Abstract
The genetically defined point centromeres of budding yeasts and the epigenetically specified regional centromeres of all other eukaryotes harbor a common epigenetic mark in the form of a non-standard nucleosome. Although, the composition of the protein core of the centromere specific nucleosome and the nature of the DNA wrap around it are at present controversial, there is no doubt that this specialized nucleosome harbors a variant of the standard histone H3 (cenH3). The association of cenH3, called Cse4 in Saccharomyces cerevisiae, with the partitioning locus (STB) of the high copy selfish plasmid 2 micron circle that resides in the yeast nucleus and propagates itself stably is intriguing. Recent observations are consistent with Cse4 being a nucleosome component at STB. A common nucleosome identity for the partitioning loci of the chromosomes and the plasmid of yeast support arguments based on evolutionary considerations that the origin of the unusual point centromere of budding yeasts may be traced to the STB locus of an ancestral plasmid.
Centromeres are chromosomal regions that promote nearly perfect fidelity in the segregation of eukaryotic genomes during cell division. The high-order multi-protein complex called kinetochore assembled at the centromere is responsible for the bipolar attachment of sister chromatids, paired by the cohesin complex, to microtubules emanating from opposite centrosomes (called spindle pole bodies in yeast).Citation1,Citation2 Dissolution of the cohesin bridge during anaphase splits the sisters asunder, and they are pulled away from each other by oppositely directed spindle forces.Citation3
Regional and Point Centromeres
The highly conserved function of centromeres in directing one-to-one segregation of duplicated chromosomes is contrasted by the wide range of size and complexity, as well as a lack of shared sequence motifs, among themCitation4,Citation5 (). There are two strikingly distinct classes of centromeres: the widespread regional centromeres and the extremely rare point centromeres. The former varies in length from tens of kilo bp in fungi to hundreds of kilo bp in flies, plants and animals. The human centromeres are 1–10 mega bp long with the core regions ranging from 0.5 to 1.5 mega bp.
Regional centromeres are epigenetically specified, are almost always embedded in heterochromatin and components of the heterochromatin machinery and RNA interference (RNAi) system are important for their establishment.Citation6–Citation8 The emergence of neocentromeres in chromosomal regions with no consensus sequence motifs, and without a history of having previously housed centromeres, that can be propagated through cell divisions provides a strong argument for the epigenetic nature of regional centromeres.Citation9,Citation10 The point centromere typified by the centromeres in Saccharomyces cerevisiae, though starkly different in DNA organization and mode of establishment from the regional centromere, serves the same function of promoting faithful chromosome segregation. This short ∼125 bp centromere is genetically defined, and is functional when transplanted to a non-native niche, for example, a plasmid.Citation11,Citation12 It consists of three DNA elements, CDE I (∼10 bp), CDE II (∼85 bp; AT rich) and CDE III (∼25 bp). The CBF3 complex, which interacts specifically with CEN DNA, provides the scaffold for organizing the kinetochore complex.Citation13 The point centromere is restricted to the small fungal lineage of budding yeasts-Saccharomycetaceae. Based on the preponderance of regional centromeres, it is reasonable to suppose that an ‘ancestral regional centromere’ served as the start point for the evolution of extant eukaryotic centromeres.Citation14 The point centromere appears to be an evolutionary quirk, and an explanation for its origin, as well as its rarity, poses a serious conceptual challenge.
A Conserved Epigenetic Nucleosome Signature of Centromeres
A unifying feature of regional and point centromeres is an epigenetic mark in the form of an unusual nucleosome that they both harbor. The histone H3, which is an integral component of the standard octameric nucleosome core of chromatin, is lacking in the centromeric nucleosome. Instead, it is replaced by a histone H3 variant, CenH3 (CENP-A in mammals; CID in Drosophila).Citation4,Citation15 In Saccharomyces cerevisiae, CenH3 is called Cse4. Whereas Cse4 retains the canonical histone fold in its C-terminal domain, it harbors extra amino acid sequences at its N-terminus. The N-terminal addition is variable in length among different organisms ().
