Abstract
To interpret pharmacokinetic (PK) data of biotherapeutics, it is critical to understand which drug species is being measured by the PK assay. For therapeutic antibodies, it is generally accepted that “free” circulating antibodies are the pharmacologically active form needed to determine the PK/ pharmacodynamic (PD) relationship, safety margin calculations, and dose projections from animals to humans and the eventual characterization of the exposure in the clinic. However, “total” drug may be important in evaluating the dynamic interaction between the drug and the target, as well as the total drug exposure. In the absence of or with low amounts of soluble ligand /shed receptor, total and free drug species are often equivalent and their detection is less sensitive to assay formats or reagent choices. In contrast, in the presence of a significant amount of ligand, assay design and characterization of assay reagents are critical to understanding the PK profiles. Here, we present case studies where different assay formats affected measured PK profiles and data interpretation. The results from reagent characterizations provide a potential explanation for the observed discrepancies and highlight the importance of reagent characterization in understanding which drug species are being measured to accurately interpret PK parameters.
Introduction
Monoclonal antibodies (mAbs) are a promising and rapidly growing class of targeted therapeutics for treating variety of indications including oncology, autoimmune, cardiovascular, and infectious diseases. To ensure the safety and efficacy of these molecules, robust bioanalytical pharmacokinetic (PK) assays that can reliably quantify drug concentrations are required. This data are used to assess drug exposure and safety as well as to characterize PK/pharmacodynamic (PD) relationships. There is an ongoing debate, however, as to what form of the mAb therapeutic (“free” or “total”) is most relevant to measure.Citation1 Due to the bivalency intrinsic to mAbs, multiple forms of the drug can exist simultaneously in vivo in the presence of soluble target.Citation2 These include non-complexed free drug and partially or completely complexed forms where one or both binding sites are occupied. Multiple complexed forms may also exist if the soluble target has more than one binding site.Citation3
The presence of ligand or shed receptor in sera often depends on the mechanism of action of the drug and the biology of the disease. The “total” and “free” drug species are often equivalent in the absence or at low levels of circulating target.Citation2 High concentrations of circulating target, however, could potentially have a significant effect on PKCitation1 and may result in nonlinear and linear mixed profiles. It is often difficult to predict the concentrations of a circulating target/shed receptor, especially after drug treatment because target levels may become elevated. In addition, the level of target being shed from cell membranes may be variable depending on the disease biology. Therefore, estimation of the potential effect on PK results often comes from experimental data rather than theoretical predictions.
The actual drug species being measured often depends on the tools/reagents available for assay development. The target binding enzyme-linked immunosorbent assay (direct ELISA) is the method of choice for quantifying free drug levels, but this format frequently measures a mixture of different drug species because of the bivalency of mAb therapeutics, choice of detection reagents and the dilution scheme used for samples. Other formats for PK measurements include use of polyclonal [polyclonal anti-therapeutic complementarity-determining region (CDR) assay, or PAC assay], or monoclonal antibodies (monoclonal anti-therapeutic CDR assay, or MAC assay). PAC assays are generally used to detect total drug by utilizing multiple binding epitopes of polyclonal antibodies against the analyte; however, it is difficult to predict which drug species (total or free) are detected by mAbs. As reagents, mAbs are often selected to detect the CDR region of antibody therapeutics and therefore are believed to block drug-target interactions, but this is not always the case.
In this study, we compared the effect of different assay formats and reagents on the PK profiles of three anti-CD20 molecules, chimeric rituximab and two humanized Abs, ocrelizumab and v114 (PRO131921)Citation4,Citation5, which share similar antigen-binding characteristics as rituximab. The MAC assay format for ocrelizumab in rheumatoid arthritis (RA) patients resulted in significant differences in the PK profiles compared with that of rituximab PK profiles in the same patient population; however, a PAC assay similar to that used in the rituximab studies generated comparable PK profiles for ocrelizumab and rituximab. For v114, the MAC and PAC assays generated equivalent clinical PK data in non-Hodgkin lymphoma (NHL) patients. To illustrate that our observations with ocrelizumab and rituximab PK assays were not simply due to the biology of the disease (RA vs. NHL), the MAC and the PAC PK profiles for ocrelizumab in both RA and NHL populations were evaluated. Reagent characterizations provided potential explanations highlighting the importance of assay format and choice of reagents on PK parameters.
