Abstract
Low-copy number plasmids need a segregation mechanism to assort one half of the plasmid copies to each daughter cell during cell division. This can be achieved directly by partitioning plasmid copies through a mechanism reminiscent of eukaryotic mitosis. Briefly, plasmid copies are paired around a centromere-like site, and then separated toward the daughter cells at cell division. Partition mechanisms are used by a majority of well-studied plasmids. They involve two proteins, a DNA-binding protein and a motor protein, besides the centromeric site. However, some plasmids do not encode typical partition systems, so alternative segregation mechanisms must be considered. For instance, chromosome segregation could provide the driving force for plasmid movement, through a “pilot-fish”-like mechanism. In support of this assumption, we recently demonstrated that plasmid R388 segregation, which does not involve a plasmid-encoded motor protein, requires a single plasmid-encoded DNA-binding protein. Besides, the new segregation system becomes essential when the plasmid encodes conjugation machinery, providing a new understanding of how plasmids integrate conjugative transfer with segregation.
The Vertical Inheritance of Plasmids
Plasmids are, along with integrative and conjugative elements (ICEs) and phages, the major carrier platforms for horizontal gene transfer. They can be considered as an intriguing combination of parasite and benefactor to the individual cell as well as a repository for a shared gene pool within a population. Being non-essential extra-chromosomal pieces of DNA, plasmids need to evolve a number of strategies that allow them to persist in a growing bacterial population. The faithful inheritance of plasmids does not rely extensively on host-encoded factors. Rather, it is ensured by three different classes of plasmid-encoded maintenance functions: multimer resolution systems, addiction systems, and active segregation systems (usually called partition systems, see below). Large low-copy number plasmids seem to require a combination of all three types of system to ensure sufficient level of inheritance.Citation1
Multimer resolution systems act by removing plasmid multimers, which can be produced by replication and recombination and create instability by reducing the number of segregation units.Citation2 Plasmid-encoded addiction systems are responsible for the post-segregational elimination of plasmid-free cells from the host population.Citation3,Citation4 Lastly, plasmid segregation systems ensure equitable distribution of plasmid copies to daughter cells during cell division. In the absence of such systems, plasmids likely distribute randomly inside the cell and would be eventually lost. Some plasmids are maintained at such high number of copies that the fraction of plasmid-free cells is low, even if they lack addiction or segregation systems. In contrast, plasmids with low copy number, which are maintained at less than ten copies per chromosome, cannot rely on a stochastic distribution of copies and require dedicated segregation processes.Citation5
Segregation by Active Partition
The best characterized segregation systems consist of active partition machineries (par systems), which separate and accurately position sister copies of the plasmid toward specific cellular sites in each daughter cell in a process akin to eukaryotic mitosis. Plasmid par systems are of different types and the associated mechanisms are not yet clearly understood. In any case, all appear to share the propensity to form nucleotide-driven cytomotive filaments to move and position replicated plasmid molecules.Citation6 There are evidences that such cytomotive filaments accomplish plasmid partitioning either by pushing plasmids copies attached to growing filaments or by pulling plasmids copies attached to shrinking filaments by polymerization and depolymerization.Citation7 However, how this movement achieves plasmid separation remains unclear. Notably, the ATPase activities of motor proteins of the classical par systems may be too low to provide sufficient energy to mediate plasmid movement, which suggests that they do not function as a direct driving force in plasmid movement.
