Abstract
The heterogenous nuclear ribonucleoprotein G (hnRNP G) controls the alternative splicing of several pre-mRNAs. While hnRNP G displays an amino terminal RNA recognition motif (RRM), we find that this motif is paradoxically not implicated in the recruitment of hnRNP G to nascent transcripts in amphibian oocytes. In fact, a deletion analysis revealed that targeting of hnRNP G to active transcription units depends on another domain, centrally positioned, and consisting of residues 186-236. We show that this domain acts autonomously and thus is named NTD for nascent transcripts targeting domain. Furthermore, using an RNA probe previously characterized in vitro as an RNA that interacts specifically with hnRNP G, we demonstrate a new auxiliary RNA binding domain (RBD). It corresponds to a short region of 58 residues positioned at the carboxyl terminal end of the protein, which recognizes an RNA motif predicted to adopt an hairpin structure. The fact that the NTD acts independently from both the RRM and the RBD strongly suggests that the initial recruitment of hnRNP G to nascent pre-mRNAs is independent of its sequence-specific RNA binding properties. Together, these findings highlight the modular organization of hnRNP G and offer new insights into its multifunctional roles.
Introduction
In all eukaryotic cells, nuclear pre-mRNA splicing is critical for a correct proteome expression. This fundamental step in the regulation of gene expression is controlled by cis-acting RNA elements and a large number of trans-acting factors, collectively referred to as splicing factors. In the current view, splicing factors and most other RNA processing factors are recruited to pre-mRNAs co-transcriptionally, resulting in the formation of nascent ribonucleoprotein (RNP) fibrils that are further packaged into export-competent RNP complexes.Citation1,Citation2 Prominent among the RNA-binding proteins (RBPs) recruited to nascent transcripts are the heterogenous nuclear ribonucleoproteins (hnRNPs), which form a family of more than 20 proteins originally labeled from A1 to U based on their respective 2-dimensional electrophoretic mobilities.Citation3,Citation4 These proteins have a modular structure exhibiting one or several regions that serve as RNA binding domains (RBDs) and others as auxiliary domains. Most hnRNP proteins can interact directly with RNA, and while many of them exhibit sequence specificity, it was conventionally thought that hnRNPs had only a structural role in packaging pre-mRNAs. Over the last two decades, however, several hnRNP proteins were implicated in a variety of specific functions, in particular the splicing regulation of many genes. In fact, hnRNP proteins have emerged as important splicing regulators of many pre-mRNAs.Citation5–Citation7 However, how these proteins are recruited to nascent transcripts in vivo and how they interact with distinct RNA structures/sequences to control specific splicing events remain poorly understood.
This is the case for the relatively low abundant hnRNP G protein, which is implicated in the splicing control of several premRNAs. In particular, hnRNP G can act positively or negatively on the incorporation of specific exons within mRNAs coding for the mammalian αS-tropomyosin, dystrophin, survival motor neuron SMN2, and Tau proteins.Citation6,Citation8 In addition to playing critical roles in pre-mRNA splicing, hnRNP G may also function as a regulator of transcription for two genes, SREBP-1cCitation9 and GnRH1.Citation10 More recently, hnRNP G was also proposed to be a tumor-suppressor as it upregulates the Txnip gene and promotes the fidelity of DNA end-joining activity.Citation11,Citation12 Whether hnRNP G influences Txnip expression through controlling transcription and/or pre-mRNAs splicing remains uncertain, however. Finally, hnRNP G appears to be critical for proper neural development of zebrafish and frog embryos.Citation13,Citation14
In human cells, the gene coding for hnRNP G is known as RBMX (for RNA binding motif gene, X chromosome). RBMX is located on the X chromosome and is expressed ubiquitously. There are several paralogues of RBMX in human genome.Citation15–Citation17 Two of them are exclusively expressed in testis and are critical for male fertility; RBMY is located on the Y chromosome and HnRNP G-T is a retrogene mapped on chromosome 11. Multiple processed, retroposed copies of RBMX exist on other autosomes (RBMX-Like sequences, RBMXLs). Three of the RBMXLs are transcribed in human tissues but only one of them (RBMXL1) has the full-length open reading frame.Citation18 The primary sequence of hnRNP G shares 51% and 73% of similarities with RBMY and hnRNP G-T, respectively. All three members of this small protein family were shown to interact with factors implicated in the regulation of pre-mRNA splicing, such as hTra2-β1 and T-STAR, raising the interesting possibility that they influence splicing in a cell type-dependent manner.Citation17,Citation19 Several protein motifs characterize hnRNP G and its paralogues. One of them is an RNA recognition motif (RRM), which exhibits a particularly high level of sequence conservation, with 82–97% of similarity with hnRNP G-T and RBMY, respectively. In all three proteins, the RRM is positioned at the amino-terminal (N-ter) end and possesses the two canonical RNP1 and RNP2 sequences. Despite the high similarities among the RRMs, they appear to recognize different RNA targets. The RRM from the human RBMY was recently shown to interact specifically with a stem-loop structure in which the loop is formed by the sequence CA/UCAA. In contrast, the RRM from hnRNP G fails to bind the same stem-loop structure,Citation20 and was shown to interact with single stranded RNA sequences containing a CCA/C motif.Citation21 Surprisingly, a mutated form of hnRNP G lacking its RRM retains its ability to change splice site selection, suggesting that hnRNP G may associate with transcripts and influence splicing independently of its RRM.