Results
Comparison of PK Data for Rituximab, Ocrelizumab and v114
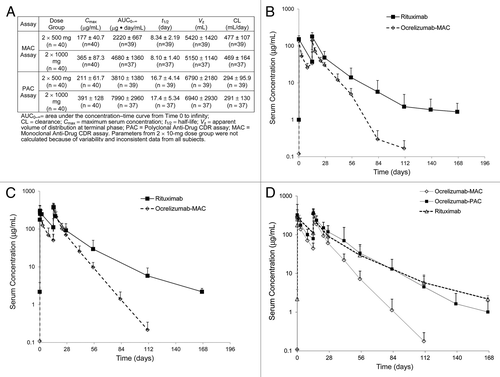
The serum concentrations of ocrelizumab in RA patients treated during the Phase1/2 ACTION trial (ClinicalTrials.gov identifier: NCT00077870)Citation6_ENREF_4 were initially quantified using the MAC assay format, the serum concentration-time profiles are presented in and . The data generated from this trial had unexpected PK differences, compared with historical data for rituximab.Citation7 Concentration-time profiles of rituximab and ocrelizumab following intravenous (IV) infusion of either 500 or 1000 mg on Day 1 and Day 15 in the RA patient population are found in and . Interestingly, the profiles suggest that ocrelizumab appeared to clear 3 times faster than rituximab at all doses tested. The ocrelizumab mean terminal half-life was calculated to be approximately 8.0 d, while the rituximab mean terminal half-life was approximately 19–22 d.Citation7
Figure 1. Pharmacokinetic Parameters of Evaluable Subjects for ocrelizumab (monoclonal anti-drug CDR (MAC) Assay and polyclonal anti-drug CDR (PAC) Assay (A). Averaged concentration-time profiles of rituximab and ocrelizumab following IV infusion of 500 mg (B) or 1000 mg (C) on Day 1 and Day 15 to human patients during phase I/II, randomized, placebo controlled, double blinded, multicenter ACTION clinical study. Averaged concentration-time profiles of ocrelizumab (PAC and MAC assay) in comparison with rituximab PK profile (PAC assay) following IV infusion of 1000 mg on Day 1 and Day 15 to human patients from the ACTION study (D).
To understand this difference, we first considered two characteristics known about these molecules: 1) rituximab is chimeric while ocrelizumab is humanized; and 2) they bind to different, but overlapping, epitopes of the extracellular domain of CD20 with similar affinities. In vitro, ocrelizumab demonstrated enhanced antibody-dependent cell-mediated cytotoxicity (ADCC) and reduced complement-dependent cytotoxicity (CDC) compared with rituximab.Citation8 Both molecules have similar binding activity to the neonatal Fc receptor (FcRn) in vitro, suggesting that the mechanism for generic IgG antibody recycling is not a factor in this faster clearance (data not shown). Furthermore, the difference in the PK terminal half-life could not be attributed to presence of anti-therapeutic antibodies (ATA). There was only one positive individual in the ocrelizumab study in the 1000 mg dose group; the data for this individual was excluded from the calculations. In addition, similar PK profiles were observed for the two anti-CD20 antibodies from pre-clinical studies in cynomolgus monkeys (binding species), when a generic polyclonal antibody based PK assay was used to quantify concentrations of rituximab and ocrelizumab (unpublished data).
These results prompted investigations of the human PK assay formats and reagents used in each assay to understand the PK differences observed. The assay format used for rituximab quantification was a polyclonal anti-CDR (PAC) assay, and for ocrelizumab, a monoclonal anti-CDR (MAC) assay. The main difference in the formats of these assays is the use of either CDR enriched polyclonal for capture and a generic anti-Hu IgG1 for detection or CDR specific mAb reagents for both capture and detection. To evaluate whether the differences observed were due to assay format, a PAC assay for ocrelizumab similar to that used for rituximab was developed, validated, and used to evaluate samples from the ACTION study. Mean concentration-time profiles of ocrelizumab derived from ACTION study patients using PAC and MAC assay formats were compared with the rituximab PAC assay profile in an RA patient population (). The data generated using the ocrelizumab and rituximab PAC assays produced similar concentration-time profiles for each drug vs. the concentration-time profiles generated using the ocrelizumab MAC assay. This same trend held regardless of the disease state (RA vs. NHL; ). The half-life of ocrelizumab ranged from 8.10 to 8.34 d (in MAC assay) and 16.7 to 17.4 d (in PAC assay) between the 2 × 500- and 2 × 1000-mg dose groups in RA patients. In comparing the PK parameters calculated from both assays (), we observed that the t1/2, AUC0-∞ increased with increasing dose and the CL decreased with increasing dose; this was expected based on target-mediated elimination. However, the maximum serum concentrations (Cmax) of ocrelizumab calculated from MAC and PAC assay were similar, indicating that the quantification of ocrelizumab by the two assays did not differ during the peak of the serum-concentration profile ().
Figure 2. Representative concentration-time profiles of ocrelizumab (A) and v114 (B) in samples collected during NHL clinical study. Concentrations were determined by ELISA based polyclonal anti-drug CDR (PAC) and monoclonal anti-drug CDR (MAC) assays. Both of the v114 assay formats (MAC and PAC) showed remarkably similar PK profiles as compared with the two ocrelizumab assay formats.
