Abstract
A dual-specific, tetravalent immunoglobulin G-like molecule, termed dual variable domain immunoglobulin (DVD-Ig™), is engineered to block two targets. Flexibility modulates Fc receptor and complement binding, but could result in undesirable cross-linking of surface antigens and downstream signaling. Understanding the flexibility of parental mAbs is important for designing and retaining functionality of DVD-Ig™ molecules. The architecture and dynamics of a DVD-Ig™ molecule and its parental mAbs was examined using single particle electron microscopy. Hinge angles measured for the DVD-Ig™ molecule were similar to the inner antigen parental mAb. The outer binding domain of the DVD-Ig™ molecule was highly mobile and three-dimensional (3D) analysis showed binding of inner antigen caused the outer domain to fold out of the plane with a major morphological change. Docking high-resolution X-ray structures into the 3D electron microscopy map supports the extraordinary domain flexibility observed in the DVD-Ig™ molecule allowing antigen binding with minimal steric hindrance.
Introduction
Monoclonal antibodies (mAbs) have emerged as an important class of therapeutics in oncology, cardiovascular disease, inflammation, transplantation and infectious disease.Citation1-Citation6 This class of therapeutics has proven successful in the clinic due to their high specificity, good safety profiles, and overall tolerability. Opportunities for innovation exist, however, in the areas of fulfilling currently unmet medical needs, as well as delivering molecules with superior efficacy and physicochemical properties. A new class of molecules, bispecific mAbs, has thus emerged, including the dual-variable-domain immunoglobulin (DVD-Ig™) molecules currently being developed at AbbVie, two-in-one molecules (Genentech), dual affinity retargeting molecules (MacroGenics), bispecific T-cell engager (Micromet), kappa-lambda antibodies (NovImmune), chemical generation (CovX/Pfizer) and tetravalent bispecific antibodies (Merrimack).Citation7,Citation8 The bispecific molecules offer potential economic and therapeutic superiority in blocking two soluble ligands or cell surface receptors, cross-linking of two receptor molecules or recruiting T-cells to aid in tumor killing. Most of these molecules are in early clinical development, although one (catuxomab) that simultaneously targets EpCAM and CD3 has been approved in the European Union.
Simultaneous targeting of multiple disease mediators using the DVD-Ig™ format was first described in 2007Citation9 and at least two are currently in clinical trials, a DVD-Ig™ molecule targeting tumor necrosis factor and interleukin IL-17, and a DVD-Ig™ molecule targeting IL-1 alpha and beta.Citation10-Citation13 These molecules, as well as a host of others in preclinical development, have demonstrated the potential of this class of molecules to retain affinity and potency for both intended targets. The molecules have good physicochemical properties and pharmacokinetics; the ability of the DVD-Ig™ molecules to simultaneously bind both antigens was shown by Biacore analysis.Citation9
The asymmetry and flexibility of IgG molecules have been suggested from the few existing solved crystal structures of intact antibodies.Citation14,Citation15 This flexibility enables IgG molecules to bind antigens of a variety of shapes and sizes and also allows the effector domain (Fc) to bind the Fc receptor or complement.Citation15,Citation16 Flexibility is mediated through the hinge region and human IgG isotypes have different amino acid composition and inter-chain disulfide bridges in their hinge regions due to the number and position of cysteine residues. Human IgG1 and IgG4 have two disulfide linkages; whereas, IgG2 and IgG3 have four and 11 respectively.Citation1 The hinge region has three structural components, i.e., the upper, core and lower hinge segments (). The core segment (CPPC) contains cysteine residues that connect the two heavy chains and paired poly (L)-proline helices that make this segment inflexible. The upper hinge region (DKTHT) of human IgG1 connects the Fab arms to the core segment and influences Fab-Fab flexibility; N-terminal residues from the upper hinge region are reported to interact with the CH1 domain.Citation17-Citation19 The lower hinge region (PAPELLGG) connects the Fc to the core region and plays a role in Fab/Fc flexibility and Fc tail wagging.Citation16,Citation20 The length of the hinge region also plays a role in flexibility; mouse IgG1, which has a shorter hinge region (12 residues and 3 inter-chain cysteines), is more compact than either IgG2a (19 residues with 3 cysteines) or human IgG1 (17 residues with 2 cysteines).Citation15 The inherent flexibility in IgG molecules has hampered crystallization efforts and only a few crystal structures of intact IgG molecules have been solved.Citation15,Citation21-Citation27 The majority of structures reported in the literature are either Fab or Fab’ fragments.
Figure 1. (A) Schematic depiction of the overall architecture of a DVD-IgTM molecule showing the outer and inner antigen binding domains in the Fab region, the hinge region and the Fc region. Class averages showing (B) an isolated DVD-Ig™ molecule (n = 761), (C) inner antigen (n = 127), (D) outer antigen (n = 234); complexes of (E) the DVD-Ig™ molecule + inner antigen (n = 750), (F) the DVD-Ig™ molecule + outer antigen (n = 1222); white circle highlights the unique “beak” shape of attached outer antigen; different conformations of the DVD-Ig™ molecule + inner antigen + outer antigen: (G) “Z” shape (n = 785), (H) “8” shape (n = 558), (I) “pretzel” shape (n = 695). The number of particles contributing to each class is shown in parentheses for each complex.