Plasmid par systems typically encode three essential segregation components: an NTPase motor protein, a cis-acting centromere-like site, and a specific DNA-binding adaptor protein, which functions as a link between the centromeric site and the motor protein (for reviews see refs. Citation8–Citation10). These two proteins are generally encoded in an autoregulated operon ().Citation11 As shown in the figure, plasmid par systems are divided into three major types, exemplified by those of plasmids F and P1 (Type I), R1 (Type II), and pBtoxis (Type III). They are differentiated by the protein family to which their motor protein belongs. Namely, they are homologous to three specific cytoskeletal protein families: Walker type, actin-like and tubulin-like. Type I par systems encode a deviant Walker type P-loop ATPase related to the chromosome segregating protein Soj, as well as to the cell division protein MinD.Citation12,Citation13 Type II systems utilize actin-like ATPases related to the chromosome segregating protein MreB, as well as the cell division protein FtsA.Citation14,Citation15 The more recently described type III systems encode a tubulin-like GTPase, related to the cytoskeleton protein FtsZ that initiates and directs bacterial cell division.Citation16
Despite their similarities in genetic organization (), these three par types use entirely different molecular mechanisms, as shown in . The type II par system, exemplified by plasmid R1, is the best characterized. It acts through a simple pushing mechanism. The ParM motor protein forms actin-like filaments, analogous to the eukaryotic mitotic spindle, that drive a bidirectional movement of plasmid copies, which are attached to both elongating tips, toward the cell polesCitation15 (). Type III par systems utilize a pulling mechanism. The tubulin-like GTPase TubZ of plasmid pBtoxis forms dynamic filaments that move rapidly along one side of the cell and grow back to the opposite side of the cell in a treadmilling-like pattern, reminiscent of eukaryotic microtubulesCitation16 (). Finally, various models have been proposed for the mode of action of the type I Walker partition ATPases. There is some evidence for a pulling mechanismCitation17 () while, more recently, a diffusion ratchet model was proposed for P1 plasmidCitation18 (). In this case, the motive force for plasmid positioning does not directly rely on the motor protein ParA polymerization, but instead is directed by a dynamic gradient of ParA in the cell. The uneven distribution of ParA molecules in the cell depends on the affinity of ParA for non-specific host nucleoid DNA when bound to ATP, and the ability of the adaptor protein ParB bound to the plasmid DNA to release ParA from the nucleoid by stimulating its ATPase activity.
Segregation without a Motor Protein
Although a lot remains to be discovered, the previous section indicates that we have reached a basic understanding of the mechanisms responsible for the three types of “classical” par systems. On the other hand, a significant number of low-copy number plasmids do not contain identifiable par systems, and the way by which they ensure stable inheritance is not understood. This strongly suggests that other as yet unknown mechanisms, which allow plasmid copies to be positioned in each daughter cell during division, have to be explored. At least two segregation systems that are significantly different from classical par systems have been described: those of plasmids pSK1 and R388. The staphylococcal plasmid pSK1 was shown to need just a single protein, Par, and no ATPase activity to achieve segregation.Citation19 Although the details by which this system might function are unknown, the pSK1 Par protein contains a coiled-coil domain that was suggested to form the basis of a molecular switch analogous to the ATPase activity identified in typical par systems.Citation20
Besides, in a recent report, we presented evidence that plasmid R388 utilizes a new type of segregation system, which involves a single plasmid-encoded DNA-binding protein, StbA, and a specific cis-acting site where StbA binds, the stbDRs.Citation21 Since StbA is neither an NTPase, nor an NTP/NDP-exchange factor, we assume that R388 segregation uses either an active motor provided by the host cell, or does not require a motor protein at all.
Plasmid R388, which is maintained at four copies per chromosome in E. coli, does not contain any typical par system.Citation22 It represents a minimal conjugative plasmid genome, containing two major regions separated by the origin of conjugative transfer (oriT): one devoted to conjugation, and one devoted to general establishment and maintenance functions. Notably, R388 and a number of other plasmids carry a cluster of two operons, which are transcribed divergently from the oriT region.Citation21,Citation22 One contains the mobility genes (MOB) involved in conjugative DNA processing, and the other includes a cluster of three genes, stbA, stbB and stbC (). Our results on plasmid R388 showed that stbA is the only gene required for R388 stability in E. coli. Moreover, we found that the defect in plasmid stability caused by deletion of stbA was not due to a decrease in copy number, but a consequence of variations in the intracellular positioning of plasmid DNA molecules. Our results further demonstrated that StbA role consists in ensuring time-averaged even distribution of plasmid copies within nucleoid-containing areas ( and ).Citation21
The way by which StbA mediates the assortment of R388 molecules along the nucleoid length is unknown. However, its role is fundamentally mediated by its binding to a specific sequence located within the stb promoter and composed of an array of DNA repeats, the stbDRs.Citation21 According to secondary structure predictions, protein StbA contains two domains, a N-terminal DNA-binding domain and a C-terminal domain of unknown function. Among all proteins showing homology to StbA, amino acid conservation is restricted to the N-terminal domain.Citation21 StbA binds specifically the stbDRs through its N-terminal DNA-binding domain (our unpublished data). The StbA-stbDRs complex may then be used to pair plasmid molecules specifically with the host chromosome. According to this view, StbA may interact either directly with chromosomal DNA, or through interactions with an unknown chromosome-associated factor. We propose that plasmid R388 segregates “passively,” without true partition but rather by taking advantage of the host chromosome segregation. The host chromosome would thus act as a “pilot-fish” for plasmid segregation, by an unknown process (). A similar strategy appears to be used by eukaryotic plasmids, which by replicating in synchrony and maintaining a stable association with replicating chromatids, are segregated with their associated sister chromatids.Citation23
Since “pilot-fish” segregation does not depend on host features such as cell length, width or shape, it may be attractive for plasmids that are able to establish themselves in a wide variety of hosts. The host range of plasmids can be inferred from their genomic signatures.Citation22 No candidate evolutionary host was detected for several plasmids from the Inc.W group, to which plasmid R388 belongs, suggesting that it probably has a broad host range.Citation24 We thus propose that the mode of segregation employed by plasmid R388 may be an important feature for broadhost-range plasmids, that is, those that are able to propagate in many different hosts.
