Abstract
The evolutionary conserved protein Sem1/Dss1 is a bona fide subunit of the 19S regulatory particle (RP) of the proteasome and in mammalian cells stabilizes the tumor suppressor protein BRCA2. A recent study from our laboratory has revealed an unexpected non-proteasomal role of Sem1 in mRNA export. We found that Sem1 is the unique subunit of the RP that is required for proper nuclear export of mRNAs, transcription elongation and preventing transcription-associated hyper-recombination. Biochemical analyses revealed that Sem1, independent of the RP, coenriched with the TREX-2 complex that is required for tethering a subset of genes to the nuclear periphery, transcription-coupled mRNA export and preventing transcription-associated genome instability. Strikingly, we found that Sem1 coenriched with a third multi-subunit protein complex namely the COP9 signalosome, which is involved in deneddylation. We propose that Sem1 is a versatile protein that regulates the functional integrity of multiple protein complexes involved in diverse biological pathways.
Introduction
In all eukaryotes, RNA polymerase II (RNAPII) initiates the first step in gene expression in the nucleus, leading to production of mRNAs. Emerging nascent pre-mRNAs are modified, processed and matured into mRNAs by several protein complexes that are loaded onto transcription sites via their interaction with the C-terminal domain of RNAPII.Citation1–Citation3 Processing and maturation events of the pre-mRNAs include the addition of a 5′-cap, splicing, 3′ cleavage and subsequent poly-adenylation. There is growing consensus that transcription, pre-mRNA processing and the export of mature mRNA to the cytoplasm are coupled.Citation2–Citation5
In budding yeast and mammalian cells the THO/transcription export (TREX) complex was shown to connect biogenesis of mRNPs with their nuclear export.Citation4 THO/TREX is thought to be recruited to the elongating RNAPII via the THO subunits (Hpr1, Tho2, Mft1 and Thp2), which function in transcription and packaging of mRNA protein complexes (mRNPs). The additional TREX factors Sub2 and Yra1 are involved in recruiting the hetero-dimeric export receptor Mex67-Mtr2 to nascent mRNPs (), thereby coupling mRNP biogenesis to nuclear export (reviewed in refs. Citation2, Citation4 and Citation6). A genetic screen with Yra1 identified an additional transcription-mRNA export complex, called TREX-2 consisting of Sac3, Thp1, Sus1 and Cdc31 (). TREX-2 potentially coordinates Spt7-Ada2-Gcn5 acetyltransferase (SAGA)-mediated trans cription of a subset of genes at the nucleoplasmic face of the NPC.Citation7 A central component of TREX-2 is Sac3, a multi-domain protein that serves as a binding platform for other members of the complex. The N-terminal and middle domain (N + M) of Sac3 bind Thp1 and Mex67-Mtr2, while the C-terminal domain mediates its NPC targetingCitation8 and recruits the centrin CdcCitation31 as well as Sus1.Citation9,Citation10 Recent works from several groups have demonstrated a requirement of TREX-2 in “gene gating,” a process involving the dynamic repositioning of a subset of gene loci from the nuclear interior to the nuclear periphery upon their activation.Citation11–Citation15
Sem1 is a Functional Component of the TREX-2 Complex
Sem1 is a small acidic protein that is highly conserved among all eukaryotic species. SEM1 was originally isolated as a multi-copy suppressor of exocyst mutants in budding yeast.Citation16 Mutations in Sem1 lead to several pleiotropic phenotypes such as defects in exocytosis, pseudohyphal growth and defects in the cell cycle.Citation16,Citation17 Genetic screens and proteomic approaches identified Sem1 as a component of the lid sub-complex of the regulatory particle (RP) of the proteasome in both budding yeast and humans.Citation18–Citation21 Loss of Sem1 impairs the functional integrity of the RP.Citation18 Consistent with this, sem1Δ mutants show impaired ubiquitin-dependent protein degradation and accumulate polyubiquitinated proteins.Citation21 The deletion of Sem1/Dss1 in fission yeast was shown to be defective in the nuclear export of mRNAs.Citation22,Citation23 Curiously, in budding and fission yeasts Sem1 was reported to purify only the RP of the proteasome, implicating the ubiquitin- proteoasome pathway in the nuclear export of mRNAs.Citation24,Citation25 Therefore, we sought to shed light on the direct role of the RP, if any, in nuclear export of mRNAs using budding yeast as a model system.
