multiple transcription factors, has technical limitations that cannot rule out distinct homodimeric complexes binding to adjacent sites. The functional relevance of heterodimers has yet to be fully elucidated, but they could exert their function through three non-mutually exclusive mechanisms ().
Figure 3. Potential mechanisms of STAT heterodimer function. (A) STAT heterodimers may act as a sink, reducing the pool of available STAT proteins able to homodimerize. (B) STAT heterodimers may act by binding a unique consensus site (green vs orange, in this model), allowing for transcription of alternative gene targets. (C) STAT heterodimers could also function through the recruitment of different transcriptional coactivators/repressors. In this model, the consensus site is not significantly altered, but rather homodimers recruit repressors while heterodimers recruit coactivators.
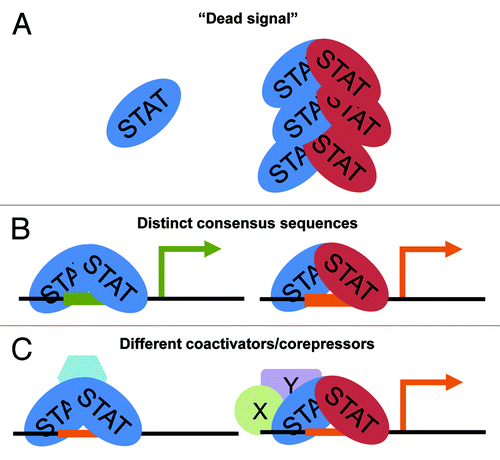
First, some groups have hypothesized that STATs heterodimerize to remove a particular STAT from the available “pool” of signaling molecules, thereby reducing the efficacy or strength of STAT homodimer signals. This has been shown notably in STAT1:STAT3 heterodimerization using overexpressed STAT3 and type I IFNs, indicating STAT3 sequesters STAT1 into heterodimers.Citation67 While overexpression of STAT3 may not be physiologically relevant, it does demonstrate a role for STAT3 in sequestering STAT1 in heterodimers. This “dead signal” hypothesis is supported by some signaling pathways, in which STAT heterodimers play a role in late signaling or diminution of signaling or transcription. However, this hypothesis doesn't explain why some cytokines elicit a majority of heterodimers, but have distinct functions. For instance, IL-23 promotes STAT3:STAT4 heterodimerization during Th17 polarization.Citation54 Given the importance of STAT3 in the generation of Th17 cells,Citation13 it is unlikely that IL-23 would generate a dead signal for STAT3 during this crucial time.
Second, given that particular STATs can bind different consensus sequences with distinct affinities, another possibility is that STAT heterodimers can bind different sequences than their homodimeric counterparts. STAT1:STAT4 induced by IL-35, for instance, can bind sequences in the promoters of Ebi3 and Il12a, but not two traditional STAT1 or STAT4 targets, Irf1 and Il18ra, respectively.Citation56 STAT3:STAT5 elicited by M-CSF can bind high affinity sis-inducible element (SIE) sites with high affinity while STAT3 or STAT5 homodimers cannot.Citation41 There are obvious caveats to this possibility, as the consensus sequences of many STAT proteins are known and appear to be quite redundant. However, the recognition of these STAT-binding sites can to be fine-tuned. For instance, STAT5a’s DNA binding specificity can be changed to STAT5b’s merely by a change of a single amino acid.Citation43 Whether or not this regulation is STAT-intrinsic or dependent on other factors has yet to be determined.
Third, it may be possible that STAT heterodimers, as tightly bound distinct subunits, can recruit different transcriptional coactivators/repressors that can influence their function. Rather than bind a distinct DNA sequence, either partner may recruit new transcriptional activators, for instance, thus allowing transcription at privileged sites. This is particularly evident in interferon signaling. IFNγ promotes STAT1 homodimers, which can bind genes like Irf1.Citation68 IFNα, in contrast, promotes STAT1:STAT2 heterodimerization, which can recruit other coactivators, such as IRF9, in the ISGF3 complex.Citation69 However, the full transcriptional networks and STAT interactome has yet to be fully explored. Experiments using proteomic identification of nuclear STAT heterodimers could help shed light on this potential method of regulation.