Earlier studies in the oocyte of the newt Pleurodeles waltlCitation22 showed that hnRNP G associates with most transcription units of RNA polymerase II (RNAPII). This finding supported for the first time the idea that hnRNP G may be a component of general transcription/processing complexes. In the nucleus of amphibian oocytes, chromatin is organized within lampbrush chromosomes (LBCs) and numerous non-chromosomal nucleoli. LBCs are highly extended diplotene bivalent chromosomes, and their characteristic resemblance to a test-tube brush comes from the fact that each homologue consists of a heterochromatin axis from which are projected numerous pairs of lateral loops.Citation23,Citation24 These loops correspond to active transcriptional sites by RNAPII and consist of euchromatin fibers surrounded by a matrix of nascent RNP fibrils, also referred to as the RNP matrix. LBCs represent, thus, a unique cellular system to analyze the interactions between RBPs and nascent transcripts. We decided to take advantage of this opportunity to investigate which domains of hnRNP G are necessary for its association with nascent RNAPII transcripts. Surprisingly, we find here that the N-ter RRM is dispensable for the recruitment of hnRNP G to the LBC loops. Instead we characterize a central short domain both necessary and sufficient for the targeting of hnRNP G to nascent RNP fibrils. We also find that this domain is critical for nuclear localization. Finally, we characterize a novel RNA binding domain in the C-terminus of hnRNP G, as well as a potential RNA target, which most likely adopts a hairpin structure recognized by this domain.
Results
The recruitment of hnRNP G to nascent RNAPII transcripts is independent from its RRM.
Using an autoimmune serum from dog that shows specificity for hnRNP G,Citation22,Citation25 we find that one major band of the correct molecular weight is detected in nuclear extracts from Xenopus laevis oocytes and HeLa cells (Suppl. Fig. 1). This distribution pattern is very similar to the one obtained with antibodies directed against other hnRNPs (our unpublished data for hnRNP Q and L) and RNP proteins such as the nuclear factor 7.Citation26 In particular, while the majority of the loops are labeled equally well, some of them present a higher level of staining (). The only other nuclear structures labeled were the Cajal bodies (CBs), which are organelles implicated in all nuclear RNA processing.Citation27
It is difficult to evaluate the contribution of the several weak cross-reacting proteins seen on western blots (Suppl. Fig. 1) to the staining of LBCs. Since our autoimmune serum sample was too small to attempt a purification against hnRNP G, we decided instead to define the sub-nuclear distribution of the full-length human hnRNP G tagged with the HA (hemagluttinin) epitope ( and ). Capped, in vitro made transcripts coding for the human HA-hnRNP G protein were injected in the cytoplasm of stage IV–V oocytes. After 18–24 hours of incubation, the newly made HA-hnRNP G was detected on nuclear spreads by indirect immunofluorescence, using the anti-HA antibody mAb 3F10. We found that the HA-hnRNP G associates with the nascent RNP fibrils of most loops transcribed by RNAPII (). These numerous RNAPII loops are readily distinguishable by phase contrast microscopy and often present a thin-tothick morphology, indicative of an active transcription.Citation23 This association can be detected as soon as 4 hours post injection as a weak labeling of most loops (data not shown). The intensity of the labeling increases overtime and reaches a plateau at ∼14 hours post injection. Like in the staining pattern obtained with the autoimmune serum, several loops appear to be more intensely labeled than all the others. Newly made HA-hnRNP G does not associate with CBs, however.
In frog oocytes, both RNAPII and RNAPIII are actively engaged in transcription on LBCs. In contrast, the activity of RNAPI is restricted to the numerous non-chromosomal nucleoli. The sites of RNAPIII transcription were previously mapped to ∼90 distinct chromosomal loci.Citation28 These sites lack the density created by nascent RNP fibrils on RNAPII loops and, thus, are not visible by phase contrast microscopy. They are, however, readily detected using anti-RNAPIII antibodies.Citation28 To test whether hnRNP G also associates with RNAPIII transcription sites, we used an antibody directed against one of the specific subunits of RNAPIII, RPC53, to localize RNAPIII on nuclear spreads prepared from oocytes expressing HA-hnRNP G (). We found that a newly expressed HA-hnRNP G is excluded from the LBC sites enriched in RNAPIII, demonstrating that hnRNP G association with LBCs is restricted to RNAPII nascent transcripts. Identical results were obtained using a different anti-RNAPIII antibody directed against the RPC15 subunit (data not shown). Finally, the same distribution patterns were obtained in a colocalization study using the autoimmune serum and the anti-RPC53 antibody (data not shown). Importantly, the staining pattern of HA-hnRNP G on LBCs is essentially identical to the pattern obtained with the autoimmune serum. This result strongly supports our conclusion that the epitope detected on LBCs by the autoimmune serum corresponds primarily to hnRNP G.
The fact that newly expressed HA-hnRNP G associates with LBC loops provided the unique opportunity to investigate the domain(s) required for the association of hnRNP G with nascent RNAPII transcripts. We generated several mutant forms of hnRNP G by either N-ter (ΔN) or/and C-ter (ΔC) deletions (). The corresponding transcripts were made in vitro and injected into the cytoplasm of stage IV–V oocytes. The distribution of the newly made HA-tagged proteins was then analyzed on nuclear spreads 18–24 hours later ( and Suppl. Fig. 2). Notably, the deletion of residues 186–236 prevents the translocation of the proteins to the nucleus and result in their accumulation in the cytoplasm (, ). The addition of a nuclear localization signal (NLS), derived from the SV40 large antigen, to these mutants was thus necessary to localize the expressed proteins to the nucleus in order to investigate their recruitment to the LBC loops (). Surprisingly, we find that the N-ter RRM is dispensable for the association of hnRNP G with nascent transcripts, as demonstrated by the fact that the ΔN82, ΔN185, ΔN82ΔC289 and ΔN82ΔC237 proteins still interact strongly with the LBC loops (, Suppl. Fig. 2 and ). Instead, the recruitment of hnRNP G to nascent transcripts requires a central domain of 51 aa (residues 186–236) since the mutants lacking this domain (NLSΔN82ΔC186, NLSΔC83, NLSΔC186) are not detected on the LBCs despite their nuclear accumulation. Interestingly, these mutants accumulate in nucleoli (: the large rounded structures in the merge panel of NLSΔN82ΔC186. They appear yellow rather than red, indicating the presence of the newly made protein; Suppl. Fig. 2A: the large rounded structures that are labeled in the ΔC186 panel). The reason for this apparent nucleolar association is unknown, but it is often encountered for mutated nuclear proteins that are expressed in frog oocytes.Citation26 Remarkably, the newly defined domain composed of residues 186–236 can target LBC loops autonomously (, panel HA-NTD). This central domain of 51 amino acids is thus necessary and sufficient for recruitment to LBC loops, and was named NTD for nascent transcripts targeting domain.