Our recent work has provided several lines of evidence that Sem1 has an additional moonlighting role in mRNA export as part of TREX-2Citation26 (). Like impaired TREX-2 mutants, sem1Δ cells were defective in nuclear export of mRNAs.Citation8 Since Sem1 was identified as a bona fide subunit of the RP we tested whether a functional RP is required for proper mRNA export. However, neither mutants impaired in RP function nor directly inhibiting proteasomal activity by the drug MG132 induced defects in nuclear export of mRNAs. Thus, Sem1 is the unique component of the RP that is required for proper nuclear export of mRNAs. Further, only Sem1, but not other components of the RP, is genetically linked to essential components of the mRNA export pathway (Mex67, Mtr2, Yra1 and Sub2). Finally, the overexpression of Sub2 strongly inhibited growth of sem1Δ, in a manner similar to TREX-2 mutants.Citation27,Citation28 Importantly, growth of other RP mutants was found not to be sensitive to Sub2 overexpression. The sensitivity of sem1Δ to Sub2 overexpression raised the possibility of a direct physical association with TREX-2. To test this, using Thp1 and Sac3 as bait proteins, we directly purified TREX-2 and showed that Sem1 specifically co-enriched in these purifications, independent of the RP. Importantly, the enrichment of Sem1 with TREX-2 was found to be RNA independent. Together these cell-biological, genetic and biochemical data suggest that, like in the 19S RP, Sem1 is a bona fide component of TREX-2. Interestingly, a combination of an unbiased large-scale genetic screen and biochemical approaches towards understanding RNA processing also identified Sem1 as a novel functional component of TREX-2.Citation29
Why was the association of Sem1 with TREX-2 not previously observed? It appears that the addition of the large TAP epitope abolishes its association with TREX-2. Consistent with this possibility Sem1-TAP is non-functional in the mRNA export pathway, as the Sem1-TAP strain showed a defect in mRNA export and, when combined with mex67 and mtr2 mutants was found to be synthetically lethal.
Sem1 Associates with Sac3_GANP and PAM Domain Containing Protein Complexes
Do the RP and TREX-2 contain common structural features that are recognized by Sem1? Indeed, both the RP and TREX-2 contain protein pairs that exhibit Sac3_GANP and PAM domains ().Citation30–Citation32 The Sac3_GANP domain is present in Rpn12 (RP) and Sac3 (TREX-2), whereas Rpn3 (RP) and Thp1 (TREX-2) contain the PAM domains ().Citation30,Citation32 Both the Sac3_GANP and PAM domains are predicted to be alpha-helical and have been proposed to mediate protein-protein interactions.Citation30 Intriguingly, bioinformatic analyses revealed that Csn12, a subunit of the COP9 signalosome (CSN), contains a PAM domain.Citation30 Further, in the Csn12-TAP purification, proteomic approaches identified a Sac3_GANP domain containing protein, Ypr045c.Citation33 We tested whether Sem1 also co-enriched with these components and found that this was indeed the case. Deletions of members of the CSN complex have been shown to accumulate neddylated cullin Cdc53.Citation34 However, we did not observe any alterations in Cdc53 de-neddyltion in sem1Δ, ypr045cΔ and csn12Δ strains. Moreover, we found that both csn12Δ and ypr045cΔ strains are not defective in mRNA export, suggesting that Sem1 has an additional role in the CSN that is independent of its function in the RP and TREX-2. Interestingly, Wilmes et al. have shown that the Csn12-Ypr045c-Sem1 sub-complex associates with the splicing machinery and that the csn12Δ and ypr045c strains were defective in pre-mRNA splicing. Thus, the Csn12-Ypr045c-Sem1 sub-complex is likely to affect a yet undetermined activity of the CSN. We propose that Sem1 specifically associates with at least three distinct protein complexes that contain known Sac3_GANP and PAM domains ().