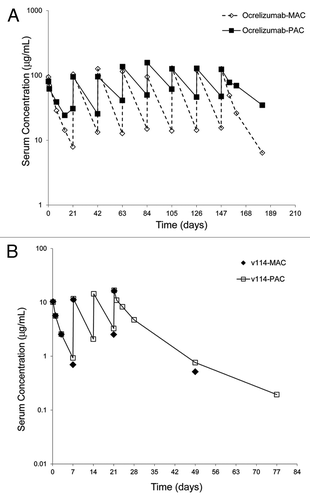
The concentration-time profiles for v114 were also evaluated using both PAC and MAC assay formats in the NHL patient population. shows a typical PK profile for a single individual from Cohort A () at the 100 mg/m2 total dose. Unlike the discrepancies observed with the data generated utilizing the MAC and the PAC assay formats for ocrelizumab, both assay formats produced remarkably similar PK profiles for v114, with similar concentration levels at evaluated time points. The mean terminal half-life for v114 was approximately 22–28 d, comparable to the PAC terminal half-life for ocrelizumab and rituximab molecules. This data (v114 terminal half-life) was generated with the PAC assay format only because of limited samples evaluated with the MAC assay format.
Table 2. Ocrelizumab and v114 study designs in the NHL patient population
CD20 Interference
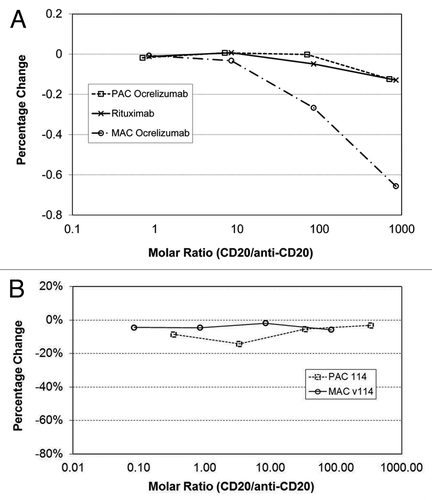
Elevated levels of circulating CD20 (cCD20) have been detected in blood of NHL, Hodgkin lymphomas,Citation9 and chronic lymphocytic leukemia (CLL) patients.Citation10 To evaluate the hypothesis that cCD20 may be interfering with the samples and causing the differences observed with the various assay formats, recombinant CD20 at various levels was used to characterize the observed effect. These experiments were conducted using the rituximab PAC assay format, ocrelizumab MAC and PAC assay formats, as well as v114 MAC and PAC assay formats, and the results are presented in and . The effect of increasing molar ratio of CD20 to rituximab or ocrelizumab () as well as to v114 () were evaluated in the anti-CD20 recovery (rituximab, ocrelizumab or v114) experiment using the various assay formats. No effect on drug recovery was observed with the rituximab (PAC) or ocrelizumab (PAC) assay formats; however, significant under-recovery was observed with the ocrelizumab MAC assay format. On the contrary, CD20 interference was not found in either v114 MAC or PAC assay formats. This data suggests that CD20 interferes with the mAbs used for detection of ocrelizumab but not with the mAb reagents used for the detection of v114 assay or the polyclonal reagents used in the PAC assay formats.
Figure 3. (A) CD20 Interference was tested in the rituximab PAC, ocrelizumab MAC and ocrelizumab PAC PK assays using samples with various molar ratios of recombinant CD20 and either rituximab or ocrelizumab. CD20 interference was only observed in the ocrelizumab MAC assay. Polyclonal anti-drug CDR (PAC) assay; monoclonal anti-drug CDR (MAC) assay; PK – pharmacokinetic. (B) CD20 interference was tested with v114 in both the PAC and MAC versions of the human PK assay. Both v114 assay formats showed remarkably similar PK profiles; thus, CD20 does not interfere in the v114 MAC or PAC assay formats.
Reagent Characterization by Fluorescence Activated Cell Sorting
Further reagent characterization was done by fluorescence activated cell sorting (FACS) analysis; ocrelizumab and v114 can bind to the CD20 molecule on the surface of WIL2-S B lymphoma cells and their binding can be detected by a phycoerythrin (PE)-labeled anti-human IgG antibody by flow cytometer. The monoclonal and polyclonal anti-ocrelizumab and anti-v114 antibodies used in various PK assays were assessed in this FACS assay to evaluate their potential effect in the binding of ocrelizumab or v114 to CD20. Briefly, v114 or ocrelizumab were co-incubated in the presence and absence of each of the mAbs used in their respective MAC assay formats or the polyclonal antibodies used in their respective PAC assay formats. Their binding to CD20 on WIL2-S cells was then compared with binding in the absence of the monoclonal and polyclonal antibody reagents. Antibodies that do not block the CD20 binding domain recognized by ocrelizumab or v114 should permit complete binding of v114 and ocrelizumab to the WIL2-S cells, as demonstrated by the curve on the right with 100% staining (). Interference that completely blocks the binding of v114 and ocrelizumab to the WIL2-S cells results in an absence of staining of the cells, as demonstrated by the curve on the left in , and partial binding should result in partial graphical shift to the left.
