Abstract
We have recently shown that GAA repeats severely impede replication elongation during the first replication cycle of transfected DNA wherein the chromatin is still at the formation stage.Citation1 Here we extend this study by showing that two GAA repeats located within the same plasmid in the direct orientation can form complexes upon transient transfection of mammalian Cos-1 cells. However, these complexes do not form in DNA that went through several replication rounds in mammalian cells. We suggest that formation of such complexes in mammalian genomes can contribute to genomic instability.
GAA repeats were shown to be the most unstable trinucleotide repeats in the primates genome evolution by comparison of orthologous human and chimp loci.Citation2 The instability of the GAA repeat in the first intron of the frataxin gene X25 is particularly well studied since it causes an inherited disorder, Friedreich ataxia (FRDA).Citation3-Citation6 In Friedreich ataxia, once the length of the GAA repeat inside the frataxin gene (FXN GAA) reaches a certain threshold, the combined probability of its expansions and deletions in progeny of affected parents is about 85%.Citation7 Deletions and contractions of the repeat in intergenerational transmissions can reach hundreds of base pairs.Citation7 However, the FXN GAA repeat is much more stable in somatic cells.Citation8 It is relatively stable in blood, but shows some instability in dorsal root ganglia,Citation9 which is responsible for some of the neurodegenerative symptoms of Friedreich ataxia.Citation5 GAA repeats were shown to be stable in FRDA fibroblasts cell lines and neuronal stem cells.Citation10
The question why the FXN GAA repeat is so much more stable in somatic cells than in intergenerational transmissions remains open. Recent studies in FRDA iPSCs that are closer to embryonic cells than somatic cells models, showed expansions of the GAA repeat with 100% probability.Citation10,Citation11 It is intriguing that all cells in the iPSC cell lines that were analyzed were synchronously adding about two GAA repeats in each replication.
The studies focused on the FXN GAA repeat provided many valuable insights; however, human genome contains many other GAA repeats: the human X chromosome, for instance, contains 44 GAA stretches with more than 100 repeats in each. About 30 GAA repeats were detected on the chromosome 4.Citation12 GAA repeats mostly originated from the 3′ end of the poly A associated with Alu elements.Citation13
It is not known what makes repeats with the GAA motif most unstable compared with other trinucleotide repeats. It is possible that GAA repeats instability is caused by their ability to form non-B DNA structures. In vitro, GAA repeats can form triplexes,Citation14,Citation15 and sticky DNA structures.Citation16 At the same time, hairpinsCitation17 and parallel duplexesCitation18 have also been observed. When transcription is going through a GAA repeat, it can also form an R-loop, a DNA-RNA complex that leaves one of the complementary strands single-stranded.Citation19 However, it is unclear whether these structures indeed form in mammalian cells. If we assume that the instability of the GAA repeat is indeed associated with the structure formation, it is still unclear why the structures would form in early embryogenesis when the GAA expansion event in Friedriech’s ataxia is believed to occur,Citation7 and do not form in somatic cells where the GAA repeat was shown to be more stable. In our recent study, we hypothesize that the differences in chromatin structure are at least partially responsible for the differences in the GAA repeats stability.Citation1
The propensity of GAA repeat to form a triplex structure may strongly depend on the structure of chromatin at the repeat and surrounding area.Citation1 Consistent with other studies, we observed that formation of chromatin at an SV40-based plasmid introduced into mammalian cells occurs gradually: 8 h after transfection there are only occasional nucleosomes at the plasmid, while by 72 h the nucleosome structure is already regular.Citation20 Our analysis of replication stalling at the repeat revealed that the repeat affects replication only in the first replication cycle, when chromatin is still at the formation stage. We believe that replication stalling at GAA is caused by a triplex structure that the GAA repeat adopts during transfection or inside the cell. In the subsequent replication cycles, replication was completely unaffected by the presence of the repeat, which is likely to be due to the inhibition of triplex formation by tight chromatin packaging.Citation1
Here we show the data that strengthen our previous observations and extend it to one more structure: a complex between two GAA stretches. Two GAA repeats has been shown to readily form complexes, such as “sticky DNA,” in vitro,Citation16 but it is not obvious whether it can also form inside the mammalian cells. The sticky DNA requires more than one GAA stretch to form.Citation21 We studied the interaction of two GAA repeats located within the same plasmid, but since the human genome contains at least several hundreds of long GAA motif repeats,Citation12,Citation13 this structure can theoretically form in the genomic environment as well. However, more experiments are needed to detect its formation in genome.
