Abstract
The generation of germline gene mutations in mice has been an invaluable tool for experimental biology. However, studying immune responses that develop in the absence of a specific protein that could alter thymic selection complicates experimental interpretations. We observed that CD8+ T cells from Stat6−/− mice displayed “autoreactivity” to STAT6-expressing cells, associated with specific STAT6 peptides binding to MHC class I molecules. These results suggest caution in interpreting experiments where STAT6-expressing cells are transferred into Stat6−/− mice, or where adoptive transfer of Stat6−/− lymphocytes is performed. Our results further highlight additional considerations when studying immune responses involving cell transfer into gene-deficient mice.
Introduction
Signal transducer and activator of transcription (STAT) family proteins are requisite molecules in the development of T helper subsets. STAT1 and STAT4 are required for the development of Th1 cells that secrete IFNγ and mediate immunity to intracellular pathogens. STAT3 promotes Th17 development and the secretion of IL-17, a cytokine required for immunity to extracellular bacterial and fungal infections. STAT6 is required for the differentiation of Th2 and Th9 cells, which contribute to extracellular parasite immunity.Citation1
The in vivo function of STAT6 has been defined largely following the generation of gene-deficient mice. Stat6−/− mice develop normally, breed as homozygotes, but have altered immune responses in vivo.Citation2-Citation5 In the absence of STAT6, mice are resistant to the development of allergic inflammation, and have exaggerated responses to inflammatory insults resulting in greater immunity to intracellular pathogens, but greater susceptibility to autoimmune inflammation, with differences in phenotype attributed to different mouse lines with targeted Stat6 alleles.Citation2,Citation6 Some of the studies that defined these functions utilized experiments involving adoptive transfer of lymphocytes, or where cells or tumors were adoptively transferred into Stat6−/− hosts. Whether cells behave normally in these transfer experiments has not been extensively examined.
In experiments to test if deficiency in STAT4 or STAT6 affected development of CTL specific for non-classical MHC molecules, we observed autoreactivity of Stat6−/− CTL for STAT6-expressing cells. The development of STAT6-specific reactivity by Stat6−/− CD8+ T cells might have important implications for the interpretation of experiments where Stat6−/− CD8+ T cells are exposed to STAT6 expressing cells in vivo.
Results
To examine the responses of BALB/c, Stat4−/− and Stat6−/− mice (H2d, Qa1b and Mtaa) as responders against NZB/B1NJ (NZB) (H2d, Qa1a and Mtab) stimulator cells, we generated CTL lines from each strain. We found no consistent differences in the ability of the Stat4−/− or Stat6−/− CD8+ T cells to develop responder CTL against Qa1a or Mtab antigens, compared with BALB/c T cells (). However, we did observe that Stat6−/− anti-NZB CTL could lyse target cells from Stat4−/− and BALB/c mice (). In contrast, BALB/c and Stat4−/− responders did not lyse BALB/c, Stat4−/− or Stat6−/− cells (). The ability of Stat6−/− CTL to lyse these target cells was unexpected because Stat4−/−, Stat6−/− and BALB/c are syngeneic, differing only in the regions adjacent to the targeted Stat4 or Stat6 alleles.
Figure 1.Stat6−/− CTL lyse Stat6+/+ targets cells. CD8+ CTL lines generated from secondary MLCs were assayed against Stat6+/+ and Stat6−/− Con A blast (CAB) target cells in a standard 51Cr release assay. (A) BALB/c, (B) Stat4−/− and (C) Stat6−/− anti-NZB responder CTL were assayed against NZB, BALB/c, Stat4−/− or Stat6−/− CAB target cells. Lysis of NZB cells in (A) and (B) represents Mtab and/or Qa1a restricted responses. Stat6−/− anti-NZB CTL (C) lyse NZB, Stat4−/− and BALB/c, but not Stat6−/− target cells, indicating lysis of Stat6+/+ cells. (D) Stat6−/− anti-BALB/c, (E) Stat6−/− anti-Stat4−/− CTL and (F) Stat4−/− anti-Stat6−/− MLC were assayed against BALB/c, Stat4−/−, Stat6−/−, H2b, B6.Tlaa or H2r (B10.RIII) CAB target cells. All data are the mean ± SD of triplicate assays and are representative of three or more experiments.
