Abstract
“Young” APE-type non-LTR retrotransposons (non-LTRs) typically encode two open reading frames (ORFs 1 and 2). The shorter ORF1 translation product (ORF1p) comprises an RNA binding activity, thought to bind to non-LTR transcript RNA, protect against nuclease degradation and specify nuclear import of the ribonuclear protein complex (RNP). ORF2 encodes a multifunctional protein (ORF2p) comprising apurinic/apyrimidinic endonuclease (APE) and reverse-transcriptase (RT) activities, responsible for genome replication and re-integration into chromosomal DNA. However, some clades of APE-type non-LTRs only encode a single ORF—corresponding to the multifunctional ORF2p outlined above (and for simplicity referred-to as ORF2 below). The absence of an ORF1 correlates with the acquisition of a 2A oligopeptide translational recoding element (some 18–30 amino acids) into the N-terminal region of ORF2p. In the case of non-LTRs encoding two ORFs, the presence of ORF1 would necessarily downregulate the translation of ORF2. We argue that in the absence of an ORF1, 2A could provide the corresponding translational downregulation of ORF2. While multiple molecules of ORF1p are required to decorate the non-LTR transcript RNA in the cytoplasm, conceivably only a single molecule of ORF2p is required for target-primed reverse transcription/integration in the nucleus. Why would the translation of ORF2 need to be controlled by such mechanisms? An “excess” of ORF2p could result in disadvantageous levels of genome instability by, for example, enhancing short, interspersed, element (SINE) retrotransposition and the generation of processed pseudogenes. If so, the acquisition of mechanisms—such as 2A—to control ORF2p biogenesis would be advantageous.
The discovery and characterization of 2A oligopeptide translational recoding sequences within the genomes of non-LTR retrotransposons (non-LTRs) reveals another interesting parallel between the molecular biology of non-LTRs and viruses. Initially, we thought the occurrence of these short 2A/2A-like sequences (2As) was confined to RNA virus genomes, but 2A sequences were discovered in L1Tc non-LTRs within the genome of trypanosome species.Citation1 From this single occurrence, it was not possible to gauge either the significance of this observation for the molecular biology, or, the evolution of non-LTRs. Recently, however, 2As have been characterized from a range of different types of non-LTR and, perhaps of equal significance, from the genomes of a wide range of species.Citation2 These new data add support to the notion that acquisition of 2As is of functional significance and, indeed, may represent a significant step in the generation of a sub-group of APE-type non-LTR retrotransposons with a different method of controlling protein biogenesis.
What Are 2A Translational Recoding Sequences?
A group of oligopeptide sequences collectively known as 2A mediate a translational recoding event known as “ribosome skipping,” “StopGo” or “Stop Carry-on” translation.Citation3-Citation5 Briefly, the model of this non-canonical form of translation proposes that when a ribosome translates an mRNA sequence encoding 2A, the nascent 2A oligopeptide interacts with the exit tunnel of the ribosome (through which the elongating polypeptide product leaves the structure) and “stalls” the progress of the ribosome. Although a stop codon has not been encountered, the nascent peptide is released (forming the C-terminus of 2A), but then translation may resume—synthesizing the polypeptide sequence downstream of 2A: the synthesis of the peptide bond at this specific point in the protein backbone is “skipped.” For the vast majority of readers who are not familiar with translational (as opposed to transcriptional) control of protein biogenesis, there are a number of essential points which should be noted—with regards the mechanism of 2As and their potential function in the molecular biology of non-LTRs. The first is that although encoded as a single ORF, proteins comprising one, or more, 2As are not produced as a full-length translation product, but synthesized as (shorter) multiple, discrete, products ()—quite distinct from the ORF being translated as a single polypeptide which is subsequently “processed” into smaller products. The second point is that there is an alternative outcome: rather than resumption (pseudo-reinitiation) of the translation of mRNA sequences downstream of 2A, translation may