The method of two-dimensional electrophoresisCitation22,Citation23 allowed us to analyze the replication progression through a DNA fragment containing two GAA repeats. In this method, the replication intermediates are isolated under non-denaturing conditions, digested by a restriction enzyme, and separated on two consecutive gel runs in perpendicular directions. The first direction runs in 0.4% agarose (that separates mostly by mass), and the second direction runs in 1% agarose (that separates by both mass and shape of a DNA molecule).
We studied replication of several SV40-based plasmids that contained two (GAA)57 repeats located at different positions in Cos-1 cells (–). In each case, the cells were transiently transfected with 1 μg of each plasmid, and replication intermediates were isolated after about 30 h. This allowed us to observe replication arcs that resulted from the plasmids that replicated for more than two rounds. However, some residual amount of the first replication cycle arcs can also be registered. Replication stalling at GAA repeats only occurs during the first replication cycle of an SV40-based plasmid,Citation1 hence we did not observe it in our system.
Figure 1. A complex between two (GAA)57 repeats within the same plasmid. Plasmid replication intermediates were isolated from Cos-1 cells 30 h after transient transfection. Intermediates were digested by restriction enzymes indicated in the plasmid maps, and separated by two-dimensional neutral-neutral agarose gel electrophoresis as described previously.Citation1 The gel was transferred to a nylon membrane and hybridized to one of the probes indicated in the plasmid maps as green lines. A position of the complex at the 2D gel pattern depends on the restriction digest of the replication intermediates (indicated by a red arrow). (A) Two-dimensional electrophoresis of replication intermediates digested by PvuII that places each repeat within a separate fragment. The membrane was hybridized with probe 1 indicated in (C). The names of the plasmids GAAGAA, GAACTT, etc., reflect the orientation of the GAA repeats within the plasmid (GAAGAA means that the two GAA stretches are in the direct orientation). (B) The same membrane was stripped of probe 1, and re-hybridized with probe 2 (C). (C) The scheme of the plasmid that was used in the experiment in (A and B). Two different fragments that resulted from the PvuII digest are shown in red and blue. The positions of GAA repeats are shown in black. They can be in the direct or the reverse orientations in this plasmid. (D) The scheme of the 2D gel in (A). Spot 1: unreplicated blue fragment, spot 2: unreplicated red fragment that appears because of the cross-contamination of probe 1 with probe 2 due to their preparation from the same plasmid with a restriction digest. Spot 3: a complex between the red and the blue fragments. Spikes 3–2 and 3–4 may result from double-stranded breaks in the plasmid during transfection that make one of the arms of the complex in spot 3 shorter.
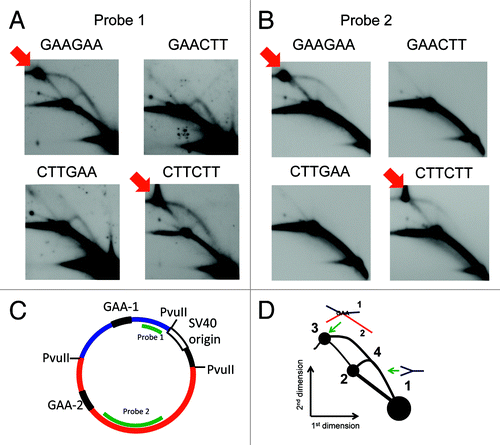
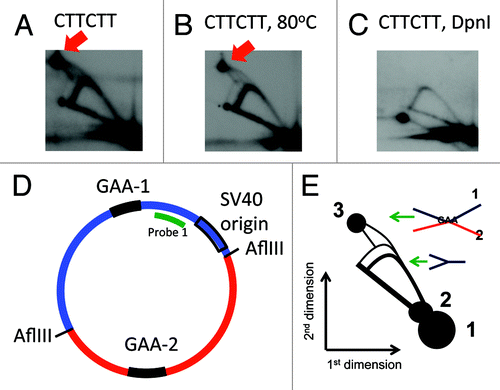
Figure 3. A complex between the two GAA repeats does not form in plasmids that went through more than two replication rounds in mammalian cells. Two-dimensional gels of replication intermediates of a plasmid containing two (GAA)57 repeats were obtained as described in the legend. (A) Two-dimensional gel of replication intermediates digested by AflIII (placing the two repeats at two different fragments). A red arrow indicates the position of the complex between the two GAA-containing fragments. (B) The same replication intermediates were incubated at 80°C in the presence of 10 mM EDTA for 10 min; the pattern of the 2D gel did not change. (C) The same intermediates were additionally digested with 10 units of DpnI for 2 h prior to loading. The spot at the position indicated by an arrow in A is not present in this picture. An additional spot that appeared in this pattern is likely not a part of the pattern, and is probably due to some contamination. (D) A map of the plasmid that was used in 2D gel in (A-C). (E) A scheme of the 2D gel shown in . Spot 1: unreplicated blue fragment, spot 2: unreplicated red fragment (which appeared due to contamination of probe 1 with other plasmid sequences). Spot 3: a complex between the blue and the red fragments. A very faint duplicate Y arc from replication of the second fragment originates from spot 2. The spikes originating from spot 3 can be interpreted the same way as in .