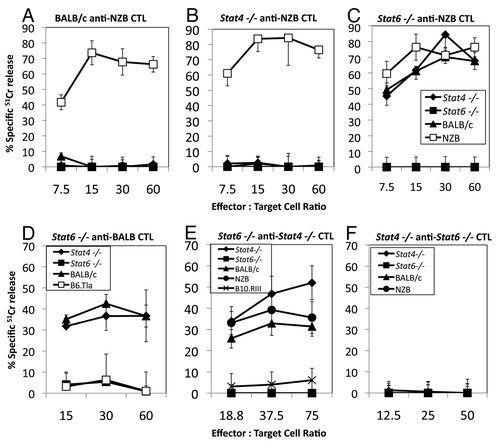
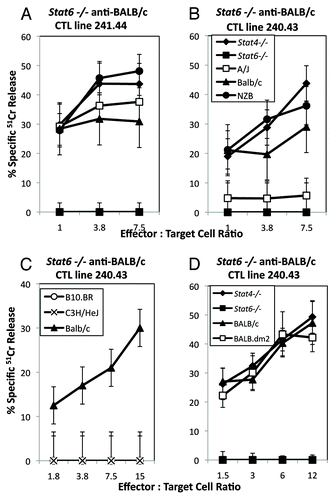
To confirm these data and to rule out problems due to the use of one specific mouse strain (NZB), we examined responses of BALB/c, Stat4−/− and Stat6−/− as stimulators and responders of CTL in similar assays. Stat6−/− anti-BALB/c and Stat6−/− anti-Stat4−/− responder CTL also lyse Stat4−/− and BALB/c target cells (), whereas Stat4−/− anti-Stat6−/− CTL do not lyse Stat4−/−, Stat6−/− or BALB/c target cells ().
To determine the restricting MHC molecule involved in these responses, we examined mice expressing different MHC haplotypes and found the responses were restricted to H2d. Stat6−/− anti-Stat4−/− or anti-BALB/c CTL lysed target cells from H2d (BALB/c, Stat4−/− and NZB) mouse strains ( and ), but not cells expressing H2b (B6.Tlaa), H2r (B10.RIII), or H2k (C3H/HeJ and B10.BR) ( and ), suggesting that the CTL were not alloreactive. To determine whether the restricting element(s) are H2-Kd, -Dd and/or -Ld molecules, we further compared Stat6−/− CTL responses against A/J (H2-Kk, -Dd and -Ld), BALB/c (H2-Kd, -Dd and -Ld) and BALB.dm2 (H2-Kd, -Dd and L null) target cells using two different Stat6−/− CTL lines. illustrates that either H2-Kd, -Dd or -Ld () are recognized by the polyclonal responder CTL. Since H2k cells (C3H.HeJ or B10.BR) were not lysed by Stat6−/− CTL (), recognition of cells from A/J (H2-Kk, -Dd and -Ld) by the 241.44 line shows restriction to H2-Dd and/or -Ld (). In contrast, the lack of response of the 240.43 line demonstrates the failure of these cells to recognize H2-Dd, suggesting the reactivity against another H2d class I molecule, most likely H2-Kd (). H2-Kd restricted CTL were also able to lyse BALB.dm2 cells, which express H2-Kd and H2-Dd, but not H2-Ld, further suggesting that H2-Ld restriction is not a prominent reactivity in these CTL lines (). Thus, Stat6−/− CTL can develop with specificities restricted to H2-Kd, H2-Dd or H2-Ld.
Figure 2.Stat6−/− anti-Stat6+/+ responses are H2-Kd and H2-Dd or -Ld restricted. Stat6 −/− anti-BALB/c CTL lines (A) 241.44, (B–D) 240.43 were incubated with target cells from H2d, or H2k expressing mouse strains in a standard 51Cr release assay. CTL line 241.44 (A) (H2-Dd and -Ld-restricted) lyses A/J (H2-Kk, -Dd and -Ld) target cells, whereas CTL line 240.43 (B) (H2Kd-restricted) did not lyse A/J or (D) BALB.dm2 (H2KdDd). Neither line lysed H2k target cells (C, data not shown). Data are the mean ± SD of triplicate assays and are representative of two or more experiments.