A new RNA motif targeted by human hnRNP G.
The fact that some of the LBC loops accumulate a higher concentration of hnRNP G than most others could reflect a higher density of transcripts on these transcription units. Equally likely, such a preferential association could stem from a higher affinity of hnRNP G for specific transcripts. Interestingly, hnRNP G was recently shown to bind RNA in a sequence-specific way.Citation21 In addition, we recently showed that human hnRNP G and its different amphibian orthologues interact strongly with an in vitro synthesized cRNA probe (WEc RNA) of 250 nucleotides (manuscript in preparation), possibly through recognition of specific RNA motif(s), as proposed earlier by Penrad-Mobayed et al.Citation29 The species-conserved interaction between hnRNP G and WEc RNA prompted us, therefore, to search for the RNA motif(s) targeted by human hnRNP G and embedded within WEc.
Initially, we checked the specificity of interaction of the human hnRNP G protein with WEc using several other RNA probes issued from the same DNA clone.Citation30 We used a mono-dimensional Northwestern assay on nuclear HeLa cells proteins, and obtained data suggesting that human hnRNP G interacts preferentially with the radiolabeled WEc RNA. The hnRNP G protein was identified using the anti-hnRNP G autoimmune serum. While hnRNP G was found to bind two of the control RNAs, these interactions were substantially weaker than the one observed with WEc RNA (). To identify the targeted motif(s) present within the WEc RNA, several smaller and overlapping RNAs (50 nt), which together span the entire WEc sequence, were synthesized and used as 32P-labeled probes on 1D-Northwestern assays (). Residues 93–142 (RNAB) of the 250 nucleotides long WEc RNA appear to be the primary binding site for human hnRNP G. Other regions of the WEc RNA show very little or no significant interaction. Surprisingly, while RNA2 and RNA3 contain the first and second half of RNAB, respectively, they both display very weak interactions with hnRNP G. The recognition of RNAB by hnRNP G was further confirmed by 2D-Northwestern assays, where RNAB is recognized as well as the WEc RNA by the major pI isoforms of hnRNP G (). Interestingly, an analysis of the RNAB structure using the mfold program reveals the very high probability that the 50 nucleotides forming RNAB adopt a stable hairpin structure formed of a long stem and a short loop of 5 residues (GGAAA) (). This stem-pentaloop structure is also predicted in the WEc RNA, but is absent from RNA2 and RNA3, which together form the primary structure of RNAB. Although the GGAAA sequence is also present in the RNA controls RNA3 and RNA4, the conformation of the predicted secondary structure of this sequence is different from the one present in RNAB (). Together, these data suggest that hnRNP G recognizes a hairpin structure (RNAB) present within the WEc RNA.
A new RNA binding domain in human hnRNP G.
With the characterization of the NTD and a specific RNA substrate, one attractive possibility was that the NTD might target hnRNP G through interaction with this substrate. To test this idea, we expressed several deleted forms of HA-tagged hnRNP G in amphibian oocytes and tested their ability to bind RNAB on 1D-Northwestern assays ( and B). Capped, in vitro-synthesized transcripts coding for the polypeptides of interest were injected into the cytoplasm of stage V-VI oocytes. After 24 h of incubation, nuclear protein extracts were prepared and used for a 1D-Northwestern assay, using RNAB as an RNA probe. The newly made proteins were detected using the anti-HA antibody 16B12, and in all cases a single protein band with the expected molecular weight was observed. As expected, full-length HA-hnRNP G shows a strong RNA binding activity. Surprisingly, mutated proteins lacking the RRM or the NTD are still able to interact with the RNA probe as well as the wild type hnRNP G. Notably, the same is also true for all mutated proteins with an intact C-ter region (of 103 residues). However, when the C-ter region is deleted, as it is the case for the two mutants ΔN82ΔC289 and ΔC186, a total loss of affinity for RNAB is observed, strongly suggesting that this region bears an RNA binding activity.
To confirm these data and further characterize the RNA binding domain present in the C-ter region, two purified polypeptides spanning the carboxyl terminal region of hnRNP G were tested on 1D-Northwestern assays ( and C). These polypeptides were bacterially expressed in fusion with the glutathione-S-transferase (GST) protein and purified by affinity chromatography. Unfortunately, the full-length hnRNP G could not be used as a standard positive control because it is unstable in E. coli.Citation31 Of the two polypeptides, only the one corresponding to the last 58 residues of hnRNP G exhibits a high affinity for the WEc hairpin. Collectively, these results demonstrate a new RBD, which is distinct from the RRM and the NTD. This RBD is located at the C-ter region of hnRNP G, and displays a strong affinity for RNAB.
Conservation of the newly identified domains NTD and Cter-RBD.