Concluding Remarks
The role of STAT heterodimerization in the modulation of cytokine function has yet to be fully explored. Furthermore, the prevalence of STAT heterodimers vs. homodimers needs to be established. Rigorous interrogation of how individual STAT heterodimers interact, as well as how they function in contrast to STAT homodimers, may elucidate the roles of these complexes in general immunity, and how they may help pleiotropic cytokines deliver complex messages. This is likely to require detailed structure-function analysis coupled with examining the full spectrum and kinetics of STAT post-translational modification, as well as a proteomic analysis of STAT binding partners, in response to a particular cytokine.
The use of chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) has expanded the horizon for determining the genomic targets of STAT proteins.Citation70,Citation71 However, a massively parallel ChIP-Seq experiment from the same cell type, done using distinct cytokine stimulations and ChIP-Seq for all seven STAT proteins, could reveal much. Overlaying the genomic locations of different STAT proteins in response to individual cytokines could impart fundamental insight into the targets of different STAT complexes, while paving the way for the discovery of novel heterodimeric STAT complexes. This machinery could be further verified by the use of ChIP-reChIP-Seq, using sequential antibody immunoprecipitations to restrict analysis to STAT heterodimers. Finally, a detailed understanding of the molecular basis for STAT heterodimer formation and function could reveal novel therapeutic approaches to specifically and surgically modulate their downstream phenotypic consequence in vivo.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Powell JD, Ragheb JA, Kitagawa-Sakakida S, Schwartz RH. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol Rev 1998; 165:287 - 300; http://dx.doi.org/10.1111/j.1600-065X.1998.tb01246.x; PMID: 9850868
- Ross JA, Nagy ZS, Cheng H, Stepkowski SM, Kirken RA. Regulation of T cell homeostasis by JAKs and STATs. Arch Immunol Ther Exp (Warsz) 2007; 55:231 - 45; http://dx.doi.org/10.1007/s00005-007-0030-x; PMID: 17659375
- Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity 2010; 33:301 - 11; http://dx.doi.org/10.1016/j.immuni.2010.09.002; PMID: 20870173
- Adamson AS, Collins K, Laurence A, O’Shea JJ. The Current STATus of lymphocyte signaling: new roles for old players. Curr Opin Immunol 2009; 21:161 - 6; http://dx.doi.org/10.1016/j.coi.2009.03.013; PMID: 19362457
- Delgoffe GM, Murray PJ, Vignali DA. Interpreting mixed signals: the cell’s cytokine conundrum. Curr Opin Immunol 2011; 23:632 - 8; http://dx.doi.org/10.1016/j.coi.2011.07.013; PMID: 21852079
- Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol 2007; 178:2623 - 9; PMID: 17312100
- Shuai K, Schindler C, Prezioso VR, Darnell JE Jr.. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 1992; 258:1808 - 12; http://dx.doi.org/10.1126/science.1281555; PMID: 1281555
- Schindler C, Shuai K, Prezioso VR, Darnell JE Jr.. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 1992; 257:809 - 13; http://dx.doi.org/10.1126/science.1496401; PMID: 1496401
- Müller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature 1993; 366:129 - 35; http://dx.doi.org/10.1038/366129a0; PMID: 8232552
- Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE Jr.. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 1994; 76:821 - 8; http://dx.doi.org/10.1016/0092-8674(94)90357-3; PMID: 7510216
- Lütticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science 1994; 263:89 - 92; http://dx.doi.org/10.1126/science.8272872; PMID: 8272872