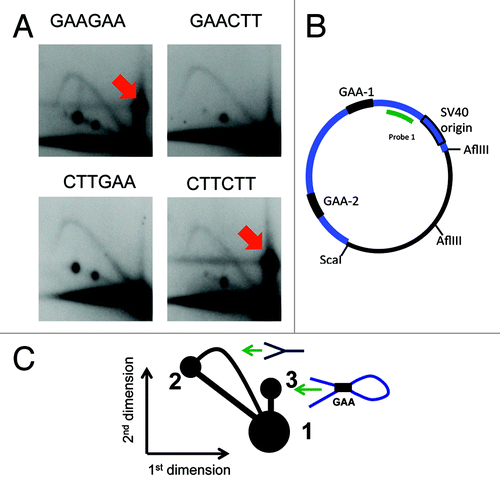
For each of the plasmids, and for all patterns of restriction digests that we studied, we observed complexes between the two (GAA)57 repeats (indicated by red arrows in each of the figures). The migration of those complexes was different depending on the digest pattern, so the complexes were at different positions in 2D gel patterns, in agreement with our expectations based on their shapes.
We did not observe replication stalling associated with these complexes. When this complex is formed, it migrates significantly slower on 2D gels; the replication of plasmids that contain such complexes should result in an extra replication arc originating from the complex position. However, the number of molecules that form this complex is significantly lower than the overall number of plasmids, and there may be not enough material to observe their replication.
For the situation when the two GAA repeats were located in two different fragments upon PvuII digest (), we showed that the spot 3 () (also indicated by an arrow in ) contains both fragments: the spot hybridized to the probes corresponding to either of them (). However, the complex appears at the position that migrates slower than unreplicated plasmid in the second dimension upon AflIII and ScaI digest when both repeats belong to the same fragment (). We suggest that this fragment contains a loop generated by the interaction between the two GAA repeats () that slows it down in the second dimension, and has little effect on the mobility in the first dimension since it has the same mass as the unreplicated fragment.
Figure 2. A complex between the two repeats within the same fragment slow down its progression in the second dimension of the 2D gel. The same plasmid as in the was used in this experiment, however, they were digested with different enzymes. (A) Replication intermediates were digested with AflIII and ScaI, placing both repeats within the same fragment. The complex of two GAA repeats results in a slowly migrating structure that is shown by a red arrow. (B) A map of the digest of the same plasmid as in with restriction enzymes ScaI and AflIII. Here both of the repeats are located within the same fragment shown in blue. (C) The scheme of the 2D gel in . Spots 1 and 2 are the same as in . Spot 3: a looped intermediate that resulted from the interaction of the two GAA repeats.
The complexes formed only when the two GAA stretches were positioned on a plasmid as direct repeats (GAAGAA and CTTCTT in the plasmid names). The inverted repeats GAACTT and CTTGAA did not form complexes as shown in and . This is in agreement with the sticky DNA formation in supercoiled plasmids containing two GAA repeats that has been previously shown in vitro.Citation16 In sticky DNA, the two GAA strands that are in the antiparallel orientation, and the CTT strand, form a stable complex stabilized by Mg 2+-dependent reverse-Hoogsteen triads. However, the sticky DNA complex fell apart upon heating in the presence of EDTA, which removes the Mg 2+ ions necessary for its stability,Citation16 while we did not detect any changes in spot 3 upon heating the intermediates with EDTA (). We suggest that in our case the complex may be different from the canonical sticky DNA. It may be based on Hoogsteen base pairing where the Mg 2+ is not needed and a slightly acidic pH has a stabilizing effect.Citation24 It has been shown that this type of structure forms within long GAA stretches in vitro even at a pH that is close to neutral.Citation15 We also cannot exclude that the complexes are hemicatenated molecules connected at GAA repeats with Watson-Crick pairing.Citation25
The complexes between the two GAA repeats persisted only until the plasmids went through one replication round. DpnI restriction enzyme is a frequent-cutter that digests all DNA that contains strands synthesized in bacteria: it cleaves DNA that is methylated at GATC by dam methylase, which is only present in bacteria, but not in mammalian cells. Extensive DpnI digest that we performed, cleaved the initial DNA used in transfection, as well as the products of the first replication cycle because they contain one strand synthesized in bacteria.