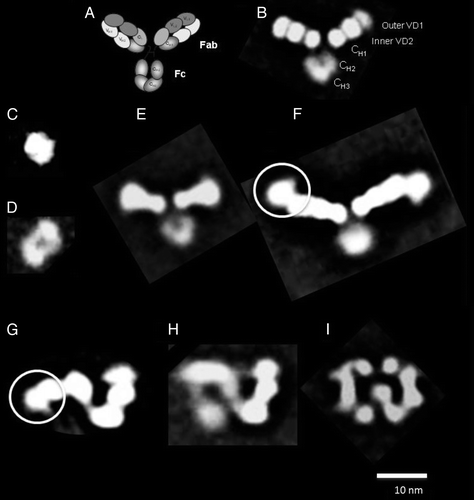
The structure of the Fab fragment of the anti-IL-12/IL-18 DVD-Ig™ molecule has been solved at 2.1 Å resolution by X-ray crystallography.Citation28 Here, we report on the overall architecture of the intact anti-IL-12/IL-18 DVD-Ig™ molecule and the DVD-Ig™ molecule bound to both inner and outer antigens determined using single particle electron microscopy (EM).Citation29 Two-dimensional class averages calculated from thousands of individual particles of the DVD-Ig™ molecule show that the molecule assumes a “Y” shape similar to that of antibodies. Analysis of the range of motion of each of the DVD-Ig™ arms showed that the average angle between the Fab arms was comparable to angles obtained for the inner binding domain (VD2) of the parental mAb, but the arms traverse a wider range of motion. Assembling the entire set of possible class averages into a sequential series reveals rotational dynamics of the outer binding domain (VD1) and translational dynamics of the Fc region, in addition to hinge region folding. Random conical tilt (RCT) reconstruction methodsCitation30 were used to reconstruct three-dimentional (3D) models of the DVD-Ig™ molecule bound to the inner antigen and to both inner and outer antigens. These models show that binding of antigen to VD2 alters the overall conformation of the molecule, causing VD1 to fold out of the plane of VD2 and CH1/CL domains. This in turn results in a major morphological change in the overall shape of the complex formed by the DVD-Ig™ molecule bound to both inner and outer antigen. These results clearly demonstrate the extraordinary domain flexibility observed in the DVD-Ig™ molecule, which allows both antigens to bind the DVD-Ig™ molecule with minimal steric hindrance. Comparable affinity of both antigens to the parental mAbs was thus retained.Citation28
Results
Structural analysis of the DVD-Ig™ molecule alone and bound to antigens
While it is challenging to discern the structure of an individual DVD-Ig™ molecule and parental mAbs from unprocessed transmission EM images (Fig. S1), the signal to noise ratio can be greatly improved by identifying, aligning, classifying and averaging particles in similar conformations. The resultant two-dimensional (2D) class averages of the DVD-IgTM molecule show ~20 nm “Y” shaped particles consistent with that of antibodies (). The DVD-Ig™ molecule has two ~10 nm arms (Fabs) that are each composed of three ~3 nm lobes (VH1/VL1 or VD1, VH2/VL2 or VD2, and CH1/CL as shown schematically in ) and one ~7 nm arm that resembles the Fc loop where the CH2 domain has less density than the CH3 domain (). The overall density of the Fc region in the 2D class averages is typically significantly reduced compared with the Fab arms, which indicates a high degree of flexibility of the Fc region (this is also corroborated in the assembled images across a number of class averages as shown in Fig. S4). The parental mAb molecules (Fig. S3) of the DVD-Ig™ molecule appear as ~15 nm “Y” shaped particles with two ~7 nm arms composed of two ~3 nm lobes (VH1/VL1 and CH1/CL).
2D class averages of EM images of the antigens to the inner and outer domains of the DVD-Ig™ molecule show the inner antigen (IL-18, molecular weight 18 kDa) as a ~5 nm particle () and the outer antigen (IL-12, a hetero-dimer of molecular weight 65 kDa) as a ~7 nm oblong particle with two domains separated by an indentation in the center ().
Binding of the inner antigen (IL-18) to the DVD-Ig™ molecule () results in a ~24 nm “Y” shaped particle, with one visible ~7 nm Fc loop and two longer ~10 nm Fab “arms” whose somewhat spatulate ends, compared with the DVD-Ig™ molecule by itself, indicate the site of the bound antigen. Binding of the outer antigen (IL-12) to the DVD-Ig™ molecule () results in ~35 nm “Y” shaped particles with one visible ~7 nm Fc loop and two longer ~10 nm Fab “arms” with distinct ~5 nm densities extending from the ends of the arms, representing the bound antigen. The particles formed by the DVD-Ig™ molecule bound to the outer antigen (IL-12) appear more flexible than the DVD-Ig™ molecule bound to inner antigen (IL-18), as indicated by the presence of more and varied conformations of the class averages (Fig. S5).