Figure 4. (A) Evaluation of the ability of the drug molecules (ocrelizumab/ v114) to bind to the CD20 on the surface of WIL2-S cells in the presence of antibodies used as assay reagents in the PK measurements. No anti-CD20 binding = 100% Stained, complete inhibition of anti-CD20 binding = Unstained, partial interference with anti-CD20 binding = Partially stained. (B) FACS characterization of the reagents used in the ocrelizumab PK assay. At higher concentrations both 8A3 and 2B3 mAbs inhibit ocrelizumab from binding to CD20 on WIL2-S cells. The polyclonal antibody only inhibits CD20 binding at the highest concentration (260 nM). This data indicates that the mAbs and CD20 are binding to the same epitope of ocrelizumab. If ocrelizumab is bound to soluble CD20 in serum the MAC assay will be unable to detect this complex resulting in a “free” PK assay. (C) FACS characterization of the reagents used in the v114 PK assay. Neither 7G7 nor 2D3 mAbs inhibit v114 from binding to CD20 on WIL2-S cells. The polyclonal antibody inhibits v114 binding most significantly at the highest concentration (1000 nM). This data suggests that both the MAC and PAC assay should measure both free and complexed drug equally (i.e., “total” drug).
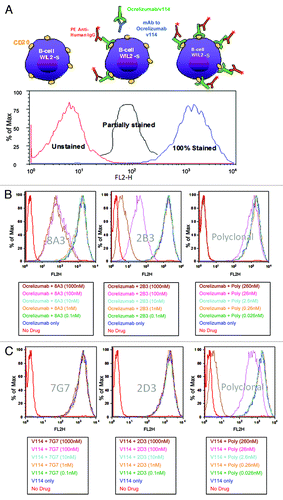
At higher concentrations evaluated (1000 nM), both 8A3 and 2B3 mAbs inhibited ocrelizumab from binding to CD20 on the surface of WIL2-S cells as evident by a curve shift to the left (). This data suggests that 8A3 and 2B3 bind to the paratope on ocrelizumab that is specific for CD20 binding. However, there appears to be a difference in the 8A3 and 2B3 binding as there is a nonlinear dose response observed with 8A3. One possible explanation for the nonlinear dose response observed with 8A3 at 1000 and 100 nM could be due to where this mAbs binds to 2H7. This data may suggest that 8A3 binds 2H7 near instead of directly on the overlapping 2H7/CD20 paratope and sterically hinders this interaction only at higher concentrations. The polyclonal antibody raised against ocrelizumab inhibits ocrelizumab binding to CD20 on WIL2-S cells only at the highest concentrations tested (260 nM) as would be expected ().
Characterization of v114 MAC and PAC Reagents
Neither 7G7 nor 2D3 mAbs inhibit the binding of v114 to CD20 on WIL2-S cells () at as high as1000 nM. As with the ocrelizumab polyclonal antibody, the polyclonal antibody raised against v114 inhibits its binding to CD20 on the surface of WIL2-S cells only at the highest concentrations tested (260 nM).
Discussion
To interpret PK data and evaluate the relationship between drug exposure and clinical outcomes (e.g., efficacy, safety), it is critical to understand what form of the drug is being measured. Although assay reagents and formats can be designed to measure the “total” or “free” forms of a drug, factors such as reagent limitations and sample dilutions can add uncertainty regarding the actual form of the drug being measured. Therefore, it is critical to ensure that all stakeholders understand what is being measured to accurately interpret the data during drug development. Total and free drug species are often equivalent in the absence or presence of low amounts of soluble ligand or shed receptor and are less sensitive to assay format and reagent choices. The presence of high levels of soluble ligand, however, can have a significant effect on the observed PK profile depending on the assay format/reagents used. Therefore, understanding what the PK assays is measuring can be crucial in dose selection and understanding of the exposure, efficacy and toxicity of therapeutic mAbs.Citation11 In this paper, we evaluated the effect of assay format and reagents on the PK profiles of three anti-CD20 molecules, a chimeric mAb rituximab, as well as two humanized mAbs, ocrelizumab and v114, which have similar binding characteristics to rituximab.
In the ocrelizumab clinical study of RA patients, faster clearance was observed for ocrelizumab compared with the historical rituximab PK data (, ). Because ocrelizumab and rituximab have similar binding characteristics to CD20, this result was unexpected. In this study, the terminal elimination half-life of ocrelizumab was observed to be approximately 3 times shorter than rituximab (t1/2 ~8.34 d for 500 mg and ~8.10 d for 1000 mg vs. approximately 19–22 d for rituximab) ( and ). Further evaluation revealed that the difference observed may be due to different assay formats or reagents used in each case. The rituximab assay was a PAC format that used a polyclonal anti-CDR reagents capture reagent and a polyclonal anti-Hu IgG as detection, and the ocrelizumab assay used a MAC format that included monoclonal anti-CDR antibodies as capture and detection reagents. Subsequently, a PAC version of the ocrelizumab assay was developed and results generated using this format were comparable to those for rituximab (). These results suggested that PAC- and MAC-based assays may detect different forms of the drug (free vs. bound). To confirm that this observation is not unique to the RA population, clinical study samples from a NHL population treated with ocrelizumab were evaluated in the MAC and PAC versions of the ocrelizumab assays. A significantly faster clearance using the MAC vs. PAC based assay formats was observed for ocrelizumab in both disease states ( and ).