terminate at the C-terminus of 2A (). We have proposed that translational (cellular) stress promotes termination rather than pseudo-reinitiation and that 2A may act to regulate the synthesis of downstream sequences, determining the ratio of translation products up- and down-stream of 2A. The third point is that for some 2As, the activity outlined above is not complete. Depending upon the nature of the 2A sequence in question, the interaction of 2A with the ribosome exit tunnel is weaker: a proportion of the ribosomes do not pause at the C-terminus of 2A and the peptide bond is formed in the normal manner (). Lastly, the study of 2A and its extensive use in biotechnological and biomedical applications shows that these sequences work in all eukaryotic cell-types tested to date, but not in bacteria.Citation6-Citation9,Citation11,Citation12
Figure 1. Sequences encoding protein domains (or complete genes) can be concatenated via 2As to create a single ORF.Citation6-Citation10 The mRNA encodes a single ORF, translation potentially giving rise to three, alternative, products—depending upon the recoding activity of the 2A sequence in question. Outcome (i) corresponds to the production of two, individual, translation products—protein 1 with a C-terminal extension of 2A plus protein 2. Outcome (ii) arises from termination of translation at the C-terminus of 2A. Outcome (iii) produces a full-length translation product (A). The genomic organization of the picornavirus foot-and-mouth disease virus is shown (~8,500 nts). The 5′ terminus of the vRNA is covalently bound to an oligopeptide (VPg), rather than a 7meG mRNA cap structure. The long 5′NCR comprises an internal ribosome entry sequence (IRES) which initiates translation of the long ORF in a cap-independent manner. The capsid proteins polyprotein domain is separated from the replication protein domains by the recoding activity of 2A. The structure of the L1Tc transcript RNA is shown with the position of 2A in the N-terminal region of the ORF. For comparison, the equivalent RNA organisations are shown for the “ancient” type of non-LTR (a single ORF comprising RT and REL domains), together with the canonical “young” non-LTRS encoding 2 ORFs, in which the REL domain in ORF2 is supplanted by the APE domain (B).
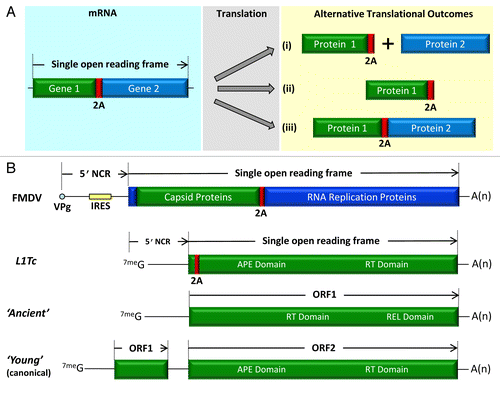
“2A” derives from the systematic nomenclature of protein domains within the polyprotein encoded by the single ORF of a group of positive-stranded (+ve strand; mRNA sense) RNA viruses—the family Picornaviridae (). Much of the early characterization of 2A arose from the study of the picornavirus foot-and-mouth disease virus (FMDV; genus Aphthovirus). As sequence databases expanded it became apparent that 2A-like sequences were present in the genomes of many other genera of the Picornaviridae, and, in other families of +ve and double-stranded RNA viruses.Citation10 Interestingly, 2As were also detected within L1Tc non-LTR retrotransposons (non-LTRs) in the genomes of Trypanosoma spp.Citation1 Naturally, similarity with such a short sequence could occur by chance, but analyses of 40 L1Tc sequences (representatives of over 100 such elements detected in the genome) showed all contained a 2A-like sequence in the same N-terminal region of the long ORF encoding the apurinic/apyrimidinic DNA endonuclease (APE) and reverse-transcriptase (RT) domains (). Furthermore, mutations were detected within a motif conserved at the C-terminus of 2A: -DxExNPG↓P- (vertical arrow indicating the site of the recoding event). This motif is very important for recoding activity, and the mutations observed among the different L1Tc non-LTRs correlated with changes in translational recoding activity.Citation1
A single ancestral form of L1Tc could have encoded this 2A prior to proliferation of these elements in the genome of Trypansosma cruzi, with the subsequent accumulation of mutations. As more genome sequences became available, however, it became clear that 2As were encoded by non-LTRs within (i) a number of different clades of non-LTRs and (ii) genomes of a wide range of different species. The notion that these 2As have a functional significance in the biology of these elements is supported by their occurrence in the same position within ORF2p. The genomes of the “ancient” non-LTRs encode a single ORF comprising a multi-functional protein with reverse transcriptase (RT) and restriction enzyme-like endonuclease (REL-endo) domains: here, re-integration is sequence-specific. The REL-endo domain is lost in “young” non-LTRs and replaced with an APE domain, integration now being sequence independent (; reviewed in refs Citation13-Citation15). These young non-LTRs have also acquired another ORF (ORF1) upstream of the long ORF (ORF2, comprising the APE and RT domains: ), with ORF2 of some elements also encoding RNaseH domains. Our bioinformatic analyses indicated a correlation between young non-LTRs encoding 2A and the apparent loss of ORF1. It should be noted, however, that the low processivity of RT is thought to be responsible for the frequent 5′ truncation of non-LTR genomes during retrotransposition—which may be a factor in our bioinformatic analyses.Citation16
The Functions and Biogenesis of ORF1p
ORF1 encodes a protein (ORF1p) with low sequence similarity among different non-LTRs. Functional studies have shown that ORF1p is (i) a high-affinity RNA-binding protein which forms a ribonucleoprotein (RNP) particle together with non-LTR transcript RNA, (ii) that ORF1p contains signals required for the nuclear import of this RNP complex, (iii) there is stringent cis-requirement for ORF1p during L1 retrotransposition, (iv) ORF1p was shown to possess nucleic acid chaperone activity in the case of a long, interspersed, element (LINE)-like transposable element in Drosophila (the I factor) and (v) deaminase-independent restriction of L1 by APOBEC3C requires an RNA-dependent interaction between human L1 ORF1p and APOBEC3C dimers.Citation17-Citation20
Our analyses showed the majority of non-LTRs encoding a 2A-like sequence did not possess an ORF1 (comprising the functions outlined above), but that non-LTRs sequences encoding 2A, and lacking ORF1, clustered alongside elements from other species which did encode an ORF1—but not a 2A.Citation2 Indeed, all elements within the Ingi clade (Ingi, Tcoingi, Tvingi, and L1Tc) do not appear to encode an ORF1 and the related Vingi elements only encode a single ORF.Citation1,Citation2,Citation15,Citation21 It should also be noted, however, that “ancient” non-LTRs do not possess an ORF1 and the retrotransposition of SINEs is mediated by LINEs in trans. It has been shown that although L1 ORF1p is not required for Alu SINE retrotransposition, this process is enhanced by supplementation with L1 ORF1p.Citation22 By analogy, it seems plausible that ORF1p functions could be supplied in trans from those non-LTRs in the genome encoding and expressing ORF1p, to form a functional RNP from those non-LTR transcript RNAs only encoding ORF2: ORF2p being supplied in cis. The few non-LTRs we identified which encode both ORF1 and a 2A-like sequence within ORF2 (e.g., CR1-26_BF, CR1-53_BF, and CR1-1_LG) may represent an intermediate evolutionary stage in the loss of ORF1.Citation2
The non-LTR RNA transcript comprises an atypical RNA polymerase II (or pol III) promoter in the 5′ non-coding region (NCR) such that transposition of a full-length non-LTR element does not require chance integration adjacent to a Pol II promoter (reviewed in refs. Citation23 and Citation24). Eukaryotic translation requires the assembly of the cap-binding protein complex at the 5′ cap structure followed by “scanning” of the 5′NCR until an initiating AUG start codon in a suitable Kozak consensus sequence is encountered. There must be, therefore, a high selective pressure upon the 5′NCR of non-LTRs against the occurrence of AUGs—at least in the context of a favorable Kozak consensus sequence: ORF1 can be translated in the “normal” (cap-dependent) manner. Once the ORF1 stop codon is encountered, translation terminates. The question arises, therefore, how is the translation of ORF2 initiated?
The Initiation of Translation of Non-LTR ORFs
Positive-stranded RNA viruses such as picornaviruses and non-LTR transcript RNAs face a common problem: how to generate multiple functions from a single mRNA. In the case of picornaviruses, a single ORF encodes a (multifunctional) polyprotein, although the full-length translation product is not observed within infected cells due to the action of virus-encoded proteinases and the 2A-mediated ribosome skipping mechanism (reviewed in ref. Citation25) Here, polyproteins are “processed” into functional domains in a series of co- and post-translational cleavage events. In the case of APE-type non-LTRs, the strategy is to encode two ORFs, both translation products are multifunctional, but neither are processed.