Binding of both the inner and outer antigens () to the DVD-Ig™ molecule results in particles with morphologies not obviously related to the characteristic “Y” shape associated with a DVD-Ig™ molecule alone or bound to each individual antigen. Consistently identified 2D class averages showed ~23 nm particles adopting either a “Z” (), “8” () or “pretzel” () shaped morphology, with some regions that resemble the Fab arms of a DVD-Ig™ molecule. Since no class averages were seen to resemble the particles found in a DVD-Ig™ molecule bound to either IL-18 (inner antigen) alone, or IL-12 (outer antigen) alone, or a DVD-Ig™ molecule by itself, it is likely that the observed structures represent a DVD-Ig™ molecule binding to both antigens. It thus appears that binding of a DVD-Ig™ molecule to both antigens causes a significant conformational change in the overall molecule. These morphological changes were elucidated using 3D reconstruction.
3D reconstruction using random conical tilt
3D reconstructions using random conical tilt (RCT) methods were obtained for the DVD-Ig™ molecule bound to inner antigen and two of the classes of the DVD-Ig™ molecule bound to both antigens (). Interpretations of the structures are based on these 3D models as well as the 2D class averages ().
Figure 2. (A) class average of the DVD-IgTM molecule+ inner antigen. (B) RCT reconstruction of the class average in (A) shows that binding of the inner antigen causes a conformational change in the morphology of the DVD-Ig™ molecule, folding down the third domain (VD1) out of the plane of the other two domains. The Fc domain is only visible at a lower density threshold (green mesh), presumably because of its flexibility relative to the other domains. (C) An interpretation of the RCT maps showing the positions of the Fc domain, the inner antigen and the three domains along one of the DVD-Ig™ molecule arms. (D) class average of the “Z” shaped class average of the DVD-Ig™ molecule bound to both inner and outer antigen. (E) RCT reconstruction of the class average in (D). At a lower threshold, density appears that we interpret as the Fc domain. (F) Interpretation of the RCT map is supported by the correspondence between the appearance of the end of the arm of the RCT model and the end of the arm on the 2D class average of the DVD-IgTM molecule bound to outer antigen (). (G) class average of the “pretzel” shaped class average of the DVD-Ig™ molecule bound to both inner and outer antigen. (H) An interpretation of the domain arrangements of the structure overlaid onto (G) the class average and (I) the RCT reconstruction of the class average in (G). The “pretzel” can be explained as an alternative conformation of the “Z” shape is the DVD-Ig™ arms rotate as shown by the arrows in (F). Also see Figure S2.
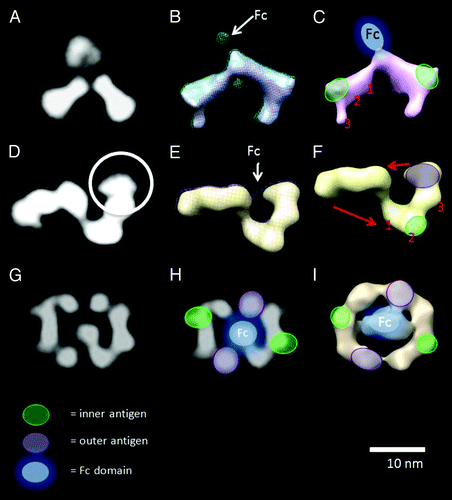
The 3D reconstruction of the DVD-Ig™ molecule bound to inner antigen (, top row) shows that binding of the inner antigen results in a marked conformational change to the structure, with the third domain (VD1) of the DVD-Ig molecule folding down out of the plane of the other two. The Fc domain is not well-defined in this map, presumably because it is highly flexible, and only appears when the density threshold of the map is lowered () so as to show weaker densities. Interpretation of the RCT map is depicted in .
The 3D reconstruction of the “Z” shaped class average calculated from images of the DVD-Ig™ molecule bound to both antigens is shown in , middle row. The 3D structure shows that the DVD-Ig™ arms are bent at an angle of almost 90° as a result of binding of inner antigen, and the outer antigen is bound to the outer ends of the DVD-Ig™ arms. This interpretation is supported by the correspondence between the appearance of the end of the arm of the RCT model and the end of the arm on the 2D class average of the DVD-Ig™ molecule bound to outer antigen (compare regions within white circles in and ). Once again, the Fc domain is poorly defined and only appears when the map is rendered at a lower density threshold. The flexibility of the Fc domain is further corroborated by the reduced density of this region in the 2D class averages (e.g., ). The 3D structure of the DVD-Ig™ molecule bound to both antigens is interpreted as depicted in . This model provides an explanation of several of the predominant 2D class averages as alternative conformations of a consistent structure (, bottom row; Fig. S2).