The replication intermediates digested with DpnI did not contain the spot 3, corresponding to the complex between two GAA stretches (). We suggest that the absence of the complex is due to the chromatin coverage of the plasmid that accompanies replication. This is similar to our observation that GAA repeats only block replication during the first replication round, until the chromatin is formed. The replication blockage that we have previously observed is consistent with formation of a triplex that occurs in transfected DNA only prior to nucleosome coverage.Citation1 Here, the complex between the two (GAA)57 repeats also occurred only without the chromatin structure. The absence of the complex in replicated DNA also shows that the complexes that we observe are not an artifact of the isolation and subsequent treatment of our intermediates, since then they would exist in at least some fraction of the replicated DNA as well.
The question remains whether the non-B DNA structures can form within GAA repeats in mammalian cells since their formation requires DNA stretches that are not folded in chromatin. A window when these complexes can form during development is the spermatogenesis when the maturing sperm chromatin changes from nucleosome- to protamine-bound assembly.Citation26 Another opportunity to form complexes comes when the chromatin of a sperm and an egg restructure after the fusion of the gamets.Citation26,Citation27 This is associated with degradation of protamines and nucleosome deposition, as the zygote DNA may lack a compact chromatin structure.Citation28 It should be noted that the expansions in Friedreich ataxia were traced to the early divisions of the zygote.Citation7
An opportunity for the complexes to form may also exist in cancer cells. It is known that some regions of their genome are overmethylated and convert in heterochromatin, while other regions are undermethylated, which may promote a loose chromatin packaging.Citation29,Citation30
It is not clear whether the two-repeats complexes would compete with triplex structure formation within each individual (GAA)57 repeat. It is possible that both of them exist and contribute to overall genomic instability. However, a separate study is necessary to determine whether these structures indeed have a biological role.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by NIH grant GM087472, and research grant from Friedreich’s Ataxia Research Alliance to MMK.
References
- Chandok GS, Patel MP, Mirkin SM, Krasilnikova MM. Effects of Friedreich’s ataxia GAA repeats on DNA replication in mammalian cells. Nucleic Acids Res 2012; 40:3964 - 74; http://dx.doi.org/10.1093/nar/gks021; PMID: 22262734
- Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res 2008; 18:30 - 8; http://dx.doi.org/10.1101/gr.7113408; PMID: 18032720
- Sharma R, Bhatti S, Gomez M, Clark RM, Murray C, Ashizawa T, et al. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum Mol Genet 2002; 11:2175 - 87; http://dx.doi.org/10.1093/hmg/11.18.2175; PMID: 12189170
- Pandolfo M. The molecular basis of Friedreich ataxia. Adv Exp Med Biol 2002; 516:99 - 118; http://dx.doi.org/10.1007/978-1-4615-0117-6_5; PMID: 12611437
- Pandolfo M. Friedreich ataxia. Arch Neurol 2008; 65:1296 - 303; http://dx.doi.org/10.1001/archneur.65.10.1296; PMID: 18852343
- Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996; 271:1423 - 7; http://dx.doi.org/10.1126/science.271.5254.1423; PMID: 8596916
- De Michele G, Cavalcanti F, Criscuolo C, Pianese L, Monticelli A, Filla A, et al. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich’s ataxia. Hum Mol Genet 1998; 7:1901 - 6; http://dx.doi.org/10.1093/hmg/7.12.1901; PMID: 9811933
- De Biase I, Rasmussen A, Monticelli A, Al-Mahdawi S, Pook M, Cocozza S, et al. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics 2007; 90:1 - 5; http://dx.doi.org/10.1016/j.ygeno.2007.04.001; PMID: 17498922
- De Biase I, Rasmussen A, Endres D, Al-Mahdawi S, Monticelli A, Cocozza S, et al. Progressive GAA expansions in dorsal root ganglia of Friedreich’s ataxia patients. Ann Neurol 2007; 61:55 - 60; http://dx.doi.org/10.1002/ana.21052; PMID: 17262846