Whereas regional centromeres contain multiple copies of the CenH3 containing nucleosomes, there is a single Cse4 containing nucleosome positioned at each centromere in S. cerevisiae.Citation16 This stoichiometry is consistent with the limited number of microtubules in yeast, restricting the association of a single kinetochore microtubule with each chromosome.Citation17 The amount of Cse4 in the nucleus is tightly regulated by its rapid turnover via the ubiquitin mediated protein degradation pathway.Citation18,Citation19 Only Cse4 associated with a centromere is stable whereas that associated elsewhere with chromatin is subject to degradation. Even free Cse4 (unbound to DNA) is likely short lived in the nucleus. By strictly limiting the pool of Cse4, the potential assembly of supernumerary kinetochores, with the consequent undesirable effects on proper chromosome segregation, can be avoided.
Acquisition of Cse4 by the Yeast Selfish Plasmid for Chromosome Coupled Segregation
The multi-copy plasmid of S. cerevisiae, 2 micron circle, present in the yeast nucleus at 40–60 copies per cell, poses a rather challenging conundrum with respect to the cellular economy of Cse4. The plasmid is an example of a highly optimized selfish DNA element that persists successfully in host populations by coupling its segregation to that of chromosome segregation.Citation20 This efficient mode of equal segregation is accomplished by two plasmid coded proteins (Rep1 and Rep2) channeling several chromosome segregation factors, including Cse4, to the partitioning locus of the plasmid STB.Citation21 Current evidence is consistent with the plasmid, apparently organized as a cluster, forming two sister clusters following DNA replication, and segregating one-to-one to daughter cells.Citation22 It is unlikely that plasmid segregation is mediated by direct attachment to the mitotic spindle. More likely, sister plasmid clusters tethered to sister chromatids hitchhike on them, thus achieving nearly chromosome like fidelity in partitioning. Mammalian viral episomes utilize a chromosome tethering strategy to ensure stable propagation during the latent phase of infection.Citation23 Chromosome attachment protects against the extrusion of viral genomes into the cytoplasm when the nuclear envelope breaks down during mitosis and the prospect of being left behind when the envelope is reformed. Such a safety mechanism does not apply to the 2 micron circle, as the closed mitosis performed by its host does not involve turnover of the nuclear envelope. Since the chromosome segregation machinery is nearly error-free, it is not surprising that diverse parasite elements have independently evolved mechanisms for chromosome associated segregation.
The Paradox between the Strict Cellular Economy of Cse4 and High Plasmid Copy Number
With 40–60 copies of the 2 micron plasmid housed in each nucleus, and Cse4 being an important host factor for their equal segregation, a key question is how the plasmid is able to cope with the cellular controls that stringently limit the availability of Cse4? Our recent study suggests that the occupancy of Cse4 at STB is limited to the origin-proximal region of STB consisting of 5 copies of a 60 bp repeated consensus element, and three of the repeat elements are necessary and sufficient for Cse4-STB associationCitation24 (). Furthermore, the available data are consistent with the occupancy of one Cse4 containing nucleosome, and not more than two such nucleosomes, at STB. Indeed, the association of centromere-specific nucleosome with the plasmid partitioning locus is intriguing. Curiously, two 124 bp sequences with a very high percent of sequence identity can be arranged as two tandem direct repeats within STB-proximal.Citation25 Recall that the length of the S. cerevisiae centromere is ∼125 bp. Even if one assumes that there is a single Cse4 containing nucleosome per STB, one is faced with the rather irksome proposition that a non-essential parasite genome (∼60 copies) garners nearly four times the amount of a sparse cellular factor as the host genome, comprising 16 chromosomes in the haploid state.