Dynamics of the DVD-Ig™ molecule
The hinge region flexibility of the DVD-Ig™ molecule relative to each parental mAb was evaluated from class averages derived from a large population of particles (; Fig. S4). Angle measurements between the Fab/Fab () or Fab/Fc arms of well-defined class averages showed that the DVD-Ig™ molecule adopts a larger number or conformations, with the Fab and Fc arms spanning a wider range of flexibility, compared with the mAb particles. While the DVD-Ig™ molecule showed the more extreme ranges, the weighted average Fab-Fc and Fab-Fab angles for parental mAb-1 (110°/135°) and the DVD-Ig™ molecule (107°/136°) were found to be similar whereas parental mAb-2 had slightly larger Fab-Fc (117°) and smaller Fab-Fab (121°) average angles. The overall hinge region flexibility of the DVD-Ig™ molecule and mAbs measured here is well within the range of those reported for engineered human IgG-1, 2, 3 and 4 with matching variable binding domains ().Citation16 The mean Fab/Fab angle measured for the DVD-Ig™ molecule (136°) matched that of the engineered IgG3 (136°). As reported in Roux et al.,Citation16 the large angular variability observed for the IgG3 molecule indicates minimal steric hindrance and maximal molecular flexibility, which permits dimer formation with the variable domain on anti-idiotypic antibodies. Dimer formation was less prevalent with other IgG1, 2 or 4 molecules because of their compact nature. The functional significance of such differences for DVD-Ig™ molecule development could be substantial, as cross-linking and downstream signaling via cell surface receptors may result in a DVD-Ig™ molecule that has agonist instead of antagonist activity. The Fab/Fc angles are also believed to play a significant role in effector function, thus modulating Fc receptor and complement binding.
Figure 3. Dynamics of the DVD-Ig™ molecule show (A) wagging of outer domain, (B) folding in the hinge region and (C) translation movement of the Fc region.
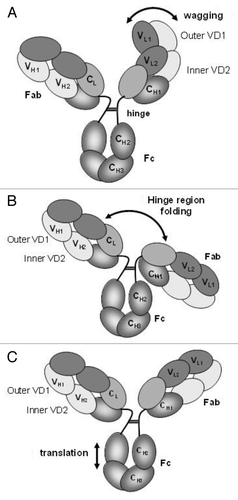
Table 1A. Description of structural flexibility of the DVD-Ig™ molecules compared with parental mAbs. Angles were measured between the Fc/Fab and Fab/Fab arms using 2D class averages.
Table 1B. Flexibility of engineered human IgG-1, 2, 3 and 4 with matched variable binding domains from Roux et al16
To further visualize DVD-Ig™ molecule flexibility, stacks of particles were classified, analyzed and sorted into sequential series and depicted as movie loops to emphasize the range of motion of the DVD-Ig™ molecules for each sample (Videos S1 and S2). In addition to flexibility about the hinge region (Fig. S4B), other dynamics recognized were wagging of the outer domain of the DVD-Ig™ molecule (Fig. S4A) and translational movement of the Fc region (Fig. S4C).
Comparison and docking with previously determined structures
The insight gained from 2D and 3D EM reconstruction of the DVD-Ig™ molecule alone and bound to antigen was corroborated by high resolution X-ray crystallography data. With IL-18 bound to VD2, the outer variable domain (VD1) was found to rest entirely on top of the heavy chain of VD2. The CDR of VD1 is positioned at right angles (~85°) from the inner variable CDR, which leaves ample room for binding outer antigen.Citation28 We docked the crystal structure of the DVD-Ig™ Fab (DFab) fragment bound to IL-18 and the Fc portion of human IgG1 (pdbcode 1HZH) into the 3D reconstructed EM density map. There is additional density in the 3D EM map that could clearly accommodate the bound IL-12, but because the orientation of IL-12 could not be uniquely defined based on the data, we did not include the docking of this antigen into the model. captures the change in the overall conformation of the DVD-Ig™ molecule when bound to both antigens. The most prominent domain arrangements occurs at the interface between the two variable domains, which are covalently held together by two linkers connecting the variable heavy and variable light antigen binding domains. The linker region is likely to be solvent-exposed and, as seen in the supplemental movie animations (Videos S1 and S2), permits extensive wagging of VD1. We postulate that VD1 can also rotate in space due to minimal steric hindrance. Other linkers of varying length and composition are under consideration in designing new DVD-Ig™ pairs. 2D class averages generated from EM images facilitates rapid understanding of the systematic effect of incorporating various types of linkers on both the dynamics of VD1 and the accessibility of different sized antigens to the binding domains.
Figure 4. The 3D RCT reconstruction of the DVD-Ig™ molecule (gray) bound to both antigens. The crystal structure of the DVD-Ig™ Fab (DFab) fragment bound to IL-18 (yellow) is shown docked on the “Z” shaped structure. Also shown are the Fc (cyan, PDB code 1HZH residues 246–478), the first constant domain (red) and the 2 variable (inner and outer) domains found in Fab arms of the DVD-Ig™ molecule. The structures were manually fitted into the 3D RCT reconstructed density map to achieve the best visual fit to the map, while avoiding steric clashes between the domains. The hinge region and IL-12 were not explicitly modeled.