Sequence comparisons of human hnRNP G and corresponding orthologues from several other species reveal a high level of conservation for the NTD domain among different species (similarity 76–100% with tetrapods, 62–68% with fish orthologues) (). Similarly, the newly identified C-ter RBD is also highly conserved (similarity 86–100% with tetrapods, 64–70% with fish orthologs) (). Such levels of similarities are consistent with the functional definition of these domains, and strongly suggest that their roles are conserved among species. Interestingly, sequence alignments of these two new domains from the human hnRNP G with the other proteins members of the RBMX/RBMY family show a striking divergence of sequences in the RBMY structure where a long stretch of intervening amino acids interrupts the NTD domain and only 35% of similarity with hnRNP G was observed for the C-ter RBD ( and D). This sequence divergence suggests that these domains may not have the same functions in the testisspecific protein RBMY.
Discussion
While hnRNP G was long ignored for a lack of function and a low nuclear concentration, it has recently seen a renewed interest with the demonstration that it can act as a pre-mRNA splicing factor implicated in specific splice site selections. In the present study, we confirm that hnRNP G associates with most nascent RNAs transcribed by RNAPII in the amphibian oocyte, and we characterize two novel functional domains: (1) a C-ter RBD, which is distinct from the RRM and for which we also identify a potential RNA interacting motif; (2) an NTD, which is a domain required for both the nuclear localization of hnRNP G and its targeting to nascent transcripts.
The recruitment of hnRNP G to nascent RNAPII transcripts.
The association of the endogenous and a newly expressed hnRNP G with most RNAPII nascent transcripts strongly suggests that hnRNP G comprises part of the general transcription/processing machinery. Remarkably, the targeting to nascent transcript is independent of the two RBDs, but instead requires the newly defined NTD. The nature of the interactions between the NTD and the nascent transcripts is not known. However, the lack of affinity for RNA, at least on Northwestern blots, suggests that the NTD binds nascent transcripts indirectly through protein-protein interactions with other RNP factors.
Nevertheless, these data support a model in which the known sequence-specific RNA binding properties of hnRNP G are dispensable for recruitment to active transcription units. One could assume that hnRNP G interacts with nascent transcripts mainly in an indirect way, using the NTD, to fulfill a general role in RNP organization, and only engages in direct RNA binding when appropriate cis-acting RNA elements emerge on specific transcripts. This model would offer an explanation to the apparent contradiction of a protein that associates with most RNAPII transcription units, but that plays a role in the splicing regulation of specific transcripts.
Interestingly, several loops were consistently better labeled than all the others. When observed by phase contrast or DIC microscopy, these loops are indistinguishable from all the other labeled loops. In particular, they do not correspond to the socalled landmark or complex loops, which usually display bulkier RNP matrices. Instead these loops have a regular fine RNP matrix, often showing a thin-to-thick morphology. It is unlikely, therefore, that the higher level of staining observed for these loops is the result of a complex folding of the RNP matrix. It is tempting to speculate that the nascent RNAs of these transcription units present a higher affinity for hnRNP G, perhaps by displaying specific cis-acting RNA elements targeted by the N-ter RRM and/or the C-ter RBD. However, in absence of information about the RNAs transcribed on these intensely labeled loops, other interpretations cannot be ruled out. For instance, an elevated density of RNAs on these loops would almost certainly result in a higher concentration of hnRNP G. Another possibility is that these loops serve as preferential sites of storage for hnRNP G for later use during early development. The idea of specialized “storage loops” was suggested previously for the protein ADAR1, which is proposed to accumulate at high concentrations to specific loops through interactions with noncoding RNAs.Citation32
Nevertheless, our model joins others where splice site selection requires the formation of dynamic RNA/protein complexes in which the cooperation among several trans-splicing factors is required to achieve specific RNA recognition.Citation33,Citation34 Interestingly, several known RNA processing factors, such as hnRNP C, SAM68, YT521-B, SAFB and Tra2β, were previously shown to interact with hnRNP G.Citation17,Citation21,Citation35 Whether some of these factors are implicated in recruiting hnRNP G to nascent transcripts, perhaps as part of a preassembled complex, is not known, however.
The NTD also appears critical for nuclear accumulation of hnRNP G. Whether it harbors a genuine nuclear localization signal (NLS) is not clear, however. Indeed, sequence analysis shows that the NTD is rich in arginine residues, but it does not reveal the presence of a short peptide with the consensus sequence of a classical NLS. One possibility is that the NTD acts in a similar way to the acidic M9 domain, which is involved in regulating the nucleo-cytoplasmic exchanges of hnRNP A1.Citation36 The M9 domain does not contain a classical NLS, but it was shown to act in nuclear targeting through a direct interaction with the transportin receptor Karyopherin β2.Citation37 Another possibility stems from the fact that hnRNP G is theoretically small enough to diffuse through the channel of nucleopore complexes. Thus, hnRNP G may not have an NLS. Rather it could simply be retained in the nucleus through its interactions with nascent transcripts. More puzzling is the observation that some truncated forms of hnRNP G were primarily cytoplasmic upon expression in Xenopus oocytes. Additional work will be needed to test whether such distribution is due to an aberrant behavior of these mutated proteins or whether it is indicative of a specific cytoplasmic retention.
HnRNP G exhibits two distinct RBDs.