What could be the Role of Sem1 in the RP, TREX-2 and CSN?
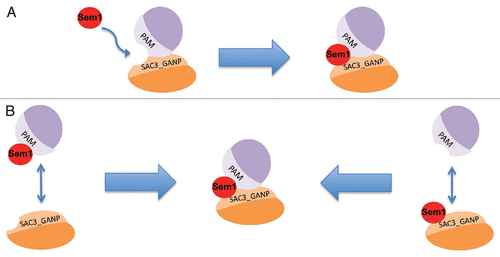
Two scenarios can be envisioned for the formation of Sem1 containing complexes (). In one scenario Sem1 might stabilize interactions between the Sac3_GANP containing subunit and its PAM domain counterpart (). Another possibility could be that Sem1 stabilizes the PAM domain which then would facilitate a robust interaction with the Sac3_GANP containing subunit or vice versa (). Consistent with the first possibility, we found that the tethering of Thp1 to the NPC was severely affected in sem1Δ and that the association of Sem1 with TREX-2 required the presence of both Sac3 and Thp1. Whether Sem1 directly “glues” the Sac3_GANP and PAM domains together still remains to be determined. Evidence favoring the second possibility is the finding that Sem1/Dss1 maintains the correct conformation of BRCA2, as BRCA2 is largely insoluble in absence of Dss1.Citation35,Citation36 Structural analyses of Sac3_GANP and PAM domains in complex with Sem1 will provide further insights into how Sem1 might stabilize protein complexes.
Can Sem1 associate with other protein complexes? Here the crystal structure of Sem1/Dss1 in complex with a part of BRCA2 can provide possible insights. Sem1/Dss1 was shown to bind the helix turn helix motif (helical domain) and an oligonucleotide-binding domain (OB) of BRCA2 in an extended conformation. Sem1/Dss1 appears to make contacts with numerous residues on the helical domain and the OB surface that is rich in basic, aromatic and hydrophobic residues.Citation36 The Sac3_GANP and PAM domains contain periodic hydrophobic patches followed by conserved positive residues found typically in α-helical structures.Citation31,Citation32 Interestingly, Sac3_GANP and PAM domains have neither structural nor sequence homology with the region of BRCA2 that interacts with Sem1. In addition to the proteasome and BRCA2, in human cells, Dss1 was shown to associate with the Integrator complex (involved in snRNA processing).Citation37 Interestingly, the Integrator complex appears not to contain an obvious Sac3_GANP and PAM domain protein pair. Hence, it seems possible that Sem1 targets multiple folds/surfaces present in several unrelated protein complexes. Genetic approaches in budding yeast need to be further exploited to reveal additional biological processes that Sem1 might regulate.
A Non-Proteasomal Role for Sem1/Dss1 in Preventing Transcription-Associated Genomic Instability
A large body of work in mammalian cells has implicated mutations in the tumor suppressor protein BRCA2 in genomic instability. BRCA2 functions in the repair of double-strand breaks (DSBs).Citation38–Citation40 Sem1/Dss1 was discovered to be a BRCA2-binding proteinCitation17 that specifically interacts with the C-terminal portion of BRCA2.Citation36 Sem1/Dss1 has been shown to stabilize BRCA2 and consistently, its depletion in mammalian cells was shown to induce phenotypes similar to those seen in BRCA2-deficient cells.Citation27,Citation40,Citation41 Although budding yeast lacks an obvious homologue of BRCA2, Sem1, as part of the RP was shown to be important for the repair of DSBs by both homologous recombination and non-homologous end joining pathways.Citation20
In addition to their direct function in mRNA export, both THO/TREX and TREX-2 play an important role in preventing transcription-associated genomic instability.Citation6 THO/TREX and TREX-2 mutants induce co-transcriptional formation of RNA:DNA hybrids (R-loops) between the emerging RNA and the transcribed single stranded DNA (ssDNA).Citation42 R-loops are believed to become obstacles for subsequent elongating RNAPIIs, thus impairing transcription elongation or generating mRNA-RNAPII-DNA tertiary structures that can obstruct replication, leading to genome instability.Citation43 TREX-2 mutants induce transcription- associated hyper-recombination phenotypes that are very similar to those seen in sem1Δ cells. Thus, our work has uncovered a non-proteosomal role of Sem1 in preventing transcription-associated genomic instability by contributing to the functional integrity of TREX-2.