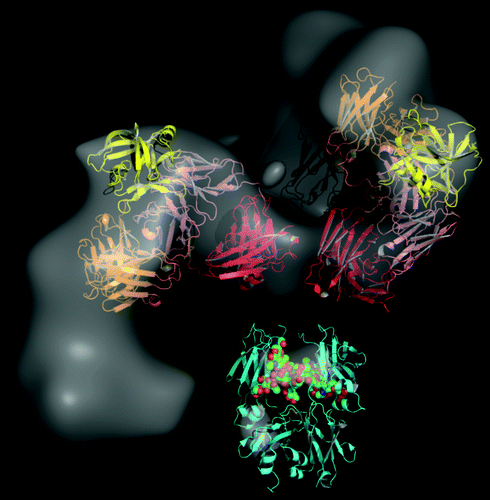
Discussion
Segmental flexibility of IgG molecules has been reported by a variety of methods, including nanosecond fluorescence depolarization assays,Citation31-Citation33 immunoelectron microscopy,Citation16,Citation20,Citation34-Citation36 X-ray crystallography,Citation26 nuclear magnetic resonance spectroscopy,Citation17 small angle X-ray scattering and sedimentation studiesCitation37 and quasi-elastic light scattering.Citation38 These methods are useful for comparing different molecules, but are inadequate for quantifying the many different modes of flexibility in the molecule. In this study, we use single particle EM to describe the architecture and segmental flexibility of the DVD-Ig™ molecule. In movie files created from 2D class average images, we directly observed hinge region flexibility, wagging of the outer binding domain and translational movement in the Fc region. To the best of our knowledge this is the first study where different modes of flexibility in an IgG-like molecule have been clearly described and quantified. Two significant findings were also made. First, that measurement of the range of motion around the hinge region of the DVD-Ig™ molecule closely matched the parental mAb to the inner binding domain (anti-IL-18). This suggests that the internal variable domain, in addition to the length of the hinge region and the CH1 domain, plays a significant role in hinge region flexibility. These results have important implications for which variable domain is placed internally, as the Fc/Fab angles, through steric hindrance, can influence Fc effector function. The second finding was that binding of IL-18 (inner antigen) to the DVD-Ig™ molecule resulted in significant conformational change in the outer binding domain (VD1). 3D reconstruction data showed that VD1 folds out of the plane of VD2 and CH1/CL domains. These observations were remarkably consistent with the crystal structure of the DVD-Ig™ Fab (DFab) fragment bound to IL-18 (inner antigen).Citation28 Structures were interpreted from both 2D class averages and 3D reconstruction data and showed that the configuration of these complexes is quite different from the original “Y” shaped structure due to binding of both inner and outer antigen. The most prominent domain movement was found between the variable domains held together by linkers. Understanding the structure and dynamics of this complex molecule will facilitate optimal linker design and point the way to stabilization of the outer variable domain.
Materials and Methods
Materials
The parental mAbs (mAb-1 and mAb-2) and DVD-Ig™ molecules used in this study were produced at AbbVie Bioresearch Center. mAb-1 is a human anti-IL-18 mAb and mAb-2 is a human anti-IL-12 mAb. The anti-IL-12/IL-18 DVD-Ig™ molecule was constructed to bind both IL-18 (VD2) and IL-12 (VD1). Construction, expression and purification of the DVD-Ig™ protein and its ability to neutralize both targets were previously described.Citation9
Sample preparation and electron microscopy
Samples were prepared over a layer of continuous carbon supported by nitrocellulose on a 400-mesh copper grid. Samples were prepared by applying 3 μL of sample suspension to a freshly plasma cleaned grid, blotting away with filter paper, and immediately staining with 3 μL uranyl formate (2%). Stain was allowed to sit on the grid for 1 min before blotting away with filter paper. The grid then sat under a lamp for 5–10 min to dry. Sample DVD-Ig™ molecules were diluted to 1 uM with phosphate-buffered saline (PBS). Samples of inner antigen (IL-18) and outer antigen (IL-12) were diluted to 3 uM with PBS. DVD-Ig™ molecule-antigen incubations were mixed at 1:1 volume and 1:3 molarity of DVD-Ig™ molecules to antigen. Mixtures were incubated for at least 3 h at room temperature. Prior to imaging, incubations of the DVD-Ig™ molecule + inner antigen + outer antigen were diluted 1:33 in PBS.
EM was performed using an FEI Tecnai T12 electron microscope operating at 120keV equipped with an FEI Eagle 4k × 4k CCD camera. Images were obtained using LeginonCitation39 at nominal magnifications of 110,000× (0.10 nm/pixel), 67,000× (0.16 nm/pixel) and 52,000× (0.21 nm/pixel). The images were acquired at a nominal underfocus of −1.5μm (110,000×), −2μm (67,000×) and −2μm to −3μm (52,000×) and electron doses of ~24–45 e/Ε2. Random conical tilt (RCT) experiments were performed using the RCT node of Leginon,Citation40 with image pairs taken at 0 and 55 degrees using a defocus of −2μm and a dose per image of ~40e−/Å2.