- Zhong Z, Wen Z, Darnell JE Jr.. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994; 264:95 - 8; http://dx.doi.org/10.1126/science.8140422; PMID: 8140422
- Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol 2007; 179:4313 - 7; PMID: 17878325
- Jacobson NG, Szabo SJ, Weber-Nordt RM, Zhong Z, Schreiber RD, Darnell JE Jr., et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med 1995; 181:1755 - 62; http://dx.doi.org/10.1084/jem.181.5.1755; PMID: 7722452
- Beadling C, Guschin D, Witthuhn BA, Ziemiecki A, Ihle JN, Kerr IM, et al. Activation of JAK kinases and STAT proteins by interleukin-2 and interferon alpha, but not the T cell antigen receptor, in human T lymphocytes. EMBO J 1994; 13:5605 - 15; PMID: 7988557
- Burchill MA, Yang J, Vang KB, Farrar MA. Interleukin-2 receptor signaling in regulatory T cell development and homeostasis. Immunol Lett 2007; 114:1 - 8; http://dx.doi.org/10.1016/j.imlet.2007.08.005; PMID: 17936914
- Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL. An interleukin-4-induced transcription factor: IL-4 Stat. Science 1994; 265:1701 - 6; http://dx.doi.org/10.1126/science.8085155; PMID: 8085155
- Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol 1995; 155:1079 - 90; PMID: 7543512
- Lai CF, Ripperger J, Morella KK, Jurlander J, Hawley TS, Carson WE, et al. Receptors for interleukin (IL)-10 and IL-6-type cytokines use similar signaling mechanisms for inducing transcription through IL-6 response elements. J Biol Chem 1996; 271:13968 - 75; http://dx.doi.org/10.1074/jbc.271.24.13968; PMID: 8662928
- Weber-Nordt RM, Riley JK, Greenlund AC, Moore KW, Darnell JE, Schreiber RD. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J Biol Chem 1996; 271:27954 - 61; http://dx.doi.org/10.1074/jbc.271.44.27954; PMID: 8910398
- Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science 1995; 267:1990 - 4; http://dx.doi.org/10.1126/science.7701321; PMID: 7701321
- Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene 2000; 19:2628 - 37; http://dx.doi.org/10.1038/sj.onc.1203481; PMID: 10851062
- Maiti NR, Sharma P, Harbor PC, Haque SJ. Serine phosphorylation of Stat6 negatively controls its DNA-binding function. J Interferon Cytokine Res 2005; 25:553 - 63; http://dx.doi.org/10.1089/jir.2005.25.553; PMID: 16181056
- Shirakawa T, Kawazoe Y, Tsujikawa T, Jung D, Sato S, Uesugi M. Deactivation of STAT6 through serine 707 phosphorylation by JNK. J Biol Chem 2011; 286:4003 - 10; http://dx.doi.org/10.1074/jbc.M110.168435; PMID: 21123173
- Wang Y, Malabarba MG, Nagy ZS, Kirken RA. Interleukin 4 regulates phosphorylation of serine 756 in the transactivation domain of Stat6. Roles for multiple phosphorylation sites and Stat6 function. J Biol Chem 2004; 279:25196 - 203; http://dx.doi.org/10.1074/jbc.M313668200; PMID: 15069079
- Weaver AM, Silva CM. S731 in the transactivation domain modulates STAT5b activity. Biochem Biophys Res Commun 2007; 362:1026 - 30; http://dx.doi.org/10.1016/j.bbrc.2007.08.087; PMID: 17822672
- Komyod W, Bauer UM, Heinrich PC, Haan S, Behrmann I. Are STATS arginine-methylated?. J Biol Chem 2005; 280:21700 - 5; http://dx.doi.org/10.1074/jbc.C400606200; PMID: 15826948
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 2001; 104:731 - 41; http://dx.doi.org/10.1016/S0092-8674(01)00269-0; PMID: 11257227
- Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res 2006; 16:196 - 202; http://dx.doi.org/10.1038/sj.cr.7310027; PMID: 16474434
- Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem 2003; 278:34133 - 40; http://dx.doi.org/10.1074/jbc.M304531200; PMID: 12832402