A hint to how the apparent paradox of the high plasmid copy number versus a low supply of Cse4 may be reconciled is perhaps provided by our findings that immunoprecipitation of a high copy STB reporter plasmid by a Cse4-directed antibody is quite inefficient.Citation24 It is possible that the plasmid is able to satisfy its functional needs by utilizing a sub-stoichiometric amount of Cse4. We have previously noted this less than one-to-one stoichiometry for the assembly of the yeast cohesin complex, also utilized by the plasmid for its segregation, at STB.Citation26 The plasmid partitioning strategy might engender a mechanism by which the host is not deprived of critical factors that it needs for normal chromosome segregation. What the molecular basis of such a strategy might be is the topic for future investigation. It is possible that the plasmid may be able to limit its need for Cse4 through cooperative interactions among individual plasmid molecules. Moderating its selfishness, so as not to impose a significant fitness penalty on its host, would confer an advantage on any selfish element, especially a multi-copy one, whose survival is intricately linked to that of its host.
The Functional Topology of STB Chromatin
Although current evidence is most consistent with the presence of a Cse4 containing nucleosome at STB, a non-nucleosomal mode of Cse4 association with STB cannot be completely ruled out. A recent observation suggests that the occupancy of a Cse4 containing nucleosome induces positive DNA super-coiling at the centromere chromatin of S. cerevisiae in vivo.Citation27 The precise interpretation of this finding, whether, for example, DNA is wrapped in a right handed fashion around a Cse4 containing nucleosome core, is under debate at this time.Citation4 Whereas in vitro assembly conditions can promote either right handed (positive writhe) or left handed (negative writhe) wrapping of DNA in CenH3 containing nucleosomes,Citation27–Citation29 current evidence favors positive writhe sequestered by Cse4 nucleosomes at yeast centromeres in vivo.Citation27 The core composition of the CenH3 containing nucleosome is also not resolved: is it an octamer, or a tetramer or an unusual hexamer lacking histones H2A and H2B but containing a Cse4 chaperone in their place? While one awaits the answer to these questions, it is worthwhile to ask whether the STB chromatin also harbors a positive writhe in vivo in its functional state. This rare DNA topology, a positive supercoil refractory to relaxation by topoisomerases, contrasted by negative supercoiling of DNA nearly everywhere else in chromatin would be a strong indicator that a Cse4 containing nucleosome is located at STB. A shared, but non-standard, DNA writhe between CEN and STB would be consistent with a possible evolutionary connection between these two loci (see below).
A Shared Nucleosome Mark between the Partitioning Loci of the 2 Micron Plasmid and Yeast Chromosomes: Evolutionary Implications
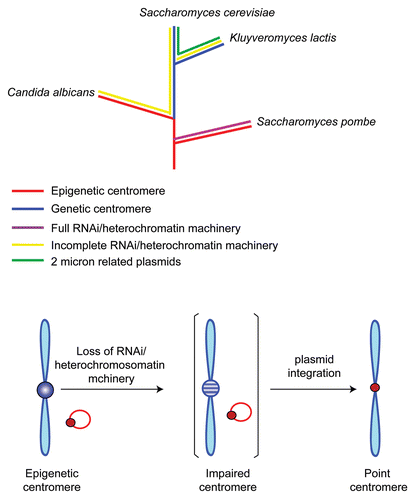
The presence of a centromeric nucleosome at the partitioning locus of a selfish plasmid has important implications in answering the perplexing connection, or lack of it, between the regional and point centromeres referred to earlier. It is striking that the budding yeast lineage has undergone a complete or almost complete loss of the protein components of the machineries for implementing RNA interference and pericentric heterochromatin.Citation30 Perhaps the resultant incapacitation of the regional centromere in the ancestor of this lineage was compensated for by the domestication of the partitioning locus of an endogenous plasmid as a ‘neo-centromere’Citation5 (). The present day chromosome segregation and plasmid segregation pathways in S. cerevisiae mediated by the point centromere and STB, respectively, likely represent two divergent, but non-conflicting, solutions that were arrived at for faithful chromosome propagation on the one hand and stable plasmid persistence on the other. The cellular factors shared between the plasmid and chromosomes might then denote vestiges of their evolutionary kinship. The notion of the plasmid origin of the chromosome centromere in Saccharomycetaceae is further strengthened by the fact that this is only lineage among fungi, as well as other eukaryotes, that harbor plasmids related to the 2 micron circle.