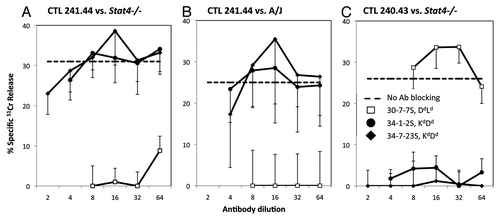
Confirming these data, in we show that pre-incubating A/J or Stat4−/− target cells with monoclonal antibodies reactive with H2-Kd, -Dd and H2-Ld molecules blocks target cell recognition and lysis by these CTL. Lysis of Stat4−/− or A/J target cells is blocked by monoclonal antibody 30-5-7S that reacts with H2-Dd or -Ld but not H2-Kd or H2-Kk, confirming restriction of this line to H2-Ld (). In , lysis of Stat4−/− targets by the Stat6−/− anti-BALB 240.43 line is not blocked by 30-5-7S, but rather is blocked by the two antibodies which bind H2-Kd molecules, 34-1-2S and 34-7-23S, demonstrating restriction to H2-Kd. Taken together, the data clearly demonstrate that Stat6−/− anti-BALB/c or anti-Stat4−/− CTL are restricted to either H2-Kd,-Dd or -Ld.
Figure 3.Stat6−/− anti-BALB/c or Stat4−/− CTL are H2-Kd, -Dd or Ld restricted. (A–C) Stat6−/− anti-BALB/c CTL lines 241.44 (A and B), and 240.43 (C) were incubated with CAB target cells from Stat4−/− (A and C) or A/J (B) mice and cytotoxicity was measured in a standard 51Cr release assay. The ability of anti-MHC I antibodies to block lysis was tested. Lysis in the absence of blocking antibodies is indicated as a dashed line. Data are the mean ± SD of triplicate assays and are representative of two or more experiments.
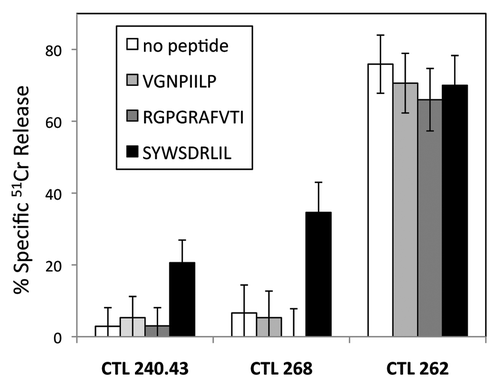
Since the primary difference between BALB/c or Stat4−/− and Stat6−/− mice is the Stat6 gene product, we tested whether Stat6−/− CTL recognized peptides derived from the STAT6 protein. We tested a STAT6 peptide previously shown to be presented by tumor cellsCitation7 and observed the H2-Kd restricted CTL recognize Stat6−/− target cells incubated with the SYWSDRLIL peptide (), which fits the H2-Kd consensus motif of XYX6(I/L/V)L. We additionally tried several STAT6 peptides that fit the H2-Dd or H2-Ld consensus motifs, (XGPX(K/R)X3(L/I/F) and (XPX6(L/M/F), respectively, but none conferred reactivity.
Figure 4. Kd-restricted Stat6−/− CTL recognize a peptide derived from the STAT6 protein. Stat6−/− anti-BALB/c CTL 240.43 (left) or Stat6−/− anti-Stat4−/− CTL 268 (middle) were incubated with Stat6−/− CAB target cells at an effector:target cell ratio of 12:1 in a standard 51Cr release assay. Cytotoxicity was assessed in the absence or presence of 9-mer peptides from H-2Kd binding peptides from HIV gp160 (RGPGRAFVTI, VGNPIILP), or STAT6 (SYWSDRLIL). The alloreactive B10 (H2b) anti-BALB/c (H2d) CTL 262 (right) did not require peptide for lysis and was not affected by target cell incubation with peptides. Data are the mean ± SD of triplicate assays and are representative of three or more experiments.
Discussion
These data demonstrate responses by Stat6−/− CTL against H2d cells from Stat6-expressing mouse strains including BALB/c, NZB and Stat4−/−. The polyclonal CTL populations generated from the Stat6−/− responder spleen cells initially recognized Kd, -Dd and -Ld restricted antigens, although with weekly passage in vitro some populations eventually self-selected for a predominantly H2-Kd, -Dd or -Ld restricted phenotype. Use of target cells from recombinant inbred mouse strains and blocking by antisera specific for H2d class I molecules confirms their MHC restriction. The H2-Kd restricted CTL recognized the STAT6-derived peptide SYWSDRLIL.