Interestingly, no differences in PK profiles were observed with the MAC and the PAC assay formats for another anti-CD20 antibody, v114, in the NHL patient population (). We suspected the differences between the ocrelizumab PAC and MAC assay results might be due to interference from circulating CD20 (cCD20) in the MAC assay format.
Because CD20 is a trans-membrane protein that crosses the surface membrane four times,Citation12 it is considered unlikely to be shed from the surface of cells. What then is the source of the cCD20? Several papers have reported the presence of elevated levels of CD20 in blood of NHL, Hodgkin lymphomas,Citation9 and chronic lymphocytic leukemia (CLL) patients.Citation10 Furthermore, results from western blot suggest that the cCD20 is full-length, rather than a cleaved immunoreactive fragment of the protein. It is postulated that the CD20 in circulation is part of a membrane complex or fragment originating from cell breakdownCitation10 and therefore the term circulating CD20 rather than soluble CD20 is being used.
To investigate the potential effect of cCD20 on each of the anti-CD20 assays recoveries, recombinant CD20 was used at increasing concentrations in each assay to evaluate its ability to interfere in the anti-CD20 mAb therapeutic recoveries. Recombinant CD20 interference was only observed utilizing the MAC-based assay for ocrelizumab and not with the PAC assay format for rituximab or with either the MAC or the PAC assay formats for v114 ().
To further evaluate the effect of CD20 on the ocrelizumab MAC and PAC assay formats vs. the v114 MAC and PAC assay formats, FACS experiments were performed to evaluate the ability of ocrelizumab and v114 to bind to CD20 on the surface of WIL2-S cells in the presence of the monoclonal and polyclonal antibody reagents used in each assay format (). These experiments demonstrated that at concentrations as high as 100–1000 nM, both of the mAbs 8A3 and 2B3 used in the ocrelizumab MAC assay format inhibited ocrelizumab from binding to CD20 on the surface of WIL2-S cells. The polyclonal antibody only inhibited binding of ocrelizumab to CD20 at the highest concentration tested (260 nM) (). This data suggests that the mAbs 8A3 and 2B3 bind near or overlap the paratope on ocrelizumab specific for CD20. If ocrelizumab is bound to cCD20 in serum, the mAbs used in the MAC format will not be able to detect ocrelizumab, resulting in a “free” drug assay. Similar FACS experiments were performed on the v114 MAC and the PAC assays reagents (). Neither of the mAbs (7G7, 2D3) used in v114 MAC assay inhibited v114 from binding to CD20 on the surface of WIL2-S cells at concentrations as high as 1000 nM. The polyclonal antibody, similar to the ocrelizumab polyclonal antibody, inhibited v114 binding most significantly only at the highest concentration tested (260 nM). This data suggests that the v114 MAC and the PAC assays measure both free and complexed drug equally and therefore both assay formats measure “total” drug.
It is known that anti-CD20 antibodies such as rituximab have nonlinear PK profiles in oncology indications during initial dosing. This is attributed to antigen-mediated clearance mechanism. Following multiple doses of rituximab in NHL patients, using a PAC assay format, a 2.5 times increase in mean terminal half-life was observed following administration of the fourth weekly dose. This is attributed to membrane-associated antigen-mediated clearance pathway that occurs through binding of the antibody (rituximab) to the antigen (CD20-positive B-cells) and subsequent internalization of the antibody–antigen complex, which is followed by degradation and the reduction in tumor burden. This difference in terminal half-life between the first and subsequent doses is not observed with rituximab in a non-oncology indication (i.e., the RA population). One possible reason is that the baseline CD20-positive B cells in RA patients are much lower than those observed in NHL patients, and therefore, the B cells do not contribute significantly to the clearance of the rituximab in RA patients.Citation13
The PK profile observed for ocrelizumab (with the MAC assay) in RA population, however, is consistent with soluble antigen interference attributed to increases in cell depletion and release of cCD20 into the serum post-ocrelizumab treatment. In this case, different PK profiles were observed depending on the assay reagents used, and were not due to clearance of the therapeutic through the antigen-mediated elimination mechanism discussed earlier.
In conclusion, a significant difference in PK profiles was observed using the ocrelizumab MAC vs. PAC assay formats, but not between the PK profiles generated with the v114 MAC and PAC assays. Circulating CD20 was identified as the potential cause for the observed differences. Our data suggest that the drug species detected in a PK assay depends on the assay reagents and formats used, and that this information is crucial for data interpretation and for making accurate conclusions. Therefore, reagent characterization is critical to understanding the drug species being measured and the strengths and limitations of the data generated.