Facing the Tradeoff between Segregation and Conjugation
Plasmids often come with the genetic machinery needed to transmit themselves from host cells to new recipients by conjugation. This process requires intimate contact between the donor and the recipient cell and involves the assembly of a DNA transport channel at the cell membrane, where transfer occurs (for a review see ref. Citation25). Hence, precise cellular localization is not only a major determinant for proper plasmid segregation, but also for conjugation.
R388 segregation relies exclusively on the StbA protein, which constrains plasmid molecules within nucleoid-containing areas of the cell.Citation21 This localization is not adequate for conjugative transfer, since R388 conjugation apparatus localizes at the cell poles.Citation26 This idea is reinforced by the observation that stbA mutations, which shift plasmid localization to the cell poles, strongly enhanced conjugation (). It was a big surprise when our results revealed that deletion of the product of the second stb gene, StbB, led to an absolute conjugation defect, which was correlated with the absence of plasmid foci at the cell poles (). Remarkably, inactivation of both stbA and stbB had no effect on conjugation, demonstrating that StbA and StbB activities are functionally connected, showing opposite effects to modulate conjugation. The main role of StbB might thus be to counteract the action of StbA, by recruiting a fraction of plasmid molecules from a nucleoid-associated pool to the poles, where conjugative transfer occurs.
StbB contains a deviant Walker A nucleotide triphosphate-binding motif related to that found in the Soj/MinD superfamily of ATPases, including type I par motor proteinsCitation27 (see above). The stb operon presents therefore striking similarities with type I par systems: it contains a DNA-binding protein, StbA, a centromere-like site, the stbDRs, and a putative ATPase protein (). However, no homology of StbA to par DNA-binding adaptor was detected, and the putative ATPase StbB is not required for R388 faithful segregation, but instead is essential for conjugation when StbA is present. We propose that the StbAB system constitutes a new type of segregation system intimately linked to conjugation, possibly derived from type I partition systems. In this view, StbB may be a motor protein forming nucleotide-driven cytomotive filaments, analogous to that of par systems but dedicated to drive plasmid molecules to nucleoid-free areas ().
Conjugation of plasmid R388 involves a DNA transfer machinery, encoded by the MOB operon, coupled to an MPFT type IV secretion system (T4SS).Citation28,Citation29 The DNA transfer machinery is composed of three proteins, the regulatory protein TrwA, the coupling protein TrwB, and the relaxase TrwC. Inactivation of either the coupling protein or the relaxase did not affect plasmid R388 stability nor cellular localization (our unpublished data). This indicated that the StbB-dependent mechanism for R388 recruitment to the cellular poles does not depend on relaxosome formation. Furthermore, our comparative genomics studies revealed that the Stb system, which is widespread among a wide variety of plasmids of different mobility groups (MOBF11, MOBP11, MOBP6 and MOBP13/P14), is always linked to MPFT T4SS systems. This suggests that this type of T4SS, rather than the DNA transfer machinery, may create the requirement for the Stb system.