In picornaviruses the long 5′NCR contains multiple AUGs, but translation of the single long ORF is initiated not in the canonical mRNA 7meG-cap-dependent manner, but by a non-canonical mechanism. The picornavirus 5′NCR comprises an RNA secondary structural feature—an Internal Ribosome Entry Sequence (IRES)—which mediates initiation in a cap-independent mechanism: ribosomes do not scan through the 5′NCR, but are “delivered” directly to the correct AUG. Along with ribosome “shunting,” leaky scanning, initiation at non-AUGs, and re-initiation, viruses employ a range of different mechanisms to initiate translation (reviewed in ref. Citation26). In essence, 2A mediates a “pseudo-reinitiation,” although in this special case only for ribosomes already engaged in the elongation cycle of translation.
How translation initiates for ORF2 remains an interesting—and important—question. In the case of the SART1 element an overlapping stop-start codon (-UAAUG-) links ORFs 1 and 2. Interestingly, an RNA secondary structure downstream of this site has been shown to affect the efficiency of the initiation of translation of ORF2.Citation27 In this specific case there is no intergenic region separating ORFs 1 and 2. For the vast majority of non-LTRs, however, an intergenic sequence is present, and in some cases appears to be hundreds of bases long. Following termination of translation of upstream ORFs (uORFs), it is known that certain post-termination events can lead to re-initiation of translation of a downstream ORF. The efficiency of this re-initiation is dependent upon the length of the uORF (time taken to translate) and the length of the intergenic region (reviewed in ref. Citation28). By these criteria, is seems highly improbable that this mechanism can be at play in ORF2p biogenesis. In most cases non-LTR intergenic regions are long enough to comprise an IRES. Although first characterized in picornaviruses, IRESes are also found in the 5′NCR of certain cellular genes (e.g., c-Myc, Apaf-1, Bcl-2, XIAP, DAP5). It is thought that their expression (translation) is controlled by these elements—particularly under conditions of cellular stress, or, in response to specific stimuli (reviewed in refs. Citation29–Citation31). In the insect dicistroviruses the intergenic region internal ribosome entry site (IGR IRES) can directly assemble 80S ribosomes and initiate translation at a non-AUG codon from the ribosomal A-site. These activities arise from two independently folded domains of the IGR IRES. The first domain, composed of two overlapping RNA pseudoknots (PKII/III), mediates recruitment of the ribosome while the second domain, composed of a single RNA pseudoknot (PKI), mimics a tRNA anticodon–codon interaction thereby positioning the non-AUG codon at the ribosomal A-site.Citation32 If such IGR IRESes were present in the genomes of non-LTRs, then the notion of what comprises ORF2—and bioinformatic analyses—would need to be revisited.
Whichever of the mechanisms mentioned above mediates the translation of ORF2, the translation of the second ORF will be much less efficient than the first. Presumably, it takes a substantially greater quantity of ORF1p to form an RNP complex than of the associated ORF2p required to perform target-primed reverse transcription/integration. This is consistent with the genomic organization of non-LTRs and the report that even though relatively high levels of L1Tc mRNAs are detected within trypanosome cells, only low levels of ORF2p protein could be detected.Citation1 In this case, however, L1Tc does not encode an ORF1 and it may be that if translational “downregulation” of ORF2 is required, that this is now being achieved not by the presence of a uORF (ORF1), but by the 2A translational recoding element in the N-terminal region of the ORF.
Concluding Remarks
The study of the molecular biology of viruses is greatly simplified by the ability to infect virtually all susceptible tissue-culture cells to produce a strong, synchronous, “signal” from the object of study. Sequencing of virus genomes is straightforward and, generally, the sequence represents a biologically active entity. Due to the biology of non-LTRs, the equivalent experimental analyses are very much more complex, or, simply impossible. In 2002 a biologically active genome of the picornavirus, poliovirus, was synthesized from synthetic oligonucleotides.Citation33 Using the same approach, this was soon followed by the creation of a synthetic mouse L1 non-LTR genome (ORFeus-Mm) and subsequently the human L1 counterpart—ORFeus-Hs.Citation16,Citation34 Synthetic biology was used to optimise codon usage within ORFs 1 and 2 in a step-wise, incremental, manner. Only when the synthetic, optimised, portion of the genome was extended to comprise the entire ORF1 and the N-terminal two-thirds of ORF2 was L1 expression and retrotransposition frequency markedly enhanced, consistent with the notion that control over ORF2 protein biogenesis is key in downregulating the activities of these elements.Citation34 This strategy also allows manipulation of non-LTR genomes at will, and can be used to resolve many of the outstanding questions surrounding the molecular biology of this fascinating group of mobile genetic elements: the non-LTR sequence databases should provide a rich resource for synthetic/molecular biologists to exploit. Alexander Pope wrote that “The proper study of Mankind is Man”: these exciting new tools can now be used to study the ~17% of the human genome that are LINEs and the many diseases which arise from retrotransposition in humans (reviewed in refs. Citation35 and Citation36).