Concluding Remarks
Sem1 was previously identified as a bona fide subunit of the RP and a stabilizer of the tumor suppressor protein BRCA2. Our recent work has identified Sem1 as a component of two other protein complexes TREX-2 and the CSN, which are involved in distinct biological pathways. The precise role of Sem1 in the CSN still remains to be unraveled. It is intriguing that despite their functional diversity the RP, TREX-2 and CSN contain common structural elements: Sac3_GANP and PAM domains a shared functional component: Sem1/Dss1. Determining at the molecular level the common principle by which Sem1 modulates the stability and hence the functioning of these important biological machines remains a future challenge.
Figures and Tables
Figure 1 mRNA export and gene gating in S. cerevisiae. (A) The THO complex is co-transcriptionally recruited and associates with nascent transcripts. Together with Sub2 and Yra1 the trEx complex is formed. Mex67-Mtr2 binds to the mrNPs through adaptor proteins such as Yra1 and facilitates their nuclear export. (B) Interactions between the SAGA transcription initiation complex and the nuclear pore associated TREX-2 complex result in tethering of activated genes to the nuclear periphery.
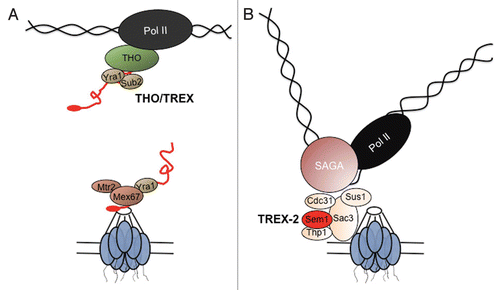
Figure 2 Association of Sem1 with target complexes. The RP of the proteasome, TREX-2 and the CSN are the three complexes in budding yeast, which contain PAM and Sac3_GANP domains and feature Sem1 as a component.
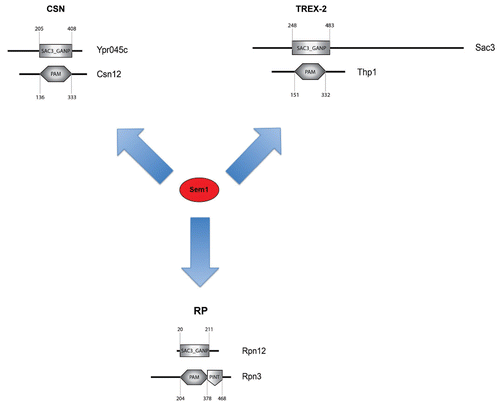
Figure 3 Models for interaction of Sem1 with Sac3_GANP and PAM domains. Depending on the ability of Sem1 to interact with PAM and Sac3_GANP domains individually, we propose two distinct models: in (a) Sem1 is not able to interact with PAM or Sac3_GANP domains individually and therefore association of Sem1 necessitates an interaction of these domains prior to binding. In model (B), Sem1 can interact with PAM and Sac3_GANP domains individually and thereby bridge these domains.
Acknowledgements
V.G. Panse is supported by grants from the Swiss National Science Foundation and Swiss Federal Institute of Technology.