These data clearly illustrate the importance of running negative control experiments, particularly when working with genetically altered strains of mice. In some situations the responses described here may not obviously affect experimental results. For example, in evaluations of immune responses within Stat6−/− mice against exogenous antigens or allergens, or in studies of inflammation or hypersensitivity, the results may show no effects easily attributable to altered peptide presentation by MHC class I molecules on Stat6−/− cells.Citation2,Citation8 However, it is possible that results could be influenced by subtle changes in reactivity of cells maturing in the Stat6−/− environment and/or by the unusual peptide repertoire transferred on H2d molecules with Stat6−/− cells. Such changes in immune responses due to these altered peptide expression might be negligible or difficult to identify and quantitate, but should nonetheless be considered.
However, the situation could be more complicated in adoptive transfer experiments. Although most adoptive transfer experiments involving Stat6−/− mice use transferred purified CD4 T cells, in experiments where Stat6−/− splenocytes or CD8 T cells are transferred to wild type recipients,Citation9,Citation10 it is possible that STAT6-specific CD8 T cells could mediate a graft vs. host response, resulting in background inflammation, apart from the specific responses being studied. Conversely, when STAT6-expressing cells are transferred into Stat6−/− recipients, it is possible that Stat6−/− CD8 T cells reactive against STAT6 peptides might eliminate transferred cells, masking some functional activity of the transferred cells.Citation10-Citation13 The results from most of these experiments are consistent with a phenotype arising from STAT6-deficiency, suggesting that the contribution of anti-STAT6 immunity may be modest.
The contribution of anti-STAT6 CTL responses may be more important in studies of graft rejection. Two reportsCitation14,Citation15 demonstrated that Stat6−/− mice demonstrated shorter graft survival times in models of cardiac transplant with minor histocompatibility differences, and where CTLA4-Ig was used to block graft rejection. Although it is possible that some of this increase is due to the increased propensity of Stat6−/− T cells to develop into inflammatory T cells, it is likely that anti-STAT6 immunity may also be contributing to the observed phenotype.
The anti-STAT6 immunity that develops in mice lacking endogenous STAT6 clearly contributes to the increased anti-tumor immunity in Stat6−/− mice. For example, Ostrand-Rosenberg, et al.Citation16 have shown delayed and reduced primary mammary carcinoma growth in Stat6−/− mice using 4T1, a BALB/c mammary carcinoma that is usually malignant, non-immunogenic and metastatic. The authors hypothesize that the deletion of the Stat6 gene facilitates development of potent anti-tumor immunity via a CD4+-independent pathway. Our data suggest that immunization with the H2d STAT6-expressing tumor might simply stimulate CD8+ CTL specific for H2d restricted antigens from STAT6-expressing cells, which lyse the cells regardless of expression of tumor antigens. Indeed, Jensen et al. demonstrate that enhanced immunity to 4T1 cells in Stat6−/− mice is dependent upon the STAT6 peptide used in our studies.Citation7 Similarly, Kacha et al.Citation17 show that effector cells from Stat6−/− mice have increased lytic activity and produce increased levels of IFNγ in response to the H2d tumor P1.HTR. Thus, it is not clear that these studies actually demonstrate an effect of STAT6 function, but rather may reflect immune responses that are not tolerant to STAT6 peptides presented in the context of MHC I.
It is important to note that “autoreactivity” to a missing self-protein is not a ubiquitous phenomenon. Although there is clearly an anti-STAT6 CTL response in Stat6−/− mice, there is no corresponding anti-STAT4 CTL response in Stat4−/− mice. There could be multiple reasons for the restricted nature of these observations. First, not all proteins have MHC class I binding peptides. It is possible that STAT4 peptides do not compete effectively for binding to MHC class I such that no reactive cells are positively selected in the thymus. Alternatively, MHC class I binding peptides from STAT4 might be homologous enough to other endogenous peptides that reactive T cells are negatively selected in the thymus. The differences between the phenotypes of Stat4−/− and Stat6−/− CTL illustrate that the autoreactive phenomenon described here must be tested empirically for each mutant strain being studied.