Image processing and model reconstructions
Image processing and model reconstructions were performed using the Appion software package.Citation41 The contrast transfer functions (CTF) of the images was estimated and corrected using Ace2.Citation42 Individual particles in the 52,000× high magnification images were selected using automated picking protocolsCitation43-Citation45 followed by several rounds of reference-free alignment and classification using the XMIPP software package.Citation46 RCT is a robust ab initio method for obtaining structure that requires collecting pairs of images, one at a fairly high tilt angle (45–60°) followed by a second untilted image.Citation30,Citation40,Citation47 RCT reconstructions were performed using the initial model pipeline within the Appion architecture.Citation43 The resolution of the RCT 3D maps was on the order of 4–5 nm. Angle measurements were performed on class averages that contained recognizable Fab and Fc domains using Image JCitation48 to measure angles between a segmented line traced from the outermost edge of one arm to the center of the particle and then to the outermost edge of the other arm. Two Fab-Fc angles and one Fab-Fab angle were measured per class. Angle ranges and weighted average angles were calculated from these measurements. Fab-Fc and Fab-Fab measurements were weighted according to the number of particles in the class average as a percent of the total number of particles over all classes.
Further analysis of the flexibility of the structures was performed using an interactive masking and classification program. Briefly, stacks of particles were evaluated using a coarse reference-free maximum-likelihood refinementCitation49 to identify and remove incorrectly boxed particles from the data sets. The filtered particles stacks were then iteratively aligned to a common reference, and the Maskiton package was used to identify particle variations within specific regions.Citation50 The resulting variations were then assessed to remove misalignment errors before being converted to a movie format to facilitate interpretation of the movements in each domain.
| Abbreviations: | ||
| 2D and 3D | = | two- and three-dimensional |
| DVD-IgTM | = | dual variable domain immunoglobulin molecule |
| CH and CL | = | constant heavy and constant light chain domains |
| VH and VL | = | variable heavy and variable light chain domains |
| VD1 and VD2 | = | outer and inner variable domain |
| RCT | = | random conical tilt |
| TEM | = | transmission electron microscopy |
| Fab | = | fragment of an immunoglobulin molecule that binds the antigen |
| Fc | = | fragment crystallized that contains effector function |
| IL-12 and IL-18 | = | Interleukins-12 and 18 |
| CCD | = | charged coupled device |
| XMIPP | = | X-window-based microscopy image processing package |
| KerDenSOM | = | kernel probability density estimator self-organizing map |
| CTF | = | contrast transfer function |
| Anti-Id | = | anti-idiotypic |
Additional material
Download Zip (17.3 MB)Acknowledgments
The authors are grateful to Gary Welch, George Avgerinos and Jochen Salfeld for their support.
Disclosure of Potential Conflicts of Interest
The design, study conduct and financial support for the study were provided by AbbVie (formerly Abbott) and from NIH NCATS grant 5R44TR000182 to Joyce Sung and Bridget Carragher. AbbVie participated in the interpretation of data, review and approval of the publication; all authors contributed to the development of the publication and maintained control over the final content. Bridget Carragher and Joyce Sung received funding support from AbbVie. Ivan Correia, Randall Burton, Czeslaw Radziejewski, Tariq Ghayur and Clarissa Jakob are AbbVie employees and may hold AbbVie stock and/or options
Supplemental Material
Supplemental materials may be found here:
http://www.landesbioscience.com/journals/mabs/article/24258
References
- Correia IR. Stability of IgG isotypes in serum. MAbs 2010; 2:221 - 32; http://dx.doi.org/10.4161/mabs.2.3.11788; PMID: 20404539
- Reichert JM. Which are the antibodies to watch in 2012?. MAbs 2012; 4:1 - 3; http://dx.doi.org/10.4161/mabs.4.1.18719; PMID: 22327425
- Reichert JM. Antibody-based therapeutics to watch in 2011. MAbs 2011; 3:76 - 99; http://dx.doi.org/10.4161/mabs.3.1.13895; PMID: 21051951
- Reichert JM. Antibodies to watch in 2010. MAbs 2010; 2:84 - 100; http://dx.doi.org/10.4161/mabs.2.1.10677; PMID: 20065640
- Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC. Monoclonal antibody successes in the clinic. Nat Biotechnol 2005; 23:1073 - 8; http://dx.doi.org/10.1038/nbt0905-1073; PMID: 16151394
- Salfeld JG. Isotype selection in antibody engineering. Nat Biotechnol 2007; 25:1369 - 72; http://dx.doi.org/10.1038/nbt1207-1369; PMID: 18066027