References
- Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol 2006; 18:299 - 306
- Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 2005; 17:269 - 273
- Sommer P, Nehrbass U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr Opin Cell Biol 2005; 17:294 - 301
- Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 2007; 8:761 - 773
- Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly and function. Dev Cell 2003; 4:775 - 789
- Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol 2005; 17:242 - 250
- Rodríguez-Navarro S, Fischer T, Luo MJ, Antúnez O, Brettschneider S, Lechner J, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 2004; 116:75 - 86
- Fischer T, Strässer K, Rácz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, et al. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 2002; 21:5843 - 5852
- Fischer T, Rodríguez-Navarro S, Pereira G, Rácz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol 2004; 6:840 - 848
- Jani D, Lutz S, Marshall NJ, Fischer T, Kohler A, Ellisdon AM, et al. Sus1, Cdc31 and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell 2009; 33:727 - 737
- Brickner J, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. Plos Biol 2004; 2:342
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 2006; 441:770 - 773
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004; 117:427 - 439
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, et al. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 2007; 26:4956 - 4965
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 2006; 441:774 - 778
- Jäntti J, Lahdenranta J, Olkkonen VM, Söderlund H, Keränen S. SEM1, a homologue of the split hand/split foot malformation candidate gene Dss1, regulates exocytosis and pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA 1999; 96:909 - 914
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol 1999; 19:4633 - 4642
- Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1 the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci 2004; 117:6447 - 6454
- Jossé L, Harley ME, Pires IM, Hughes DA. Fission yeast Dss1 associates with the proteasome and is required for efficient ubiquitin-dependent proteolysis. Biochem J 2006; 393:303 - 309
- Krogan N, Lam M, Fillingham J, Keogh M, Gebbia M, Li J, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 2004; 16:1027 - 1034
- Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem 2004; 279:28807 - 28816
- Mannen T, Andoh T, Tani T. Dss1 associating with the proteasome functions in selective nuclear mRNA export in yeast. Biochem Biophys Res Comm 2008; 365:664 - 671
- Thakurta A, Gopal G, Yoon J, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J 2005; 24:2512 - 2523
- Duncan K, Umen JG, Guthrie C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr Biol 2000; 10:687 - 696
- Neumann S, Petfalski E, Brugger B, Grosshans H, Wieland F, Tollervey D, et al. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep 2003; 4:1156 - 1162
- Faza MB, Kemmler S, Jimeno S, Gonzalez-Aguilera C, Aguilera A, Hurt E, et al. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol 2009; 184:833 - 846
- Gallardo M. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J Biol Chem 2003; 278:24225 - 24232
- Gonzalez-Aguilera C, Tous C, Gomez-Gonzalez B, Huertas P, Luna R, Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 2008; 19:4310 - 4318
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 2008; 32:735 - 746
- Ciccarelli FD, Izaurralde E, Bork P. The PAM domain, a multi-protein complex-associated module with an all-alpha-helix fold. BMC Bioinformatics 2003; 4:64
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci 1998;
- Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics 2005; 6:71
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 2006; 440:637 - 643
- Maytal-Kivity V, Piran R, Pick E, Hofmann K, Glickman MH. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep 2002; 3:1215 - 1221
- Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination and genome stability in Ustilago maydis. Mol Cell 2003; 12:1043 - 1049
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y. BRCA2 Function in DNA Binding and Recombination from a BRCA2-DSS1-ssDNA Structure. Science 2002; 297:1822 - 1823
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005; 123:265 - 276
- Gudmundsdottir K, Lord C, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene 2007; 26:7601 - 7606
- Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep 2004; 5:989 - 993
- Li J, Zou C, Bai Y, Wazer D, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene 2006; 25:1186 - 1194
- Gudmundsdottir K, Lord C, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene 2007; 26:7601 - 7606
- Huertas P, Aguilera A. Cotranscriptionally formed DNA: RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 2003; 12:538 - 539
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204 - 217