The reader may have noted that the interesting questions as to “the how” and the “from where” these 2A sequences have been acquired by non-LTRs were not discussed above. Although first characterized in virus genomes, we recently identified 2As in a range of cellular genes where 2A appears to play a role in controlling the sub-cellular localization of the translation products (refs. Citation8 and Citation37, and unpublished observations). Furthermore, given that 2A translational recoding elements comprise only some 18–30 amino acids, it may well be that these sequences have a number of independent evolutionary origins. The “how?” and “from where?” questions most probably will never be answered, but these new experimental approaches to study non-LTR retrotransposons ultimately will answer the “why?”
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors gratefully acknowledge the long-term support of our research by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the Wellcome Trust.
Citation: Luke GA, Roulston C, Odon V, de Felipe P, Sukhodub A, Ryan MD. Lost in translation: The biogenesis of non-LTR retrotransposon proteins. Mobile Genetic Elements 2013; 3:e27525; 10.4161/mge.27525
References
- Heras SR, Thomas MC, García-Canadas M, de Felipe P, García-Pérez JL, Ryan MD, López MC. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell Mol Life Sci 2006; 63:1449 - 60; http://dx.doi.org/10.1007/s00018-006-6038-2; PMID: 16767356
- Odon V, Luke GA, Roulston C, de Felipe P, Ruan L, Escuin-Ordinas H, Brown JD, Ryan MD, Sukhodub A. APE-type non-LTR retrotransposons of multicellular organisms encode virus-like 2A oligopeptide sequences, which mediate translational recoding during protein synthesis. Mol Biol Evol 2013; 30:1955 - 65; http://dx.doi.org/10.1093/molbev/mst102; PMID: 23728794
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol 2001; 82:1013 - 25; PMID: 11297676
- Atkins JF, Wills NM, Loughran G, Wu CY, Parsawar K, Ryan MD, Wang CH, Nelson CC. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go). RNA 2007; 13:803 - 10; http://dx.doi.org/10.1261/rna.487907; PMID: 17456564
- Doronina VA, Wu C, de Felipe P, Sachs MS, Ryan MD, Brown JD. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol 2008; 28:4227 - 39; http://dx.doi.org/10.1128/MCB.00421-08; PMID: 18458056
- Halpin C, Cooke SE, Barakate A, El Amrani A, Ryan MD. Self-processing 2A-polyproteins--a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J 1999; 17:453 - 9; http://dx.doi.org/10.1046/j.1365-313X.1999.00394.x; PMID: 10205902
- de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol 2006; 24:68 - 75; http://dx.doi.org/10.1016/j.tibtech.2005.12.006; PMID: 16380176
- Luke GA, Ryan MD. The protein co-expression problem in biotechnology and biomedicine: virus 2A and 2A-like sequences provide a solution. Future Virology 2013; 8:983 - 1; http://dx.doi.org/10.2217/fvl.13.82
- http://www.st-andrews.ac.uk/ryanlab/page10.htm
- Luke GA, de Felipe P, Lukashev A, Kallioinen SE, Bruno EA, Ryan MD. Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J Gen Virol 2008; 89:1036 - 42; http://dx.doi.org/10.1099/vir.0.83428-0; PMID: 18343847
- Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J 1994; 13:928 - 33; PMID: 8112307
- Donnelly ML, Gani D, Flint M, Monaghan S, Ryan MD. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol 1997; 78:13 - 21; PMID: 9010280
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 1999; 16:793 - 805; http://dx.doi.org/10.1093/oxfordjournals.molbev.a026164; PMID: 10368957
- Kapitonov VV, Tempel S, Jurka J. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene 2009; 448:207 - 13; http://dx.doi.org/10.1016/j.gene.2009.07.019; PMID: 19651192
- Novikova OS, Blinov AG. [Origin, evolution, and distribution of different groups of non-LTR retrotransposons among eukaryotes]. Genetika 2009; 45:149 - 59; PMID: 19334608
- Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature 2004; 429:314 - 8; http://dx.doi.org/10.1038/nature02535; PMID: 15152256
- Matsumoto T, Takahashi H, Fujiwara H. Targeted nuclear import of open reading frame 1 protein is required for in vivo retrotransposition of a telomere-specific non-long terminal repeat retrotransposon, SART1. Mol Cell Biol 2004; 24:105 - 22; http://dx.doi.org/10.1128/MCB.24.1.105-122.2004; PMID: 14673147
- Matsumoto T, Hamada M, Osanai M, Fujiwara H. Essential domains for ribonucleoprotein complex formation required for retrotransposition of telomere-specific non-long terminal repeat retrotransposon SART1. Mol Cell Biol 2006; 26:5168 - 79; http://dx.doi.org/10.1128/MCB.00096-06; PMID: 16782900
- Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J Biomed Biotechnol 2006; 2006:45621; http://dx.doi.org/10.1155/JBB/2006/45621; PMID: 16877816
- Horn AV, Klawitter S, Held U, Berger A, Jaguva Vasudevan AA, Bock A, Hofmann H, Hanschmann KM, Trösemeier JH, Flory E, et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res 2013; In press http://dx.doi.org/10.1093/nar/gkt898; PMID: 24101588
- Kojima KK, Kapitonov VV, Jurka J. Recent expansion of a new Ingi-related clade of Vingi non-LTR retrotransposons in hedgehogs. Mol Biol Evol 2011; 28:17 - 20; http://dx.doi.org/10.1093/molbev/msq220; PMID: 20716533
- Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene 2008; 419:1 - 6; http://dx.doi.org/10.1016/j.gene.2008.04.007; PMID: 18534786
- Han JS. Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA 2010; 1:15; http://dx.doi.org/10.1186/1759-8753-1-15; PMID: 20462415
- Finnegan DJ. Retrotransposons. Curr Biol 2012; 22:R432 - 7; http://dx.doi.org/10.1016/j.cub.2012.04.025; PMID: 22677280
- Ryan MD, Flint M. Virus-encoded proteinases of the picornavirus super-group. J Gen Virol 1997; 78:699 - 723; PMID: 9129643
- Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol 2012; 93:1385 - 409; http://dx.doi.org/10.1099/vir.0.042499-0; PMID: 22535777
- Kojima KK, Matsumoto T, Fujiwara H. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol Cell Biol 2005; 25:7675 - 86; http://dx.doi.org/10.1128/MCB.25.17.7675-7686.2005; PMID: 16107714
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 2012; 86:45 - 93; http://dx.doi.org/10.1016/B978-0-12-386497-0.00002-5; PMID: 22243581
- Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem 2005; 280:23425 - 8; http://dx.doi.org/10.1074/jbc.R400041200; PMID: 15749702
- Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem 2010; 285:29033 - 8; http://dx.doi.org/10.1074/jbc.R110.150532; PMID: 20576611
- Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 2011; 10:229 - 40; http://dx.doi.org/10.4161/cc.10.2.14472; PMID: 21220943
- Jang CJ, Jan E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA 2010; 16:1182 - 95; http://dx.doi.org/10.1261/rna.2044610; PMID: 20423979
- Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 2002; 297:1016 - 8; http://dx.doi.org/10.1126/science.1072266; PMID: 12114528
- An W, Dai L, Niewiadomska AM, Yetil A, O’Donnell KA, Han JS, Boeke JD. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob DNA 2011; 2:2; http://dx.doi.org/10.1186/1759-8753-2-2; PMID: 21320307
- Hancks DC, Kazazian HH Jr.. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 2012; 22:191 - 203; http://dx.doi.org/10.1016/j.gde.2012.02.006; PMID: 22406018
- Grandi FC, An W. Non-LTR retrotransposons and microsatellites: Partners in genomic variation. Mob Genet Elements 2013; 3:e25674; http://dx.doi.org/10.4161/mge.25674; PMID: 24195012
- Brown JD, Ryan MD. Ribosome “Skipping”: “Stop-Carry On” or “StopGo” Translation. in ‘Recoding: Expansion of Decoding Rules Enriches Gene Expression’. 2010; pp 101-122. Atkins JF, Gesteland RF (eds) Springer ISBN 978-0-387-89382-2