- Dhimolea E, Reichert JM. World Bispecific Antibody Summit, September 27-28, 2011, Boston, MA. MAbs 2012; 4:4 - 13; http://dx.doi.org/10.4161/mabs.4.1.18821; PMID: 22327426
- Xu L, Kohli N, Rennard R, Jiao Y, Razlog M, Zhang K, et al. Rapid optimization and prototyping for therapeutic antibody-like molecules. MAbs 2013; 5; In press PMID: 23392215
- Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, et al. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat Biotechnol 2007; 25:1290 - 7; http://dx.doi.org/10.1038/nbt1345; PMID: 17934452
- DiGiammarino E, Ghayur T, Liu J. Design and generation of DVD-Ig™ molecules for dual-specific targeting. Methods Mol Biol 2012; 899:145 - 56; http://dx.doi.org/10.1007/978-1-61779-921-1_9; PMID: 22735951
- Gu J, Ghayur T. Generation of dual-variable-domain immunoglobulin molecules for dual-specific targeting. Methods Enzymol 2012; 502:25 - 41; http://dx.doi.org/10.1016/B978-0-12-416039-2.00002-1; PMID: 22208980
- Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov 2012; 11:763 - 76; http://dx.doi.org/10.1038/nrd3794; PMID: 23023676
- Wu C, Ying H, Bose S, Miller R, Medina L, Santora L, et al. Molecular construction and optimization of anti-human IL-1alpha/beta dual variable domain immunoglobulin (DVD-Ig) molecules. MAbs 2009; 1:339 - 47; http://dx.doi.org/10.4161/mabs.1.4.8755; PMID: 20068402
- Burton DR. Antibody: the flexible adaptor molecule. Trends Biochem Sci 1990; 15:64 - 9; http://dx.doi.org/10.1016/0968-0004(90)90178-E; PMID: 2186517
- Saphire EO, Stanfield RL, Crispin MD, Parren PW, Rudd PM, Dwek RA, et al. Contrasting IgG structures reveal extreme asymmetry and flexibility. J Mol Biol 2002; 319:9 - 18; http://dx.doi.org/10.1016/S0022-2836(02)00244-9; PMID: 12051932
- Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. J Immunol 1997; 159:3372 - 82; PMID: 9317136
- Kim H, Matsunaga C, Yoshino A, Kato K, Arata Y. Dynamical structure of the hinge region of immunoglobulin G as studied by 13C nuclear magnetic resonance spectroscopy. J Mol Biol 1994; 236:300 - 9; http://dx.doi.org/10.1006/jmbi.1994.1136; PMID: 8107111
- Schneider WP, Wensel TG, Stryer L, Oi VT. Genetically engineered immunoglobulins reveal structural features controlling segmental flexibility. Proc Natl Acad Sci U S A 1988; 85:2509 - 13; http://dx.doi.org/10.1073/pnas.85.8.2509; PMID: 3128789
- Tan LK, Shopes RJ, Oi VT, Morrison SL. Influence of the hinge region on complement activation, C1q binding, and segmental flexibility in chimeric human immunoglobulins. Proc Natl Acad Sci U S A 1990; 87:162 - 6; http://dx.doi.org/10.1073/pnas.87.1.162; PMID: 2296577
- Roux KH, Strelets L, Brekke OH, Sandlie I, Michaelsen TE. Comparisons of the ability of human IgG3 hinge mutants, IgM, IgE, and IgA2, to form small immune complexes: a role for flexibility and geometry. J Immunol 1998; 161:4083 - 90; PMID: 9780179
- Harris LJ, Skaletsky E, McPherson A. Crystallization of intact monoclonal antibodies. Proteins 1995; 23:285 - 9; http://dx.doi.org/10.1002/prot.340230218; PMID: 8592710
- Harris LJ, Skaletsky E, McPherson A. Crystallographic structure of an intact IgG1 monoclonal antibody. J Mol Biol 1998; 275:861 - 72; http://dx.doi.org/10.1006/jmbi.1997.1508; PMID: 9480774
- Kuznetsov YG, Day J, Newman R, McPherson A. Chimeric human-simian anti-CD4 antibodies form crystalline high symmetry particles. J Struct Biol 2000; 131:108 - 15; http://dx.doi.org/10.1006/jsbi.2000.4282; PMID: 11042081
- Larson S, Day J, Greenwood A, Skaletsky E, McPherson A. Characterization of crystals of an intact monoclonal antibody for canine lymphoma. J Mol Biol 1991; 222:17 - 9; http://dx.doi.org/10.1016/0022-2836(91)90731-K; PMID: 1942064
- Saphire EO, Parren PW, Barbas CF 3rd, Burton DR, Wilson IA. Crystallization and preliminary structure determination of an intact human immunoglobulin, b12: an antibody that broadly neutralizes primary isolates of HIV-1. Acta Crystallogr D Biol Crystallogr 2001; 57:168 - 71; http://dx.doi.org/10.1107/S0907444900017376; PMID: 11134947
- Saphire EO, Stanfield RL, Crispin MD, Morris G, Zwick MB, Pantophlet RA, et al. Crystal structure of an intact human IgG: antibody asymmetry, flexibility, and a guide for HIV-1 vaccine design. Adv Exp Med Biol 2003; 535:55 - 66; http://dx.doi.org/10.1007/978-1-4615-0065-0_4; PMID: 14714888
- Stura EA, Satterthwait AC, Calvo JC, Stefanko RS, Langeveld JP, Kaslow DC. Crystallization of an intact monoclonal antibody (4B7) against Plasmodium falciparum malaria with peptides from the Pfs25 protein antigen. Acta Crystallogr D Biol Crystallogr 1994; 50:556 - 62; http://dx.doi.org/10.1107/S0907444994001782; PMID: 15299418
- Jakob CG, Edalji R, Judge RA, et al. Structure Reveals Function of The Dual Variable Domain Immunoglobulin (DVD-Ig™) Molecule. MAbs 2013; In Press
- Frank J. Three-dimensional electron microscopy of macromolecular assemblies. Oxford, UK: Oxford University Press, 2006.