In eukaryotic cells, hnRNP G is thought to interact directly with RNAs through its N-terminal RRM, which is a protein motif widely encountered in RBPs. Supporting this model is the demonstration that the RRM of the human hnRNP G binds to specific RNAs in vitro.Citation21 The RRM may not be the only mean of RNA binding for hnRNP G, however, as we demonstrate in the present study a novel RBD in its C-terminal region. This domain, which is composed of the last 58 residues of hnRNP G, can act autonomously as it is able to bind RNA when produced bacterially as a recombinant polypeptide. This was also shown by testing the affinity of human hnRNP G and various deletion mutants synthesized in vitro or in Xenopus oocytes for RNAB. This RNA, which putatively forms a hairpin structure, interacted with hnRNP G at a higher affinity than all the other tested RNAs of the same length. Interestingly RNA2 and RNA3, which combined sequences form the primary structure of RNAB, displayed only a low affinity for hnRNP G. A similar low level of interaction was found for several other tested RNAs and could merely be a reflection of the fact that hnRNP G displays a basal level of sequence-independent affinity for single stranded nucleic acids, since it can be retained on a ssDNA affinity column.Citation22 This is the case for many RNA binding proteins. A good example is the protein U2B”, which was shown to bind the U2 small nuclear RNA (U2 snRNA) with a high affinity as part of the U2 snRNP complex, but also to bind RNA non-specifically in vitro.Citation38 It is likely, therefore, that the higher affinity of hnRNP G for RNAB than for RNA2 or RNA3 is due to the structural element, a stable stem-(GGAAA)loop, predicted to form in RNAB.
The N-ter RRM and C-ter RBD have very different primary structure, and not surprisingly, they exhibit distinct RNA binding specificity. In particular the N-ter RRM has no affinity for RNAB. This is in agreement with the recent finding that the N-ter RRM of hnRNP G does not display any preference for stable stem structures, but rather binds preferentially to CCA/C motif present in single stranded RNAs.Citation21 The C-ter RDB is rich in arginine, glycine and serine, which are residues often enriched in RNA binding domains, like the RGG box and the SR domain of many RBPs. In fact, there are three RGG repeats toward the end of the C-ter RBD. Interestingly, the activity of these domains can be regulated through post-translational modification: serine phosphorylation and arginine methylation, respectively. We recently obtained evidence that several isoforms of human hnRNP G are phosphorylated. However, treating hnRNP G with alkaline phosphatase does not inhibit the binding to RNAB (unpublished data). In addition, a bacterially expressed C-ter RBD still maintains its ability to bind RNAB. Therefore, the RNA binding activity of the C-ter RBD is unlikely to be regulated by phosphorylation. Many hnRNP proteins, including hnRNP G, are methylated on arginine residues.Citation39 Whether methylation of the C-ter RBD influences its RNA binding activity is not known, however.
The presence of several RBDs in hnRNP G suggests that it could interact with multiple RNA targets. This is the case for many other processing factors with multiple RBDs. A good example is the protein nucleolin, which has four distinct RRM motifs. Two of them are implicated in the recognition of different regions of a same stem-loop structure containing the motif U/GCCCGA, also called the nucleolin recognition element (NRE). In contrast, all four RRMs are required for interacting with a short single stranded RNA, referred to as the evolutionary conserved element (ECM),Citation40 which highlights a fundamental concept: different combinations of RBDs within one single protein may be used to specifically interact with distinct RNA targets. In the light of the role of hnRNP G in splice site selection,Citation8,Citation21 it will be of a great interest, therefore, to further investigate whether the two RBDs of hnRNP G are involved in two distinct roles or are acting in concert to regulate specific splicing events. In the latter case, it would be particularly interesting to examine whether both RBDs could be involved in bringing distant RNA sequences to close proximity. This is the case for two of the four RBDs of the polypyrimidine tract binding protein (PTB), which were recently shown to interact with each other. This interaction lead to a model in which PTB could induce RNA looping when bound to two distinct polypyrimidine tracts on the same premRNA in order to regulate splicing.Citation41
Remarkably, the C-ter RBD of hnRNP G lies within the region previously characterized as an interacting domain for the human splicing factor, transformer-2 (hTra2-β1).Citation42 HnRNP G and hTra2-β1 have antagonistic effects on the alternative splicing of several genes in a tissue specific manner.Citation8 One attractive possibility, then, is that the interaction of hTra2-β1 with hnRNP G would modulate the RNA binding specificity of hnRNP G by inhibiting the C-ter RBD. This idea is in good agreement with the recent demonstration that hnRNP G retains its role in splice site selection in absence of its RRM.Citation21 Together, these data favour a model in which the RRM and the C-ter RBD exhibit distinct functions, and where the C-ter RBD would play a critical role in the regulation of alternative splicing by hnRNP G.
Materials and Methods
Cell cultures.
Human cervical carcinoma cells HeLa (ATCC CCL-2) were cultured in RPMI 1640 medium (GIBCO, Invitrogen) supplemented with 5% (v/v) foetal bovine serum, 2 mM L-glutamine. Cell cultures were maintained at 37°C in 5% CO2 atmosphere.
Oocytes and nuclear spreads.
Ovarian biopsies were performed on adult female frogs (Xenopus laevis) that were anesthetized in 0.15% tricaine methane sulfonate (MS222, Sigma Chemical, St. Louis, MO). Oocytes were defolliculated for 1 hour in saline buffer OR2Citation43 containing 0.15% collagenase type II (Sigma Chemical, St. Louis, MO). Stage IV–VI oocytes were selected and maintained in OR2 at 18°C. Nuclear spreads were prepared as described previously,Citation44 fixed 30 minutes at room temperature in phosphate buffer saline (PBS) containing 2% paraformaldehyde and 1 mM MgCl2.
Antibodies and immunofluorescence.