- Vogt M, Domoszlai T, Kleshchanok D, Lehmann S, Schmitt A, Poli V, et al. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci 2011; 124:900 - 9; http://dx.doi.org/10.1242/jcs.072520; PMID: 21325026
- Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res 2008; 18:443 - 51; http://dx.doi.org/10.1038/cr.2008.41; PMID: 18364677
- Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol 2004; 5:208 - 15; http://dx.doi.org/10.1038/ni1032; PMID: 14704793
- Chang HC, Zhang S, Oldham I, Naeger L, Hoey T, Kaplan MH. STAT4 requires the N-terminal domain for efficient phosphorylation. J Biol Chem 2003; 278:32471 - 7; http://dx.doi.org/10.1074/jbc.M302776200; PMID: 12805384
- Wenta N, Strauss H, Meyer S, Vinkemeier U. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc Natl Acad Sci U S A 2008; 105:9238 - 43; http://dx.doi.org/10.1073/pnas.0802130105; PMID: 18591661
- Bacon CM, Petricoin EF 3rd, Ortaldo JR, Rees RC, Larner AC, Johnston JA, et al. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci U S A 1995; 92:7307 - 11; http://dx.doi.org/10.1073/pnas.92.16.7307; PMID: 7638186
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 1996; 382:174 - 7; http://dx.doi.org/10.1038/382174a0; PMID: 8700209
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature 1996; 382:171 - 4; http://dx.doi.org/10.1038/382171a0; PMID: 8700208
- Qureshi SA, Salditt-Georgieff M, Darnell JE Jr.. Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci U S A 1995; 92:3829 - 33; http://dx.doi.org/10.1073/pnas.92.9.3829; PMID: 7537377
- Ghislain JJ, Wong T, Nguyen M, Fish EN. The interferon-inducible Stat2:Stat1 heterodimer preferentially binds in vitro to a consensus element found in the promoters of a subset of interferon-stimulated genes. J Interferon Cytokine Res 2001; 21:379 - 88; http://dx.doi.org/10.1089/107999001750277853; PMID: 11440635
- Novak U, Mui A, Miyajima A, Paradiso L. Formation of STAT5-containing DNA binding complexes in response to colony-stimulating factor-1 and platelet-derived growth factor. J Biol Chem 1996; 271:18350 - 4; http://dx.doi.org/10.1074/jbc.271.31.18350; PMID: 8702476
- Feldman GM, Rosenthal LA, Liu X, Hayes MP, Wynshaw-Boris A, Leonard WJ, et al. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood 1997; 90:1768 - 76; PMID: 9292509
- Boucheron C, Dumon S, Santos SC, Moriggl R, Hennighausen L, Gisselbrecht S, et al. A single amino acid in the DNA binding regions of STAT5A and STAT5B confers distinct DNA binding specificities. J Biol Chem 1998; 273:33936 - 41; http://dx.doi.org/10.1074/jbc.273.51.33936; PMID: 9852045
- Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene 2000; 19:2566 - 76; http://dx.doi.org/10.1038/sj.onc.1203523; PMID: 10851055
- Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol 2009; 9:480 - 90; http://dx.doi.org/10.1038/nri2580; PMID: 19543225
- Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol 2011; 23:598 - 604; http://dx.doi.org/10.1016/j.coi.2011.08.003; PMID: 21889323
- Rosenthal LA, Winestock KD, Finbloom DS. IL-2 and IL-7 induce heterodimerization of STAT5 isoforms in human peripheral blood T lymphoblasts. Cell Immunol 1997; 181:172 - 81; http://dx.doi.org/10.1006/cimm.1997.1208; PMID: 9398404
- Jones LL, Vignali DA. Molecular interactions within the IL-6/IL-12 cytokine/receptor superfamily. Immunol Res 2011; 51:5 - 14; http://dx.doi.org/10.1007/s12026-011-8209-y; PMID: 21553332
- Haan S, Keller JF, Behrmann I, Heinrich PC, Haan C. Multiple reasons for an inefficient STAT1 response upon IL-6-type cytokine stimulation. Cell Signal 2005; 17:1542 - 50; http://dx.doi.org/10.1016/j.cellsig.2005.03.010; PMID: 15935617
- Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol 2012; 13:722 - 8; http://dx.doi.org/10.1038/ni.2366; PMID: 22814351