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 1987; 146:113 - 36; http://dx.doi.org/10.1111/j.1365-2818.1987.tb01333.x; PMID: 3302267
- Dangl JL, Wensel TG, Morrison SL, Stryer L, Herzenberg LA, Oi VT. Segmental flexibility and complement fixation of genetically engineered chimeric human, rabbit and mouse antibodies. EMBO J 1988; 7:1989 - 94; PMID: 3138110
- Oi VT, Vuong TM, Hardy R, Reidler J, Dangle J, Herzenberg LA, et al. Correlation between segmental flexibility and effector function of antibodies. Nature 1984; 307:136 - 40; http://dx.doi.org/10.1038/307136a0; PMID: 6690993
- Reidler J, Oi VT, Carlsen W, Vuong TM, Pecht I, Herzenberg LA, et al. Rotational dynamics of monoclonal anti-dansyl immunoglobulins. J Mol Biol 1982; 158:739 - 46; http://dx.doi.org/10.1016/0022-2836(82)90258-3; PMID: 7120418
- Roux KH. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol 1989; 178:130 - 44; http://dx.doi.org/10.1016/0076-6879(89)78010-1; PMID: 2601621
- Roux KH. Immunoglobulin structure and function as revealed by electron microscopy. Int Arch Allergy Immunol 1999; 120:85 - 99; http://dx.doi.org/10.1159/000024226; PMID: 10545762
- Roux KH, Greenspan NS. Monitoring the formation of soluble immune complexes composed of idiotype and anti-idiotype antibodies by electron microscopy. Mol Immunol 1994; 31:599 - 606; http://dx.doi.org/10.1016/0161-5890(94)90167-8; PMID: 8196670
- Gregory L, Davis KG, Sheth B, Boyd J, Jefferis R, Nave C, et al. The solution conformations of the subclasses of human IgG deduced from sedimentation and small angle X-ray scattering studies. Mol Immunol 1987; 24:821 - 9; http://dx.doi.org/10.1016/0161-5890(87)90184-2; PMID: 3657808
- Yarmush DM, Murphy RM, Colton CK, Fisch M, Yarmush ML. Quasi-elastic light scattering of antigen-antibody complexes. Mol Immunol 1988; 25:17 - 32; http://dx.doi.org/10.1016/0161-5890(88)90086-7; PMID: 3343970
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 2005; 151:41 - 60; http://dx.doi.org/10.1016/j.jsb.2005.03.010; PMID: 15890530
- Yoshioka C, Pulokas J, Fellmann D, Potter CS, Milligan RA, Carragher B. Automation of random conical tilt and orthogonal tilt data collection using feature-based correlation. J Struct Biol 2007; 159:335 - 46; http://dx.doi.org/10.1016/j.jsb.2007.03.005; PMID: 17524663
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 2009; 166:95 - 102; http://dx.doi.org/10.1016/j.jsb.2009.01.002; PMID: 19263523
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy 2005; 104:8 - 29; http://dx.doi.org/10.1016/j.ultramic.2005.02.004; PMID: 15935913
- Voss NR, Lyumkis D, Cheng A, Lau PW, Mulder A, Lander GC, et al. A toolbox for ab initio 3-D reconstructions in single-particle electron microscopy. J Struct Biol 2010; 169:389 - 98; http://dx.doi.org/10.1016/j.jsb.2009.12.005; PMID: 20018246
- Voss NR, Potter CS, Smith R, Carragher B. Software tools for molecular microscopy: an open-text Wikibook. Methods Enzymol 2010; 482:381 - 92; http://dx.doi.org/10.1016/S0076-6879(10)82016-6; PMID: 20888970
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 2009; 166:205 - 13; http://dx.doi.org/10.1016/j.jsb.2009.01.004; PMID: 19374019
- Sorzano CO, Marabini R, Velázquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 2004; 148:194 - 204; http://dx.doi.org/10.1016/j.jsb.2004.06.006; PMID: 15477099
- Orlova EV, Saibil HR. Structural analysis of macromolecular assemblies by electron microscopy. Chem Rev 2011; 111:7710 - 48; http://dx.doi.org/10.1021/cr100353t; PMID: 21919528
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671 - 5; http://dx.doi.org/10.1038/nmeth.2089; PMID: 22930834
- Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernández G, et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 2010; 171:197 - 206; http://dx.doi.org/10.1016/j.jsb.2010.03.011; PMID: 20362059
- Yoshioka C, Lyumkis D, Carragher B, Potter CS. Maskiton: Interactive, web-based classification of single-particle electron microscopy images. J Struct Biol 2013; In press http://dx.doi.org/10.1016/j.jsb.2013.02.007; PMID: 23428431