Fixed nuclear spreads were rinsed in PBS and blocked for 10 minutes with 0.5% BSA and 0.5% gelatin (from cold water fish) in PBS for 10 minutes. Nuclear spreads were then incubated with primary antibody for 1 h at room temperature, washed 3 × 10 minutes with PBS, incubated with secondary antibody for 1 h at room temperature and then washed again in PBS before they were mounted in 50% glycerol containing 1 mg/ml of phenylenediamine and 10 pg/ml of 4,6-diamidino-2-phenylindole (DAPI). The two anti-HA antibodies, mAb 3F10 (Hoffmann-La Roche Inc., Nutley, NJ) and HA.11/16B12 (Covance) were used at 50 ng/mL and 1/1,000 dilution, respectively. The anti-hnRNP G autoimmune serum from dog (reviewed in refs. 22 and 25) and used at a 1/4,000 dilution. The rabbit anti-RPC53 antibody was kindly supplied by Dr. R.G. Roeder and used at a 1/50,000 dilution. The anticoilin antibody, mAb H1 (Invitrogen Corp., Carlsbad, CA), was used at 500 ng/mL. The following two Alexa Fluor® (Invitrogen Corp., Carlsbad, CA) conjugated antibodies were used at 2.5 µg/mL: Alexa 594 goat anti-rat IgG, Alexa 568 goat anti-mouse IgG, and Alexa 488 goat anti-rabbit IgG. The Texas red conjugated goat anti-dog IgG (Abnova, Walnut, CA) was used at 2 µg/mL. Standard transmitted light and fluorescence microscopy was performed using an upright Leica DMR (Heidelberg, Germany) with a HCL FL Fluotar 100X oil objective (NA = 1.30). Images were captured using a monochrome Retiga EXI Charge-Coupled Device (CCD) camera (Qimaging, Canada) driven by the In vivo software (version 3.2.0, Media Cybernetics, Bethesda, MD). All images were processed using Adobe Photoshop CS version 8.0 and assembled with Adobe InDesign CS version 3.0.
Cloning, RNA synthesis and microinjection.
The full length human hnRNP G cDNA was used as the template for PCR-mediated mutagenesis. The hemagglutinin (HA) tag, (YPYDVPDYA) was added to the N-terminus of all constructs. A simian virus 40 nuclear localization signal (SV40 NLS; PKKKRKV) was also added upstream of the HA tag, but only in constructs where residues 186–236 were deleted. Constructs were cloned into the vector pCR-Blunt II-TOPO (Invitrogen Corp., Carlsbad, CA). In vitro transcriptions were performed as described in ref. Citation26. Stage IV–VI oocytes were microinjected in the cytoplasm with ∼20 ng of RNA/oocyte. All injections were performed under a dissecting microscope using either the nanoject II or the Nanoject Auto-Nanoliter injector from Drummond, Broomal PA. The glass needles used for microinjection were prepared using a horizontal pipette puller P-97 (Sutter Instrument, Novato CA) and capillary glass tubes (0.5 mm i.d. and 1.2 mm o.d.).
Nuclear extracts.
HeLa cell nuclear extract. Cells were washed twice with cold PBS and resuspended in buffer A: 10 mM Tris pH 8.0, 3 mM MgCl2, 25 mM NaCl, protease inhibitor (Complete Protease Inhibitor Cocktail Tablets, Roche Applied science). After 2 minutes incubation on ice, cells were disrupted using an Ultra-Turrax homogenizer and centrifuged at 1,200 g for 5 min. Nuclei were resuspended in buffer B: 0.25 M Sucrose, 10 mM Tris pH 8.0, 10 mM MgCl2, 10 mM NaCl, protease inhibitor cocktail. The suspension was layered over modified buffer B (0.88 M Sucrose) and centrifuged for 10 min. Nuclei were resuspended in the denaturing electrophoresis loading buffer.
Oocyte nuclear extract. For nuclear protein extraction, injected oocytes were incubated in MBS containing 2% TCA (W/V) for 15–20 min at 4°C to precipitate the proteins. The oocytes were then washed several times in cold MBS and kept at 4°C for 3 h before being manually dissected in MBS. Germinal vesicles (GVs) were isolated, transferred to microtubes, washed in water, then in 70% Ethanol. Air-dried GVs were resuspended in Laemmli buffer, heated for 10 min at 95°C, centrifuged for 10 min at 13,000 rpm and the supernatant was loaded on SDSPAGE gels. Nucleoplasmic and organelle protein extracts were prepared by centrifuging isolated nuclei at 22,000 g for 15 minutes at 4°C. The resulting nuclear organelle pellets (which contain LBCs) were directly resuspended in 1X Laemmli protein sample buffer. Supernatants corresponded to nucleoplasmic fractions. Cytoplasms were centrifuged at 22,000 g for 30 minutes at 4°C to discard yolk and pigment granules. The recovered supernatants were used as cytoplasmic fractions.
Gel electrophoresis and staining.
For mono-dimensional electrophoresis, samples were denatured by heating in Laemmli buffer for 5 min at 95°C and separated on a 10% SDS-PAGE gel (Mini PROTEAN II or Protean II xi from Bio-rad). For bi-dimensional electrophoresis, samples were resuspended in DeStreak rehydration solution (8 M urea, 1 M thiourea, 2.5% CHAPS, hydroxyethyl disulfide, GE Healthcare) plus 1% carrier ampholytes mixture pH 6–11 (GE Healthcare). The isoelectric focusing (IEF) conditions were optimized for focalization of basic proteins. After 12 h of strip rehydration, samples were loaded on the anodic side of 13 cm strips that had a linear pH 6–11 gradient (IPG strips, GE Healthcare). The IEF was performed on Ettan IPGphor (GE Healthcare) at 20°C using the following parameters: voltage was increased gradually to 500 V for 100 Vh then to 4,000 V for 3,400 Vh and finally to 8,000 V for 13,330 Vh. Prior to second dimension the strips were equilibrated in 6 M urea, 2% SDS, 1% DTT and 30% glycerol. In a second equilibration step the DTT was replaced by iodoacetamide to prevent proteins reoxidation. The second dimension was performed in Ettan Dalt VI (GE Healthcare) or Protean II xi (Bio-rad) vertical electrophoresis apparatuses using 12% SDS-PAGE gels. Bi-dimensional electrophoresis was performed in the proteomic platform/2D service of the Institute Jacques Monod.