In conclusion, the StbAB system not only adds more variation to the knowledge of plasmid segregation mechanisms, but also provides a new understanding of the interplay between vertical transmission (segregation) and horizontal transmission (conjugation). On one hand, plasmid R388 segregation protein StbA is essential for stable maintenance in progeny cells while, on the other hand, StbB is required for conjugation as long as the segregation system is present. Plasmid maintenance functions and conjugation appear to be connected at different levels. Several plasmids have been shown to encode a master regulator that co-regulate sets of genes required for replication and/or stable inheritance, and conjugation (RP4,Citation30 pWWO,Citation31 Ti plasmidsCitation32). To our knowledge, the StbAB system represents the first evidence for a mechanistic interplay between plasmid segregation and conjugation processes. Further studies on this system will thus allow an improved understanding of how distinct ways of plasmid propagation influence each other and facilitate stable establishment, propagation in dividing cells, and spread of plasmids in different hosts.
Figures and Tables
Figure 1 Main types of plasmid segregation systems. The figure shows the genetic organization of the main types of arrangement of plasmid segregation systems, as represented by prototype plasmids P1 (type I), R1 (type II), pBToxis (type III) and R388 (type IV?). Boxes represent genes that encode motor proteins (blue) and DNA-binding adaptor proteins (red). Cis-acting sites are shown as yellow boxes. Dashed curved arrows indicate binding of adaptor proteins to their target sites.
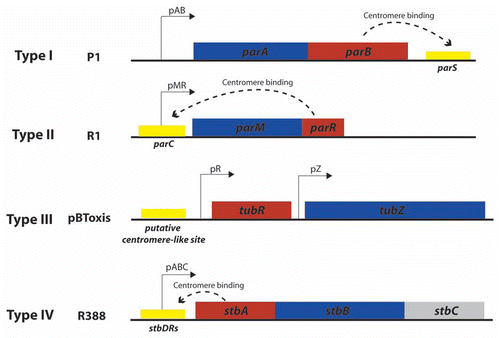
Figure 2 Proposed mechanisms of plasmid segregation. For the sake of clarity, only two newly replicated plasmid copies are represented. When necessary, light red and white regions indicate nucleoid and cytosol spaces, respectively. (A) Pushing mechanism, exemplified by R1 type II par system. Partitioning complexes are formed through specific binding of ParR proteins (red circles) to the centromere-like site parC of newly replicated plasmid molecules, and serve as a nucleation point for ParM-mediated filament formation. Continuous insertion of ParM-ATP motor proteins (blue circles) on the filament ends pushes plasmid molecules apart. Conversion of ParM-ATP to ParM-ADP (open blue circles) leads to destabilization of the filament, thus allowing the entry of another ParM-ATP. At cell division, plasmid molecules localize near opposite cell poles, thus ending in daughter cells.Citation15 (B) Pulling mechanism, as proposed for type I (pB171) and type III (pBtoxis) par systems.Citation16,Citation17 NTP-bound motor proteins (blue circles) bind cooperatively to nucleoid DNA, forming a nucleating core from which filaments form. Subsequently, a growing filament contacts a partition complex formed by the adaptor protein (red circles) bound to the plasmid centromere-like site. Stimulation of NTPase activity of the motor protein by the adaptor protein at the end of the filament leads to conversion to its NDP form (open blue circles) and its release, leaving a new filament end accessible for interactions with the partition complex. The plasmid is thus pulled in the opposite direction to the growth of the filament, and moves around its position, between two other plasmids or between a plasmid and the nucleoid end. (C) Diffusion-ratchet mechanism, as proposed for plasmid P1 type I par system.Citation18 ParB (red circles) loads onto the plasmid at the centromere-like site parS, forming the partition complex. After plasmid replication, partition complexes develop repulsive interactions. ParB stimulates ParA (dark blue circles) ATPase activity, and ParA-ADP molecules (open blue circles) are then excluded from the nucleoid. The motive force for plasmid movement is directed toward regions of high ParA concentration. Movement of the partition complex is thus constrained to one direction because of the low ParA concentration behind it, and at the nucleoid end, it changes direction. ParA-ADP molecules diffuse randomly, exchange ADP for ATP (light blue circles), and then rebind the nucleoid. (D) “Pilot-fish” mechanism, as proposed for plasmid R388 segregation, representing the prototype of a potentially new class of segregation system type IV.Citation21 In contrast to typical par systems, the StbB putative ATPase is not involved in R388 segregation, which does not need a motor protein. We propose that the partition complex, formed by StbA binding to the centromere-like site stbDRs, is used to pair plasmid molecules to the host nucleoid (or other structure associated to the nucleoid). Plasmid segregation is ensured by the host chromosome segregation system.