Materials and Methods
Antibodies
All antibodies were generated at Genentech unless otherwise stated. The chimeric IgG1 antibody rituximab, humanized IgG1 antibodies ocrelizumab and v114 were produced from Chinese hamster ovary cells (CHO). The capture reagent for the rituximab PAC assay was a purified, anti-CDR goat anti-rituximab polyclonal antibody. A goat anti-mouse IgG F(ab’)2 polyclonal antibody conjugated to horseradish peroxidase (HRP) (Jackson ImmunoResearch Labs, catalog#115–035–006) was used for detection. Capture and detection reagents for the ocrelizumab MAC assay were the 2B3 mAb and the 8A3 mAb conjugated to biotin, respectively. Capture and detection antibodies for the ocrelizumab PAC assay were purified, anti-CDR enriched goat anti-ocrelizumab polyclonal antibody and the peroxidase-conjugated AffiniPure F(ab’)2 goat anti-human IgG specific to Fc fragment (Jackson ImmunoResearch Labs, catalog# 109–006–006), respectively. The capture antibody for the v114 PAC assay was purified, anti-CDR enriched goat anti-v114 polyclonal, and the detection antibody was the biotin conjugated 14B11 anti-v114 mAb. Finally, for the v114 MAC assay, capture and detection antibodies were the anti-v114 mAbs 7G7 and 2D3 conjugated with biotin, respectively. Antibodies for FACS analysis included: anti-ocrelizumab mAbs 8A3 and 2B3, and anti-v114 mAbs 7G7 and 2D3. The FACS detection reagent was a phycoerythrin conjugated-F(ab’)2 goat anti-human IgG-Fc specific (Jackson ImmunoResearch Labs, catalog# 109–116–170).
Reagents
Sera from healthy individuals or subjects from the disease population were utilized to develop, qualify, and validate the PK assays. Pooled normal human serum (BioReclamation, catalog# HMSRM), as well as individual patient sera from normal, non-Hodgkin lymphoma (BioReclamation, catalog# HMSRM-NONHODGLYM), chronic lymphocytic leukemia (BioReclamation, catalog# HMSRM-BCLL), and rheumatoid arthritis (BioReclamation, catalog# HMSRM-RA) patients, were purchased from BioReclamation. Pooled sera used for the ocrelizumab MAC assay, was purchased from Sigma Aldrich (catalog# H3667). Maxisorp Nunc-immuno 96-well microtiter plates were purchased from NalgeNunc International (catalog# M9410). Biotin-LC-Sulfo-NHS Ester was purchased from BioVeris (catalog# 110015), and EZ-Link Sulfo NHS-LC-Biotin was purchased from Pierce (catalog# 21338). Mouse IgG was obtained from Jackson ImmunoResearch (catalog# 015–000–003). AmershamAmdex Streptavidin HRP (GE Healthcare Life Sciences, catalog# RPN4401) was used for all ELISAs except for the v114 PAC assay, for which peroxidase-conjugated Streptavidin (Zymed/Invitrogen, catalog# 43–8323) was used. HRP substrate 3,3′,5,5′-tetramethylbenzidine (TMB; catalog# 50–85–06) and hydrogen peroxide (catalog# 71–00–09) were purchased from Kirkegaard and Perry Labs.
Biotin Conjugation of Detection Reagents
Prior to conjugation, the mAbs 8A3, 14B11, and 2D3 were buffer-exchanged in PBS and then incubated with biotin using EZ-Link SulfoNHS-LC-Biotin (Pierce, catalog# 21338for 8A3) or Biotin-LC-Sulfo-NHS Ester (Bioveris, catalog# 110015 for 14B11 and 2D3) according to the manufacturers’ instructions. Briefly, the biotin was incubated with the antibodies in PBS at room temperature for 2 h, and dialyzed with PBS at 2–8°C overnight, with one buffer exchange after 2 h. The concentration of each conjugate was determined using a BCA protein assay kit (Pierce, catalog# 23225).
Clinical PK assay methods
The PAC and MAC-based PK assays used colorimetric ELISAs with very similar procedures. The buffers used in the various PK assays and a brief summary of assay conditions for each method are summarized in . Briefly, 96-well Nunc plates were coated with 100 μL of coat reagent diluted to 0.5, 1 or 2μg/mL in coating buffer (0.05 M sodium carbonate, pH 9.6) as specified in and incubated at 2–8°C for 18 to 72 h. Plates were washed and blocked with 200 μL of appropriate assay buffer for 1 h (). After washing, samples, standards, and controls were diluted at a minimum dilution in the assay buffer and incubated for 1 h at room temperature. After a wash step, HRP-or biotin-conjugated secondary antibody was added and incubated for 1 h. When biotin labeled antibody was used, streptavidin HRP was added and incubation for an additional hour was needed. The signal for all assays was detected using TMB substrate and the drug level was quantitated using absorbance spectrophotometry at 450 nm with 620 or 630 nm as reference on a plate reader. The minimum quantifiable concentration(MQC) for each assay is listed in .
Table 1. Summary of contents of assay buffers and conditions of assays
Recombinant CD20 Generation
Full length recombinant CD20 (rCD20) was generated at Genentech. Briefly, rCD20 was engineered to have a thrombin cleavage site, a His-tag and a Flag-tag, and was expressed in E.coli. Following cell homogenization, lysis and centrifugation, the supernatant containing CD20 was purified using Ni-NTA chromatography. Fractions containing CD20 were pooled and subjected to thrombin cleavage. CD20 was separated from the cleaved N-terminal sequence by Superdex 200 size-exclusion chromatography. The amino acid sequence was confirmed by Edman degradation ().