- Du J, Campau E, Soragni E, Ku S, Puckett JW, Dervan PB, et al. Role of mismatch repair enzymes in GAA·TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J Biol Chem 2012; 287:29861 - 72; http://dx.doi.org/10.1074/jbc.M112.391961; PMID: 22798143
- Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, et al. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA⋅TTC triplet repeat instability. Cell Stem Cell 2010; 7:631 - 7; http://dx.doi.org/10.1016/j.stem.2010.09.014; PMID: 21040903
- Siedlaczck I, Epplen C, Riess O, Epplen JT. Simple repetitive (GAA)n loci in the human genome. Electrophoresis 1993; 14:973 - 7; http://dx.doi.org/10.1002/elps.11501401155; PMID: 7907288
- Chauhan C, Dash D, Grover D, Rajamani J, Mukerji M. Origin and instability of GAA repeats: insights from Alu elements. J Biomol Struct Dyn 2002; 20:253 - 63; http://dx.doi.org/10.1080/07391102.2002.10506841; PMID: 12354077
- Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, et al. GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol Cell 1998; 1:583 - 93; http://dx.doi.org/10.1016/S1097-2765(00)80058-1; PMID: 9660942
- Potaman VN, Oussatcheva EA, Lyubchenko YL, Shlyakhtenko LS, Bidichandani SI, Ashizawa T, et al. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res 2004; 32:1224 - 31; http://dx.doi.org/10.1093/nar/gkh274; PMID: 14978261
- Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, et al. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol Cell 1999; 3:465 - 75; http://dx.doi.org/10.1016/S1097-2765(00)80474-8; PMID: 10230399
- Heidenfelder BL, Makhov AM, Topal MD. Hairpin formation in Friedreich’s ataxia triplet repeat expansion. J Biol Chem 2003; 278:2425 - 31; http://dx.doi.org/10.1074/jbc.M210643200; PMID: 12441336
- LeProust EM, Pearson CE, Sinden RR, Gao X. Unexpected formation of parallel duplex in GAA and TTC trinucleotide repeats of Friedreich’s ataxia. J Mol Biol 2000; 302:1063 - 80; http://dx.doi.org/10.1006/jmbi.2000.4073; PMID: 11183775
- McIvor EI, Polak U, Napierala M. New insights into repeat instability: role of RNA•DNA hybrids. RNA Biol 2010; 7:551 - 8; http://dx.doi.org/10.4161/rna.7.5.12745; PMID: 20729633
- Chandok GS, Kapoor KK, Brick RM, Sidorova JM, Krasilnikova MM. A distinct first replication cycle of DNA introduced in mammalian cells. Nucleic Acids Res 2011; 39:2103 - 15; http://dx.doi.org/10.1093/nar/gkq903; PMID: 21062817
- Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem 2001; 276:27171 - 7; http://dx.doi.org/10.1074/jbc.M101879200; PMID: 11340071
- Krasilnikova MM, Mirkin SM. Analysis of triplet repeat replication by two-dimensional gel electrophoresis. Methods Mol Biol 2004; 277:19 - 28; PMID: 15201446
- Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol 1995; 262:613 - 27; http://dx.doi.org/10.1016/0076-6879(95)62048-6; PMID: 8594382
- Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu Rev Biochem 1995; 64:65 - 95; http://dx.doi.org/10.1146/annurev.bi.64.070195.000433; PMID: 7574496
- Lucas I, Hyrien O. Hemicatenanes form upon inhibition of DNA replication. Nucleic Acids Res 2000; 28:2187 - 93; http://dx.doi.org/10.1093/nar/28.10.2187; PMID: 10773090
- McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction 2003; 125:625 - 33; http://dx.doi.org/10.1530/rep.0.1250625; PMID: 12713425
- Spinaci M, Seren E, Mattioli M. Maternal chromatin remodeling during maturation and after fertilization in mouse oocytes. Mol Reprod Dev 2004; 69:215 - 21; http://dx.doi.org/10.1002/mrd.20117; PMID: 15293223
- Imschenetzky M, Puchi M, Gutierrez S, Montecino M. Sea urchin zygote chromatin exhibit an unfolded nucleosomal array during the first S phase. J Cell Biochem 1995; 59:161 - 7; http://dx.doi.org/10.1002/jcb.240590205; PMID: 8904310
- Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem 2010; 52:145 - 67; http://dx.doi.org/10.1016/S0065-2423(10)52006-7; PMID: 21275343
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010; 70:27 - 56; http://dx.doi.org/10.1016/B978-0-12-380866-0.60002-2; PMID: 20920744