The oddities shared by the point centromere and STB also extend to the proteins that orchestrate the assembly of the kinetochore complex and the plasmid partitioning complex at these loci. The inner kinetochore proteins Ndc10 and Ctf13, are integral components of the CBF3 complex which provides the basic platform for the kinetochore complex. Yet they have no homologues among kinetochore proteins outside of Saccharomycetaceae.Citation5 Similarly the Rep1 and Rep2 proteins, central to plasmid partitioning, also lack homologues outside the family of 2 micron circle related yeast plasmids. Even within this small group of proteins, strong divergence among the Rep2 proteins has all but erased the amino acid sequence evidence for their relatedness. Even if Ndc10 and Ctf13 did share common ancestors with Rep1 and Rep2, it is not surprising that they now carry no trace of this family history. Since the host must bear a fitness cost for harboring plasmids, however small it might be, chromosome segregation would have evolved away from the original pathway that it shared with the plasmid. The plasmid must have countered by evolving strategies by which it could still exploit the chromosome segregation machinery for its efficient propagation. The near absence of sequence homology among the Rep2 proteins may denote the distinct evolutionary paths followed by individual plasmids in response to the evolution of their respective hosts. The atomic structures of Ndc10-Ctf13 and Rep1-Rep2 proteins would indeed be useful in revealing possible shared evolutionary paths among these proteins in the form of conserved three dimensional peptide motifs.
Epilogue
The 2 micron plasmid has been extensively exploited as a tool for introducing genes into yeast and shuttling genes between bacteria and yeast. Furthermore, the Flp recombination system, harbored by the plasmid for its copy number maintenance, has been put to good use for directed genome engineering in a variety of organisms. Oddly though, the plasmid has been a highly neglected entity from the standpoint of its utility for addressing fundamental biological questions of wide interest. The provocative intellectual issues raised by the interactions of Cse4 and other chromosome segregation factors with the plasmid call for a change of heart in the general perception of its scientific value and significance.
Abbreviations
| CBF3 | = | centromere binding factor |
| CDE | = | centromeric DNA element |
| CEN | = | centromere |
| CenH3 | = | centromere specific histone H3 variant |
| STB | = | stability conferring partitioning locus of the yeast 2 micron plasmid |
Figures and Tables
Figure 1 The variation in centromere sizes among eukaryotes is schematically illustrated. The simple point centromere of the budding yeast is only 125 bp long, and is organized into three well defined centromere DNA elements (CDE I, II and III). The fission yeast centromere is roughly 600 times larger, and is comprised of a ∼25 kbp central region that includes a ∼5 kbp core flanked immediately by two large inverted repeats (imrs; innermost repeats). The distal borders of the centromere harbor smaller repeats (otrs; outer repeats). The human centromere is roughly 60–70 times the size of the fission yeast centromere. Its repeating motif is a 171 bp element known as the α-satellite (alphoid) DNA. Also included within this array is a regularly repeated 17 bp consensus sequence to which the CENP-B protein binds.