- Trinchieri G, Pflanz S, Kastelein RA. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity 2003; 19:641 - 4; http://dx.doi.org/10.1016/S1074-7613(03)00296-6; PMID: 14614851
- Lankford CS, Frucht DM. A unique role for IL-23 in promoting cellular immunity. J Leukoc Biol 2003; 73:49 - 56; http://dx.doi.org/10.1189/jlb.0602326; PMID: 12525561
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem 2007; 282:9358 - 63; http://dx.doi.org/10.1074/jbc.C600321200; PMID: 17277312
- Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol 2002; 168:5699 - 708; PMID: 12023369
- Guzzo C, Che Mat NF, Gee K. Interleukin-27 induces a STAT1/3- and NF-kappaB-dependent proinflammatory cytokine profile in human monocytes. J Biol Chem 2010; 285:24404 - 11; http://dx.doi.org/10.1074/jbc.M110.112599; PMID: 20519510
- Collison LW, Delgoffe GM, Guy CS, Vignali KM, Chaturvedi V, Fairweather D, et al. The composition and signaling of the IL-35 receptor are unconventional. Nat Immunol 2012; 13:290 - 9; http://dx.doi.org/10.1038/ni.2227; PMID: 22306691
- Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 2007; 450:566 - 9; http://dx.doi.org/10.1038/nature06306; PMID: 18033300
- Collison LW, Chaturvedi V, Henderson AL, Giacomin PR, Guy C, Bankoti J, et al. IL-35-mediated induction of a potent regulatory T cell population. Nat Immunol 2010; 11:1093 - 101; http://dx.doi.org/10.1038/ni.1952; PMID: 20953201
- Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 1998; 394:145 - 51; http://dx.doi.org/10.1038/28101; PMID: 9671298
- Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE Jr., Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 1998; 93:827 - 39; http://dx.doi.org/10.1016/S0092-8674(00)81443-9; PMID: 9630226
- Heim MH, Kerr IM, Stark GR, Darnell JE Jr.. Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science 1995; 267:1347 - 9; http://dx.doi.org/10.1126/science.7871432; PMID: 7871432
- Gupta S, Jiang M, Pernis AB. IFN-alpha activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J Immunol 1999; 163:3834 - 41; PMID: 10490982
- Eriksen KW, Sommer VH, Woetmann A, Rasmussen AB, Brender C, Svejgaard A, et al. Bi-phasic effect of interferon (IFN)-alpha: IFN-alpha up- and down-regulates interleukin-4 signaling in human T cells. J Biol Chem 2004; 279:169 - 76; http://dx.doi.org/10.1074/jbc.M310472200; PMID: 14559900
- John S, Vinkemeier U, Soldaini E, Darnell JE Jr., Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol 1999; 19:1910 - 8; PMID: 10022878
- Murphy TL, Geissal ED, Farrar JD, Murphy KM. Role of the Stat4 N domain in receptor proximal tyrosine phosphorylation. Mol Cell Biol 2000; 20:7121 - 31; http://dx.doi.org/10.1128/MCB.20.19.7121-7131.2000; PMID: 10982828
- Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur Rasheed M, et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 2012; 36:586 - 99; http://dx.doi.org/10.1016/j.immuni.2012.02.017; PMID: 22520852
- Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem 2006; 281:14111 - 8; http://dx.doi.org/10.1074/jbc.M511797200; PMID: 16571725
- Briken V, Ruffner H, Schultz U, Schwarz A, Reis LF, Strehlow I, et al. Interferon regulatory factor 1 is required for mouse Gbp gene activation by gamma interferon. Mol Cell Biol 1995; 15:975 - 82; PMID: 7823961
- Hilkens CM, Schlaak JF, Kerr IM. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J Immunol 2003; 171:5255 - 63; PMID: 14607926
- O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol 2011; 11:239 - 50; http://dx.doi.org/10.1038/nri2958; PMID: 21436836
- O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 2012; 36:542 - 50; http://dx.doi.org/10.1016/j.immuni.2012.03.014; PMID: 22520847