Figure 5. Red highlight, thrombin cleavage site; Blue highlight, His-tag; Green highlight, Flag-tag.

The identity of CD20 was confirmed using N-terminal sequencing and MALDI-TOF peptide mass fingerprinting.
CD20 Interference
CD20 interference was tested in the rituximab PAC assay, the ocrelizumab MAC and PAC assay, and the v114 MAC and PAC assay formats, using samples with various molar ratios of rCD20 and either rituximab, ocrelizumab or v114.For the rituximab and ocrelizumab assays, varying concentrations (2, 20, 200 and 2000 ng/mL) rCD20 was spiked into buffer samples containing either 6 ng/mL ocrelizumab to be assayed in the PAC assay, 10 ng/mL ocrelizumab to be assayed in the MAC assay, or 40 ng/mL rituximab for assaying in the PAC assay. To assess recovery of v114 in the PAC assay, buffer samples containing 1 ng/mL of v114 in the presence of 0.008, 0.08, 0.8, and 8 µg/mL rCD20 was tested; for the MAC assay, 400 ng/mL of v114 spiked into serum was assessed in the presence of 0.008, 0.08, 0.8, and 8 µg/mL of rCD20. Spiked samples were incubated with rCD20 for 1 h at room temperature before run in the respective assays as described.
Reagent Characterization Using Flow Cytometry
Competitive FACS assays were used to assess the effect of the coat and detection ELISA assay reagents on the ability of humanized IgG1 anti-CD20 mAbs ocrelizumab and v114 to bind to cell surface expressed CD20 on the surface of WIL2-S cells. Anti-ocrelizumab mAbs 8A3 and 2B3, and anti-v114 mAbs 7G7 and 2D3, were serially diluted 1/10 to provide a final concentration of 1000, 100, 10, 1.0, and 0.1 nM. Similar dilutions were also made with affinity-purified polyclonal anti-ocrelizumab and anti-v114 antibodies to provide a final concentration of 260, 26, 2.6, 0.26, and 0.026 nM. All dilutions were performed in a 96-well U-bottom plate containing 100 µL of assay buffer D. To the anti-ocrelizumab and anti-v114 dilutions, 100 µL of a 100 nM solution (in assay buffer D) of ocrelizumab and v114 were added, respectively. After the mixtures were incubated for 30 min at room temperature, 100µL of assay buffer containing 0.5 x 106 CD20+ WIL2-S cells (ATCC, catalog# CRL-8885) was added to each well and incubated for 1.5 h at 4°C. Following the incubation, the cells were washed 3 times with assay buffer D. Each well then received 200 µL of a 1/2000 dilution of phycoerythrin conjugated-F(ab’)2-goat anti-mouse IgG-Fc specific (Jackson ImmunoResearch Labs, catalog# 115–116–071) plus 10 μg/mL of murine IgG. After 30 min incubation at 4°C, the cells were washed in assay buffer D and then fixed in 500 µL of PBS containing 1% paraformaldehyde. Samples were analyzed in a FACS Calibur flow cytometer (BD Biosciences).
Pharmacokinetics
The following parameters were calculated from the serum therapeutic concentration–time data using standard non-compartmental PK methods and the computer program WinNonlin® Pro, Version 5.2.1 (Pharsight Corporation):
Cmax: maximum observed serum concentration
AUC0–∞: area under the concentration–time curve from time zero to infinity
CL: Clearance
Vz: volume of distribution
t1/2: elimination half-life
Formal statistical comparisons were not performed; PK parameters were analyzed using descriptive statistics. Representative serum concentrations vs. time profiles from MAC and PAC assays are shown in –.
Clinical Study Designs
Rituximab Study in the RA Patient Population
A Phase 2b study, with 316 RA patients who previously failed disease-modifying anti-rheumatic drug (DMARD) therapy was completed. Subjects received IV infusions of rituximab (2 x1000 mg or 2 x 500 mg), on Days 1 and 15 during a 2-week period. Blood samples were collected 30 min prior to start of the infusion and 60 min following the end of the rituximab infusion on Days 1 and 15 and on Weeks 4, 8, 12, 16, 20, and 24 following the start of the first infusion for PK analysis.