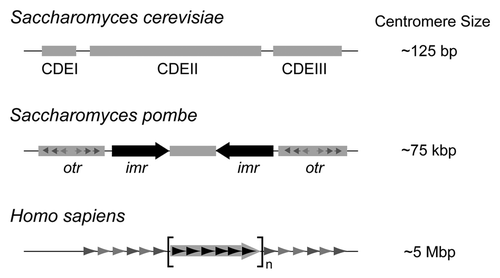
Figure 2 The standard histone H3 and centromere specific histone H3 (cenH3) organizations are compared between budding yeast and humans. The N-terminal tail of S. cerevisiae Cse4 is longer than that of human CENP-A. The sub-organizations within their conserved C-terminal histone fold domains are shown. The CENP-A targeting domain (CATD) differs from the corresponding H3 region by multiple amino acid substitutions. A subset of these changes is responsible for the reversal of surface charge within the bulged loop L1 between CENP-A and H3. The CENP-A-CENPA and CENPA-histoneH4 interfaces are thus altered significantly from H3-H3 and H3-H4 interfaces, respectively.Citation29
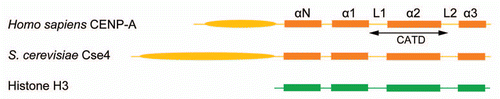
Figure 3 The partitioning locus STB of the 2 micron circle is located between the PstI and AvaI restriction sites on the plasmid genome. STB can be subdivided into two regions, one proximal to the plasmid replication origin and the other distal, by the HpaI site situated roughly half way between PstI and AvaI. STB-proximal harbors a tandem array of five copies of a 60 bp element. This is the region where Cse4 is localized on the plasmid.Citation24 STB-distal contains the termination signal for two plasmid transcripts directed towards the origin. It is likely that, analogous to the centromere, transcription through STB proximal may antagonize its partitioning competence. There is also a silencing element (shaded box) within STB-distal, which depresses the activity of a promoter in whose vicinity it is placed.
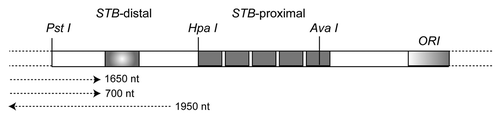
Figure 4 In a plausible scheme for the transition from regional centromere to the point centromere, a key event is the loss of the host's capacity for RNA interference and heterochromatin assembly. This would be antithetical to the epigenetic maintenance of regional centromeres. The partial or complete loss of RNAi and heterochromatin machineries in the budding yeast lineage is schematically diagrammed (after Malik and Henikoff, ref. Citation5). The integration of an endogenous plasmid into the host genome would have resulted in a salvage pathway for chromosome segregation promoted by the plasmid partitioning proteins, with the plasmid partitioning locus serving as the new centromere. Schizosaccharomyces pombe and Candida albicans harbor epigenetic centromeres, even though the latter has suffered a loss of proteins of the RNA i and heterochromatin machineries. Interestingly, Candida centromeres are relatively small (3–5 kbp), have only a limited number of flanking repeats and are rapidly evolving.Citation31
Acknowledgements
Work in the Jayaram laboratory is supported by grants from the National Institutes of Health (GM064363), the National Science Foundation (MCB-1049925) and a Robert F Welch Foundation award (F-1274).