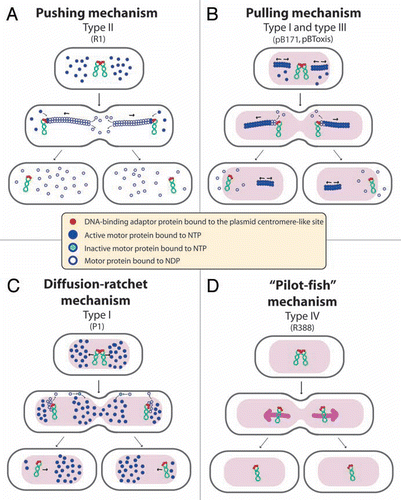
Figure 3 Schematic model to explain the role of the StbAB system.Citation21 Open and shaded regions indicate nucleoid and cytosol spaces, respectively. Blue arrows indicate segregation of the host chromosome, and red arrows represent conjugative transfer to recipient cells (light red nucleoid). Plasmid R388 molecules are evenly distributed both in nucleoid and cytosol areas. At cell division, each daughter cell contains plasmid copies. Localization of plasmid copies at the cell pole is correlated with R388 capacity to undergo conjugative transfer. In contrast, DNA molecules of plasmid R388ΔstbA are exclusively localized in cytosol spaces toward the cell poles and the cell center. This is correlated with plasmid instability, since cells containing all copies in one side of the cell give rise to plasmid-free cells (dark cytoplasm), and with high conjugation frequency, since there are more plasmid copies at the poles. DNA molecules of plasmid R388ΔstbB are distributed in nucleoid but not in cytosol spaces, which correlates with a defect in conjugative transfer.
Acknowledgments
We thank F. Cornet for helpful discussions. This work was supported by grants BFU2008-00995/BMC from Ministerio de Ciencia e Innovacion (MCINN, Spain), RD06/0008/1012 from Instituto de Salud Carlos III, and 248919/FP7-ICT-2009-4 from the European VII Framework Program. C.G. was the recipient of fellowships from Fondation pour la Recherche Medicale (SPE20080512320) and European Molecular Biology Organization (157-2008).
References
- Sengupta M, Austin S. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect Immun 2011; 79:2502 - 2509; PMID: 21555398; http://dx.doi.org/10.1128/IAI.00127-11
- Summers DK, Beton CW, Withers HL. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol Microbiol 1993; 8:1031 - 1038; PMID: 8361350; http://dx.doi.org/10.1111/j.1365-2958.1993.tb01648.x
- Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet 2005; 39:453 - 479; PMID: 16285868; http://dx.doi.org/10.1146/annurev.genet.38.072902.091252
- Yarmolinsky MB. Programmed cell death in bacterial populations. Science 1995; 267:836 - 837; PMID: 7846528; http://dx.doi.org/10.1126/science.7846528
- Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol 2010; 8:600 - 607; PMID: 20634810;; http://dx.doi.org/10.1038/nrmicro2391
- Nordström K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet 1989; 23:37 - 69; PMID: 2694936; http://dx.doi.org/10.1146/annurev.ge.23.120189.000345
- Löwe J, Amos LA. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol 2009; 41:323 - 329; PMID: 18768164; http://dx.doi.org/10.1016/j.biocel.2008.08.010
- Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell 2010; 141:927 - 942; PMID: 20550930; http://dx.doi.org/10.1016/j.cell.2010.05.033
- Salje J. Plasmid segregation: how to survive as an extra piece of DNA. Crit Rev Biochem Mol Biol 2010; 45:296 - 317; PMID: 20586577; http://dx.doi.org/10.3109/10409238.2010.494657
- Bouet JY, Nordström K, Lane D. Plasmid partition and incompatibility-the focus shifts. Mol Microbiol 2007; 65:1405 - 1414; PMID: 17714446; http://dx.doi.org/10.1111/j.1365-2958.2007.05882.x
- Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol 2000; 37:455 - 466; PMID: 10931339; http://dx.doi.org/10.1046/j.1365-2958.2000.01975.x
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 2007; 76:539 - 562; PMID: 17328675; http://dx.doi.org/10.1146/annurev.biochem.75.103004.142652
