Abstract
Horizontal Gene Transfer (HGT) is a major force in bacterial evolution. Bacteria are under a strong selection to optimize their growth rate by improving features related to their codon usage. In a recent study we have shown that these two forces are coupled: 1) the codon bias of transferred genes has a strong influence on the probability that they will become fixed in the new genome, and 2) frequent HGTs may increase the similarity in the tRNA pools of organisms within the same community. Thus, methods for inferring HGTs probably underestimate their number. We believe that that the principles that affect microorganisms may also apply to mobile genetic elements (including plasmids, viruses, and transposons).
Horizontal Gene Transfer (HGT), a process in which one organism incorporates genetic material from another without being its offspring, is a highly significant phenomenon in microbial evolution.Citation1–Citation5 A fundamental question in this context is related to what determines the success of such events.Citation4,Citation6–Citation8
Gene translation and its efficiency is a central process in all organismsCitation9–Citation12 and an important determinant of the growth rate of unicellular creatures.Citation10,Citation13–Citation16 Due to the significant correlation between the genomic copy number of tRNA species and their cellular expression levels, it is possible to estimate the translation speed of all the possible codons in different organisms.Citation10,Citation17,Citation18 Thus, it is feasible to study the effect of translation efficiency on HGT in a systems biology context.
Based on the analysis of the genomes of dozens of prokaryotes and the HGTs between them,Citation3,Citation4 we have recently suggested in reference 1 that there are bi-directional associations between horizontal gene transfer and translation efficiency.
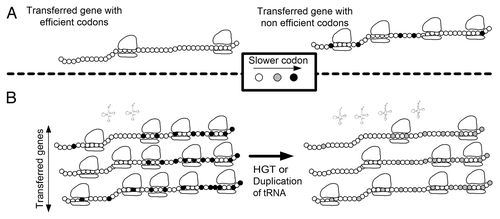
First, organisms with similar tRNA pools tend to exchange more genes. Assuming constant initiation rate and no ribosomal abortion, there should be an inverse relation between the speed of translation and the ribosomal density on a translated geneCitation10 (). Thus, transferred genes from a donor with a non-compatible tRNA pool tend to have less efficient codons and to consume more ribosomes. As a result, ribosome availability declines and the organism fitness decreasesCitation1,Citation13 ().
Second, to improve the global allocation of ribosomes and organism fitness, highly expressed genes are under stronger selection for more efficient codons.Citation13,Citation14 If the fraction of highly expressed, horizontally transferred genes is considerable,Citation1–Citation4 a change in the tRNA pool (duplication, deletion or horizontal gene transfer) that improves the translation efficiency of the highly expressed transferred genes should also increase organism fitness ().
Kudla et al.Citation13 have recently published data that demonstrate the first direction described above. In their study, they imitate the HGT of a gene, by introducing a library encoding GFP proteins to E. coli. Each of the GFP genes in this library has different codons but identical amino acid composition. They reported a strong correlation between the growth rate of the treated E. coli and the translation efficiency of the GFP version ().
Wet lab validations of the second direction pose a greater challenge. For example, it can be partially validated by introducing a set of neutral genes to E. coli at the first stage; and at the second stage, introducing tRNA gene(s) from the same origin (). The growth rate of the host should be measured before and after introducing the tRNA gene(s).
The fact the transferred genes already have codons that are adapted to the host implies that it is non-trivial to detect such genes. Specifically, methods that detect HGT based on the assumption that transferred genes have atypical codon usage or GC content (see, for example, references Citation4, Citation19–Citation20) probably underestimate the number of HGT events. Thus, the numbers of inferred HGTs reported in previous studies in references Citation3, Citation4, Citation19 and Citation20 are probably significantly lower than the actual numbers. On the more positive side, the fact that genes tend to be transferred between organisms with similar tRNA pools and genes with codons that are compatible to the tRNA pool of the host have higher probability to be transferred, can be incorporated into existing methods for HGT inference to improve their accuracy.
Finally, we believe that similar principles may hold for Mobile Genetic Elements (MGE; including plasmids, viruses and transposons). For example, it has already been suggested that in many cases viruses have a codon bias that is similar to their host.Citation21 In addition, the translation efficiency of the genes encoded in a plasmid is determined by its codon bias and the tRNA pool of the host, and should have significant impact on the success of its duplication or its transfer from one host to another. Similarly, it is possible that transposons that have transposases and additional ORFs (see, for example, reference Citation22) that are encoded by more efficient codons, have a higher transposition rate. Further research on this topic is required to establish these conjectures. The adaptation levels of different genes in a MGE to the tRNA pool of the host may be related to their function and expression levels. For example, genes that encode proteins that appear more frequently in viruses (e.g., in the envelope) are expected to have higher adaptation to the host tRNA pool.
However, one should keep in mind that there are (relatively rare) cases in which MGEs include tRNA genes (see, for example, reference Citation23). In such cases, the MGEs may include genes with codons that are not adapted to the host together with tRNA species that recognize these codons that rarely appear in the host genome.
Figures and Tables
Figure 1 Relations between translation efficiency and horizontal gene transfer. (A) Transferred genes with neutral function but with codons that are adapted to the host have a smaller effect on the fitness of the host as they recruit less ribosomes (left) than genes with codons that are not adapted to the host (slower codons; right). (B) A duplication or horizontal transfer of a tRNA that recognizes rare codons in the transferred genes may improve the translation efficiency of these genes and thus decrease their ribosomal density. After such a change in the tRNA pool, the number of available ribosomes increases, and thus the fitness of the host is augmented (right vs. left).
Acknowledgements
T.T. is supported by the Koshland center for basic research. We would like to thank Ms. Hadas Zur, Dr. Uri Gophna, Prof. Martin Kupiec and Prof. Eytan Ruppin for their helpful comments.
Addendum to:
References
- Tuller T, Girshovich Y, Sella Y, Kreimer A, Freilich S, Kupiec M, et al. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res 2011; 2011:22
- Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci USA 2007; 104:2043 - 2049
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA 2008; 105:10039 - 10044
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 2004; 36:760 - 766
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature 2000; 405:299 - 304
- Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 2007; 318:1449 - 1452
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 2005; 3:711 - 721
- Wellner A, Lurie MN, Gophna U. Complexity, connectivity and duplicability as barriers to lateral gene transfer. Genome Biol 2007; 8:156
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 2010; 12:32 - 42
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 2010; 141:344 - 354
- Shah P, Gilchrist MA. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet 2010; 6:e1001128
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 2009; 10:715 - 724
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science 2009; 324:255 - 258
- Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA 2010; 107:3645 - 3650
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 1985; 2:13 - 34
- Rocha EP. Codon usage bias from tRNA's point of view: redundancy, specialization and efficient decoding for translation optimization. Genome Res 2004; 14:2279 - 2286
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 2004; 32:5036 - 5044
- Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 1999; 238:143 - 155
- Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res 2011; 2011:26
- Garcia-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res 2003; 31:187 - 189
- Bahir I, Fromer M, Prat Y, Linial M. Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol Syst Biol 2009; 5:311
- Witherspoon DJ, Doak TG, Williams KR, Seegmiller A, Seger J, Herrick G. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol Biol Evol 1997; 14:696 - 706
- Bailly-Bechet M, Vergassola M, Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res 2007; 17:1486 - 1495