In summary, our data illustrate the importance of demonstrating maintenance of self-tolerance in mice made defective in a particular gene by homologous recombination. Particularly in genes that have immune function, where common techniques involve transplantation and adoptive transfer, it is critical to define the ability of T cells to recognize peptides from the targeted proteins as foreign. Failure to do so could result in misinterpretation of how a gene of interest contributes to immune responses.
Material and Methods
Mice
Stat4−/− and Stat6−/− mice were generated and backcrossed for at least 10 generations to BALB/c mice as described previously.Citation3,Citation18 B6.Tlaa, B10.BR, C3H/HeJ breeder mice and BALB/c.dm2 spleen cells were the kind gift of Dr James Forman. BALB/c, C57BL/6J, NZB/B1NJ, C3H/HeJ, B10.BR and A/J breeder mice were purchased from The Jackson Laboratory. All animals were bred and housed in the IUSM-Evansville animal facility and procedures were approved by the IUSM IACUC. Where indicated, mice were immunized intraperitoneally with 25 × 106 spleen cells in 0.5 mL balanced salt solution, rested for at least 7 d, euthanized, and their spleens removed for the in vitro generation of CTL.
Cell lines and assays
Using specific strain combinations as described, CTL were generated in secondary MLC as described.Citation19 Briefly, 5 × 106 responder and 5 × 106 irradiated (3000Rad) stimulator spleen cells were incubated for 6–7 d in RPMI 1640 supplemented with 7–10% fetal bovine serum, 1% L-glutamine and 50 ng/mL 2-mercaptoethanol at 37°C in 7% CO2. Some CTL lines were re-stimulated weekly with irradiated stimulator cells in supplemented Mishell-Dutton medium that also contained T cell growth factors from (NH4)2SO4-precipitated supernatants of phorbol myristic acetate-stimulated EL4.IL2 cells.Citation20 Generation of concanavalin A (ConA) blast cells (CAB), 51Cr labeling of target cells, the standard 4–6 h 51Cr release assay, controls, and the method for determination of the % specific lysis of target cells have been described.Citation21 Standard error was computed by propagation of errors as previously described.Citation21 51Cr release assays for analysis of peptide binding, peptide blocking and antibody blocking were performed using pre-incubations as previously described.Citation22 RMA-S-Dd, T2-Dd, E3 and LTKD cell lines were provided by Dr D. Marguiles. RMA-S-Ld cell lines were provided by Dr Ted Hansen. Hybridomas producing anti-MHC antibodies were purchased from ATCC. Peptides were purchased from Genemed Synthesis. Flow cytometry analyses of CTL lines was performed as previously described.Citation19 using anti-CD4 antibody GK1.4 (PharMingen) and anti-CD8α antibody YTS169.4 (kindly provided by Dr. James Forman,) The CTL were CD8+ and CD4−, as expected.
Acknowledgments
The authors thank Diana Fuqua and Lucy Shuck for animal care, and Rachel Gahagan and Daniel Walsh for help with figure generation. This work was supported in part by American Cancer Society Grant IRG 161-L.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Goenka S, Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc Natl Acad Sci U S A 2006; 103:4210 - 5; http://dx.doi.org/10.1073/pnas.0506981103; PMID: 16537510
- Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res 2011; 50:87 - 96; http://dx.doi.org/10.1007/s12026-011-8205-2; PMID: 21442426
- Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 1996; 4:313 - 9; http://dx.doi.org/10.1016/S1074-7613(00)80439-2; PMID: 8624821
- Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature 1996; 380:630 - 3; http://dx.doi.org/10.1038/380630a0; PMID: 8602264
- Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S-I, et al. Essential role of Stat6 in IL-4 signalling. Nature 1996; 380:627 - 30; http://dx.doi.org/10.1038/380627a0; PMID: 8602263