- Shih YL, Rothfield L. The bacterial cytoskeleton. Microbiol Mol Biol Rev 2006; 70:729 - 754; PMID: 16959967; http://dx.doi.org/10.1128/MMBR.00017-06
- van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J 2002; 21:6935 - 6943; PMID: 12486014; http://dx.doi.org/10.1093/emboj/cdf672
- Salje J, Gayathri P, Löwe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol 2010; 8:683 - 692; PMID: 20844556; http://dx.doi.org/10.1038/nrmicro2425
- Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev 2007; 21:1340 - 1352; PMID: 17510284; http://dx.doi.org/10.1101/gad.1546107
- Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA 2009; 106:46 19369 - 19374; PMID: 19906997; http://dx.doi.org/10.1073/pnas.0908347106
- Vecchiarelli AG, Han YW, Tan X, Mizuushi M, Ghirlando R, Biertümpfel C, et al. ATP control of dynamic ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol 2010; 78:78 - 91; PMID: 20659294
- Simpson AE, Skurray RA, Firth N. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J Bacteriol 2003; 185:2143 - 2152; PMID: 12644483; http://dx.doi.org/10.1128/JB.185.7.2143-2152.2003
- Bouet JY, Funnell BE. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J 1999; 18:1415 - 1424; PMID: 10064607; http://dx.doi.org/10.1093/emboj/18.5.1415
- Guynet C, Cuevas A, Moncalian G, de la Cruz F. The stb operon balances the requirements for vegetative stability and conjugative transfer of plasmid R388. PLoS Genet 2011; 7:e1002073; PMID: 21625564; http://dx.doi.org/10.1371/journal.pgen.1002073
- Fernández-López R, Garcillán-Barcia MP, Revilla C, Lazaro M, Vielva L, de la Cruz F. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev 2006; 30:942 - 966; PMID: 17026718; http://dx.doi.org/10.1111/j.1574-6976.2006.00042.x
- Stehle IM, Postberg J, Rupprecht S, Cremer T, Jackson DA, Lipps HJ. Establishment and mitotic stability of an extra-chromosomal mammalian replicon. BMC Cell Biol 2007; 8:33; PMID: 17683605; http://dx.doi.org/10.1186/1471-2121-8-33
- Suzuki H, Yano H, Brown CJ, Top EM. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 2010; 192:6045 - 6055; PMID: 20851899; http://dx.doi.org/10.1128/JB.00277-10
- de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 2010; 34:18 - 40; PMID: 19919603; http://dx.doi.org/10.1111/j.1574-6976.2009.00195.x
- Vecino AJ, de la Arada I, Segura RL, Goñi FM, de la Cruz F, Arrondo JL, Alkorta I. Membrane insertion stabilizes the structure of TrwB, the R388 conjugative plasmid coupling protein. Biochim Biophys Acta 2011; 1808:1032 - 1039
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol 1993; 229:1165 - 1174; PMID: 8445645; http://dx.doi.org/10.1006/jmbi.1993.1115
- Llosa M, Gomis-Rüth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 2002; 45:1 - 8; PMID: 12100543; http://dx.doi.org/10.1046/j.1365-2958.2002.03014.x
- Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev 2010; 74:434 - 452; PMID: 20805406; http://dx.doi.org/10.1128/MMBR.00020-10
- Lessl M, Balzer D, Lurz R, Waters VL, Guiney DG, Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J Bacteriol 1992; 174:2493 - 2500; PMID: 1556069
- Lambertsen LM, Molin S, Kroer N, Thomas CM. Transcriptional regulation of pWW0 transfer genes in Pseudomonas putida KT2440. Plasmid 2004; 52:169 - 181; PMID: 15518874; http://dx.doi.org/10.1016/j.plasmid.2004.06.005
- Su S, Khan SR, Farrand SK. Induction and loss of Ti plasmid conjugative competence in response to the acyl-homoserine lactone quorum-sensing signal. J Bacteriol 2008; 190:4398 - 4407; PMID: 18203831; http://dx.doi.org/10.1128/JB.01684-07