Northwestern assay.
Following gel electrophoresis, the separated proteins were transferred to a nitrocellulose membrane (Hybond-ECL, GE Healthcare) and temporarily stained with Ponceau S dye (0.1% in 1% acetic acid). Membranes were washed in PBS and incubated overnight in blocking buffer (5% fat-free dry milk, 0.1% Tween 20 in PBS) at 4°C. Membranes were then incubated with 32P-labeled RNA in blocking buffer for 1 h at room temperature. The unbound RNA was removed by washing 3 times with blocking buffer. Autoradiograhy of the airdried membranes was performed using x-ray films (Hyperfilm, GE Healthcare) or storage phosphor screen (rego x-ray GmbH), which were scanned on GS-800 calibrated densitometer (Biorad) or Typhoon Imaging System (GE Healthcare) respectively. Membranes were subsequently used for western blot analysis. 32P-labeled single stranded (ss) RNA probes were synthesized in vitro from either DNA strand of the P130B clone.Citation29 with T7 or SP6 RNA polymerases using the Riboprobe in vitro Transcription system from Promega (Madison, WI) and the [α −32P]UTP (800 Ci/mmol) from PerkinElmer (Waltham, MA). The previously described RNA probe BP6 was renamed here WEc RNA. The small 32P-labeled ssRNA probes of 50 nucleotides were transcribed as above from DNA templates that were obtained by hybridization (1:1 molar ratio) of their respective antiparallel ssDNAs (Eurofins MWG Operon, HPLC purified). All radiolabeled RNAs were purified by gel filtration chromatography using G25 or G50 Sephadex columns (Quick spin columns, Roche Applied science) followed by ethanol precipitation.
Western blots.
Membranes were incubated overnight in PBS + 0.1% Tween 20 (PBST) + 5% fat-free dry milk at 4°C. Membranes were then incubated with the primary antibody in PBS + 0.5% BSA for 1 h, washed in PBST, incubated with the secondary antibody in PBS + 0.5% BSA for 1 h, and washed in PBST. Detection was performed using the ECL or ECF kit from GE Healthcare. Membranes were exposed to x-ray films (Kodak BioMax MR films) or scanned in the LAS-3000 imaging system (FUJI). The anti-hnRNP G autoimmune serumCitation22,Citation25 was used at a 1/2,000 dilution. Two different anti-HA monoclonal antibodies were used: HA.11/16B12 (Covance) used at 1/2,000 dilution and mAb 3F10 (Hoffmann-La Roche Inc., Nutley, NJ) used at 50 ng/mL. The HRP conjugated anti-dog (Jackson ImmunoResearch) and anti-mouse (Sigma Chemical, St. Louis, MO) antibodies were diluted to 1/10,000, while the alkaline phosphatase conjugated anti-rat (Sigma Chemical, St. Louis, MO) was used at 1 µg/mL.
Production and purification of recombinant proteins.
The cDNA clones used for the production of the GST-tagged fragments of hnRNP G were kindly provided by Dr. V. Della Valle.Citation25 They consist of various regions of hnRNP G cloned into the expression vector pGEX-2T (GE Healthcare). These clones were used to transform E. coli strain BL21 (Novagen) and protein expression was induced at 23°C by 1 mM IPTG (Isopropyl-beta-D-thiogalactopyranoside, Euromedex) for 24 h. After centrifugation, bacterial pellets were washed in NaCl 0.9%, resuspended in PBS containing a protease inhibitor cocktail (Roche), and sonicated for 3 × 20 seconds on ice. DNAse (20 ng/ml, Deoxyribonuclease I) and DOC (5 mg/ml, Sodium deoxycholate) (Sigma Chemical, St. Louis, MO) were added to the bacterial lysate, which was left on ice for 30 minutes with frequent pipetting before centrifugation at 17,000 xg for 30 min at 4°C. The GST-tagged proteins were purified from the supernatant by affinity chromatography on a gluthatione column (GSTrap FF Column, GE Healthcare). Recombinant proteins were monitored on SDS-PAGE gels and quantified using the 2D Quant kit (GE Healthcare).
Abbreviations
| RNP | = | ribonucleoprotein |
| RBD | = | RNA binding domain |
| RNAPII | = | RNA polymerase II |
| LBC | = | lampbrush chromosomes |
Figures and Tables
Figure 1 Subnuclear distribution of hnRNP G in Xenopus oocytes. (A) phase contrast and corresponding fluorescent micrographs of one LBC from a nuclear spread that was stained with an autoimmune serum directed against hnRNP G (green). Note that most LBcs loops, which are visible by phase contrast (RNAPII transcription units), are labeled. arrows indicate loops with a particularly strong labeling. DNA was counterstained with DAPI, pseudo-colored in red. Note that the axes of both homologues forming an LBC appear as linear arrays of condensed chromatin granules. The lateral loops are only weakly labeled as they correspond to highly decondensed chromatin fibers. In addition, non chromosomal organelles containing DNA (nucleoli) and/or high concentration of RNAs (nucleoli and nuclear speckles) are also well labeled by DAPI. (B) Magnified view of a nuclear spread to show several non-chromosomal organelles, including nucleoli (asterix) and a Cajal body (arrow), which are readily distinguishable by DIC. In this preparation, the anti-hnRNP G serum stains CBs (green), which were identified using the monoclonal antibody mAb H1 directed against the protein coilin. scale bars are 10 µm.