- Wang Y, Evans JT, Rodriguez F, Fields P, Mueller C, Chitnis T, et al. A tale of two STAT6 knock out mice in the induction of experimental autoimmune encephalomyelitis. J Neuroimmunol 2009; 206:76 - 85; http://dx.doi.org/10.1016/j.jneuroim.2008.11.003; PMID: 19100630
- Jensen SM, Meijer SL, Kurt RA, Urba WJ, Hu HM, Fox BA. Regression of a mammary adenocarcinoma in STAT6-/- mice is dependent on the presence of STAT6-reactive T cells. J Immunol 2003; 170:2014 - 21; PMID: 12574371
- Glosson NL, Bruns HA, Kaplan MH. Wheezing and Itching: The Role of STAT proteins in allergic inflammation. JAK-STAT 2012; 1:3 - 12; http://dx.doi.org/10.4161/jkst.19086
- Chitnis T, Najafian N, Benou C, Salama AD, Grusby MJ, Sayegh MH, et al. Effect of targeted disruption of STAT4 and STAT6 on the induction of experimental autoimmune encephalomyelitis. J Clin Invest 2001; 108:739 - 47; PMID: 11544280
- Tomkinson A, Duez C, Lahn M, Gelfand EW. Adoptive transfer of T cells induces airway hyperresponsiveness independently of airway eosinophilia but in a signal transducer and activator of transcription 6-dependent manner. J Allergy Clin Immunol 2002; 109:810 - 6; http://dx.doi.org/10.1067/mai.2002.123531; PMID: 11994705
- Jin D, Takamoto M, Hu T, Taki S, Sugane K. STAT6 signalling is important in CD8 T-cell activation and defence against Toxoplasma gondii infection in the brain. Immunology 2009; 127:187 - 95; http://dx.doi.org/10.1111/j.1365-2567.2008.02935.x; PMID: 18795973
- Mattes J, Yang M, Siqueira A, Clark K, MacKenzie J, McKenzie AN, et al. IL-13 induces airways hyperreactivity independently of the IL-4R alpha chain in the allergic lung. J Immunol 2001; 167:1683 - 92; PMID: 11466392
- Yokozeki H, Ghoreishi M, Takagawa S, Takayama K, Satoh T, Katayama I, et al. Signal transducer and activator of transcription 6 is essential in the induction of contact hypersensitivity. J Exp Med 2000; 191:995 - 1004; http://dx.doi.org/10.1084/jem.191.6.995; PMID: 10727461
- Sho M, Yamada A, Najafian N, Salama AD, Harada H, Sandner SE, et al. Physiological mechanisms of regulating alloimmunity: cytokines, CTLA-4, CD25+ cells, and the alloreactive T cell clone size. J Immunol 2002; 169:3744 - 51; PMID: 12244168
- Zhou P, Szot GL, Guo Z, Kim O, He G, Wang J, et al. Role of STAT4 and STAT6 signaling in allograft rejection and CTLA4-Ig-mediated tolerance. J Immunol 2000; 165:5580 - 7; PMID: 11067913
- Ostrand-Rosenberg S, Grusby MJ, Clements VK. Cutting edge: STAT6-deficient mice have enhanced tumor immunity to primary and metastatic mammary carcinoma. J Immunol 2000; 165:6015 - 9; PMID: 11086031
- Kacha AK, Fallarino F, Markiewicz MA, Gajewski TF. Cutting edge: spontaneous rejection of poorly immunogenic P1.HTR tumors by Stat6-deficient mice. J Immunol 2000; 165:6024 - 8; PMID: 11086033
- Kaplan MH, Sun Y-L, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 1996; 382:174 - 7; http://dx.doi.org/10.1038/382174a0; PMID: 8700209
- Jenkins RN, Aldrich CJ, Landolfi NF, Rich RR. Correlation of Qa-1 determinants defined by antisera and by cytotoxic T lymphocytes. Immunogenetics 1985; 21:215 - 25; http://dx.doi.org/10.1007/BF00375374; PMID: 2580784
- Jenkins RN, Rich RR. Characterization of determinants encoded by four Qa-1 genotypes and their recognition by cloned cytotoxic T lymphocytes. J Immunol 1983; 131:2147 - 53; PMID: 6195252
- Jenkins RN, Aldrich CJ, Lopez LA, Rich RR. Oligosaccharide-dependent and independent Qa-1 determinants. J Immunol 1985; 134:3218 - 25; PMID: 2580018
- Aldrich CJ, Rodgers JR, Rich RR. Regulation of Qa-1 expression and determinant modification by an H-2D-linked gene, Qdm. Immunogenetics 1988; 28:334 - 44; http://dx.doi.org/10.1007/BF00364232; PMID: 2459056