Ocrelizumab (2H7) Study in RA Patient Population
The ACTION trial was a Phase 1/2 randomized, blinded, placebo-controlled, dose-ranging study evaluating the safety of ocrelizumab in combination with MTX in patients with RA.Citation6
Ocrelizumab Study in the NHL Patient Population
This was an open-label, multicenter, Phase 1/2 study evaluating ocrelizumab in patients with CD20+, follicular non-Hodgkin lymphoma whose disease has relapsed or progressed after prior rituximab therapy. There were 3 cohorts, each enrolling 15 patients for a total of 45 patients. Patients in each cohort received a total of eight 3-weekly infusions of ocrelizumab as shown in .
v114 Study in the NHL Patient Population
This was an open-label, multicenter, Phase 1/2 study of the safety of escalating doses of v114 (PRO131921) as a single-agent in patients with relapsed or refractory CD20-positive follicular NHL. The original study design included two phases: a Phase 1 dose escalation portion for patients with indolent NHL and a Phase 2 portion with enrollment of additional patients with follicular NHL into two expanded treatment cohorts to expand the safety database and collect preliminary anti-lymphoma activity data. The sponsor decided to stop the study during Phase 1 and no patients were enrolled in the Phase 2 cohort. For the Phase 1 portion of the trial, from which samples were utilized for analysis, each enrolled patient received four weekly infusions of v114 at the assigned dose on Days 1, 8, 15, and 22, and a total of 23 patients were enrolled (3–6 patients in each of the six cohorts, A–F) ().
References
- Kuang B, King L, Wang HF. Therapeutic monoclonal antibody concentration monitoring: free or total?. Bioanalysis 2010; 2:1125 - 40; http://dx.doi.org/10.4155/bio.10.64; PMID: 21083212
- Lee JW, Kelley M, King LE, Yang J, Salimi-Moosavi H, Tang MT, et al. Bioanalytical approaches to quantify “total” and “free” therapeutic antibodies and their targets: technical challenges and PK/PD applications over the course of drug development. AAPS J 2011; 13:99 - 110; http://dx.doi.org/10.1208/s12248-011-9251-3; PMID: 21240643
- Liu J, Lester P, Builder S, Shire SJ. Characterization of complex formation by humanized anti-IgE monoclonal antibody and monoclonal human IgE. Biochemistry 1995; 34:10474 - 82; http://dx.doi.org/10.1021/bi00033a020; PMID: 7654701
- Robak T, Robak E. New anti-CD20 monoclonal antibodies for the treatment of B-cell lymphoid malignancies. BioDrugs 2011; 25:13 - 25; http://dx.doi.org/10.2165/11539590-000000000-00000; PMID: 21090841
- Friedberg JW, Vose JM, Kahl BS, Brunvand MW, Goy A, Kasamon YL, et al. A phase I study of PRO131921, a novel anti-CD20 monoclonal antibody in patients with relapsed/refractory CD20_ indolent NHL: Correlation between clinical responses and AUC pharmacokinetics. ASH Annual Meeting. New Orleans, LA: Blood, 2009:3742.
- Genovese MC, Kaine JL, Lowenstein MB, Del Giudice J, Baldassare A, Schechtman J, et al, ACTION Study Group. Ocrelizumab, a humanized anti-CD20 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: a phase I/II randomized, blinded, placebo-controlled, dose-ranging study. Arthritis Rheum 2008; 58:2652 - 61; http://dx.doi.org/10.1002/art.23732; PMID: 18759293
- Breedveld F, Agarwal S, Yin M, Ren S, Li NF, Shaw TM, et al. Rituximab pharmacokinetics in patients with rheumatoid arthritis: B-cell levels do not correlate with clinical response. J Clin Pharmacol 2007; 47:1119 - 28; http://dx.doi.org/10.1177/0091270007305297; PMID: 17766699
- Morschhauser F, Marlton P, Vitolo U, Lindén O, Seymour JF, Crump M, et al. Results of a phase I/II study of ocrelizumab, a fully humanized anti-CD20 mAb, in patients with relapsed/refractory follicular lymphoma. Ann Oncol 2010; 21:1870 - 6; http://dx.doi.org/10.1093/annonc/mdq027; PMID: 20157180
- Giles FJ, Vose JM, Do KA, Johnson MM, Manshouri T, Bociek G, et al. Circulating CD20 and CD52 in patients with non-Hodgkin’s lymphoma or Hodgkin’s disease. Br J Haematol 2003; 123:850 - 7; http://dx.doi.org/10.1046/j.1365-2141.2003.04683.x; PMID: 14632776
- Manshouri T, Do KA, Wang X, Giles FJ, O’Brien SM, Saffer H, et al. Circulating CD20 is detectable in the plasma of patients with chronic lymphocytic leukemia and is of prognostic significance. Blood 2003; 101:2507 - 13; http://dx.doi.org/10.1182/blood-2002-06-1639; PMID: 12446458
- Keating MJ, O’Brien S, Albitar M. Emerging information on the use of rituximab in chronic lymphocytic leukemia. Semin Oncol 2002; 29:Suppl 2 70 - 4; http://dx.doi.org/10.1053/sonc.2002.30142; PMID: 11842391
- Riley JK, Sliwkowski MX. CD20: a gene in search of a function. Semin Oncol 2000; 27:Suppl 12 17 - 24; PMID: 11225995
- Ng CM, Bruno R, Combs D, Davies B. Population pharmacokinetics of rituximab (anti-CD20 monoclonal antibody) in rheumatoid arthritis patients during a phase II clinical trial. J Clin Pharmacol 2005; 45:792 - 801; http://dx.doi.org/10.1177/0091270005277075; PMID: 15951469