References
- Tanaka TU. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J 2010; 29:4070 - 4082; PMID: 21102558; http://dx.doi.org/10.1038/emboj.2010.294
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell 2010; 19:797 - 806; PMID: 21145497; http://dx.doi.org/10.1016/j.devcel.2010.11.011
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem 2005; 74:595 - 648; PMID: 15952899; http://dx.doi.org/10.1146/annurev.biochem.74.082803.133219
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 2011; 144:471 - 479; PMID: 21335232; http://dx.doi.org/10.1016/j.cell.2011.02.002
- Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell 2009; 138:1067 - 1082; PMID: 19766562; http://dx.doi.org/10.1016/j.cell.2009.08.036
- Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 2008; 319:94 - 97; PMID: 18174443; http://dx.doi.org/10.1126/science.1150944
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 2010; 20:134 - 141; PMID: 20207534; http://dx.doi.org/10.1016/j.gde.2010.02.003
- Allshire RC. RNA interference, heterochromatin and centromere function. Cold Spring Harb Symp Quant Biol 2004; 69:389 - 395; PMID: 16117672; http://dx.doi.org/10.1101/sqb.2004.69.389
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA 2004; 101:6542 - 6547; PMID: 15084747; http://dx.doi.org/10.1073/pnas.0308637101
- Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, et al. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J 2001; 20:2087 - 2096; PMID: 11296241; http://dx.doi.org/10.1093/emboj/20.8.2087
- Carbon J, Clarke L. Centromere structure and function in budding and fission yeasts. New Biol 1990; 2:10 - 19; PMID: 2078550
- Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol 2002; 157:199 - 203; PMID: 11956223; http://dx.doi.org/10.1083/jcb.200201052
- Lechner J, Carbon J. A 240 kD multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 1991; 64:717 - 725; PMID: 1997204; http://dx.doi.org/10.1016/0092-8674(91)90501-O
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 2006; 7:R23; PMID: 16563186; http://dx.doi.org/10.1186/gb-2006-7-3-r23
- Bernad R, Sanchez P, Losada A. Epigenetic specification of centromeres by CENP-A. Exp Cell Res 2009; 315:3233 - 3241; PMID: 19660450; http://dx.doi.org/10.1016/j.yexcr.2009.07.023
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA 2007; 104:14706 - 14711; PMID: 17804787; http://dx.doi.org/10.1073/pnas.0706985104
- Bouck DC, Joglekar AP, Bloom KS. Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu Rev Genet 2008; 42:335 - 359; PMID: 18680435; http://dx.doi.org/10.1146/annurev.genet.42.110807.091620
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, et al. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 2010; 40:444 - 454; PMID: 21070970; http://dx.doi.org/10.1016/j.molcel.2010.10.014
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 2010; 40:455 - 464; PMID: 21070971; http://dx.doi.org/10.1016/j.molcel.2010.09.025
- Ghosh SK, Hajra S, Paek A, Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu Rev Biochem 2006; 75:211 - 241; PMID: 16756491; http://dx.doi.org/10.1146/annurev.bio-chem.75.101304.124037
- Jayaram M, Yang XM, Mehta S, Voziyanov Y, Velmurugan S. Funnell BE, Phillips G. The 2 micron plasmid of Saccharomyces cerevisiae. Plasmid Biology 2004; Washington DC ASM Press 303 - 324
- Ghosh SK, Hajra S, Jayaram M. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc Natl Acad Sci USA 2007; 104:13034 - 13039; PMID: 17670945; http://dx.doi.org/10.1073/pnas.0702996104
- Botchan M. Hitchhiking without covalent integration. Cell 2004; 117:280 - 281; PMID: 15109488; http://dx.doi.org/10.1016/S0092-8674(04)00410-6
- Huang CC, Hajra S, Ghosh SK, Jayaram M. Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: implications in centromere evolution. Mol Cell Biol 2011; 31:1030 - 1040; PMID: 21173161; http://dx.doi.org/10.1128/MCB.01191-10
- Murray JA, Cesareni G. Functional analysis of the yeast plasmid partition locus STB. EMBO J 1986; 5:3391 - 3399; PMID: 16453734
- Ghosh SK, Huang CC, Hajra S, Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res 2010; 38:570 - 584; PMID: 19920123; http://dx.doi.org/10.1093/nar/gkp993
- Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell 2009; 138:104 - 113; PMID: 19596238; http://dx.doi.org/10.1016/j.cell.2009.04.049
- Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem 2011; 286:4021 - 4026; PMID: 21115484; http://dx.doi.org/10.1074/jbc.M110.189340
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 2010; 467:347 - 351; PMID: 20739937; http://dx.doi.org/10.1038/nature09323
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci USA 2000; 97:11319 - 11324; PMID: 11016957; http://dx.doi.org/10.1073/pnas.200346997
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci USA 2008; 105:19797 - 19802; PMID: 19060206; http://dx.doi.org/10.1073/pnas.0809770105