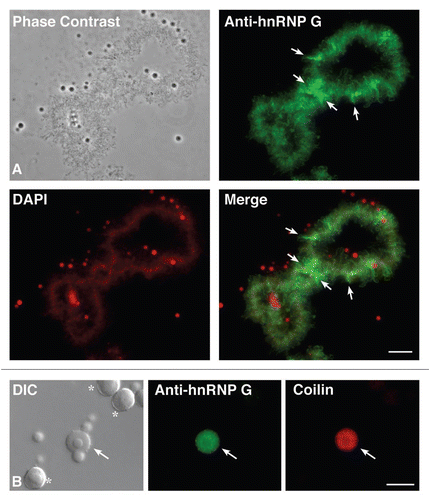
Figure 2 The NTD is necessary and sufficient for hnRNP G association with nascent RNAPII transcripts. Phase contrast and corresponding fluorescent micrographs of LBCs from oocytes expressing HA-tagged hnRNP G and different mutated forms (see ). capped, in vitro made transcripts of interest were injected into the cytoplasm of stage IV–V oocytes, and newly expressed proteins were detected 18 hours later on nuclear spreads using the anti-HA antibody mAb 3F10 (green). Full length, wild type HA-hnRNP G is found to associate specifically with the matrix of LBC loops. HA-ΔN82ΔC289 is presented as typical example of a mutant protein that associates with LBC loops as well as the wild type HA-hnRNP G; in contrast, NLS-HA-ΔN82ΔC186 fails to target LBC loops, as it is the case for all mutants lacking the NTD. Interestingly, these mutants accumulate in nucleoli, which appear yellow in the merge panel (also presented in Suppl. Fig. 2); finally, HA-NTD is recruited to LBCS loops in a pattern identical to the one observed for the full length HA-hnRNP G. DNA was counterstained with DAPI, pseudo-colored in red. Scale bars are 10 µm.
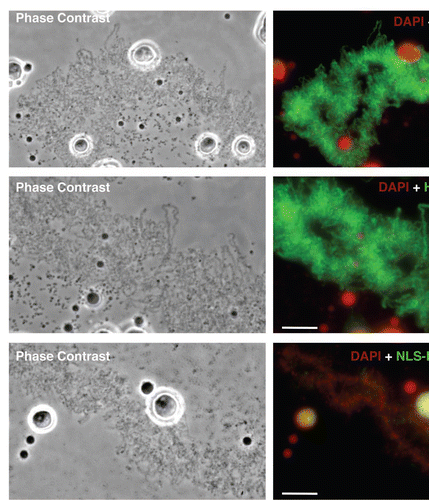
Figure 3 hnRNP G is associated with RNAPII, but not RNAPIII transcripts. Stage IV–V oocytes were injected with in vitro made transcripts coding for HA-hnRNP G. Nuclei were isolated 18 hours later for nuclear spread preparations, and the newly made HA-hnRNP G protein was detected using the mAb 3F10 antibody (green). The sites of transcription by RNAPIII were also defined on nuclear spreads using an antibody directed against RPC53 (red), one of the specific subunits of RNAPIII. A narrow region of one LBC (top three panels; scale bar is 5 µm) as well as magnified views of two distinct RNAPIII loops (the six bottom panels; scale bars are 1 µm) are presented. Most chromosomal loops were labeled with mab 3F10 (green), while only a small number of RNAPIII loci (in red) were observed.
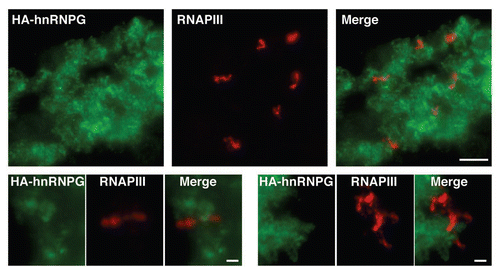
Figure 4 The NTD is required for nuclear localization. The expression of HA-tagged hnRNP G mutant proteins in Xenopus oocytes was monitored by western blot analysis, using the mAb 3F10 antibody. (A) Three representative examples are presented here. Except for ΔN82ΔC186, which lacks the NTD, proteins were found distributed between the cytoplasm and the nucleus (also see Suppl. Fig. 2 for wild-type hnRNP G and the other mutants used in this study). Within the nucleus, proteins were found associated with the organelle pellets (P) containing the LBcs and the soluble nucleoplasmic compartment (S). Proteins from 10 nuclei (organelle and nucleoplasmic fractions) and five cytoplasms, were used. Molecular masses are in kiloDaltons. (B) Comparison of the subcellular distribution of ΔN82ΔC186 and NLs + ΔN82ΔC186. While the expressed ΔN82ΔC186 is restricted to the cytoplasmic compartment, the addition of an NLS results in its nuclear accumulation. coilin, which was detected using the mab H1 antibody, was used here as a control protein that is exclusively nuclear. Proteins from 10 whole nuclei and five cytoplasms were used.
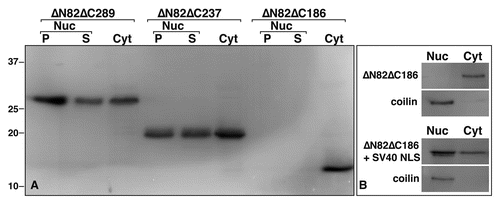
Figure 5 Specific interaction of hnRNP G protein with WEC RNA. (A) Schematic representation of the E. coli subclone from which the tested RNA probes are transcribed in vitro. Arrowheads indicate the direction of transcription; Bam HI, PST I restriction sites and the promoters for the SP6 and T7 polymerases are also indicated. (B) Mono-dimensional Northwestern assay on heLa cell nuclear proteins using WEC RNA and four other different RNAs as 32P-labeled probes. NW, Northwestern assay; ID, immunodetection using anti-hnRNP G. The arrow indicates the hnRNP G protein band at the expected molecular mass of 45 kDa. (C) Representative quantification of the autoradiography signals corresponding to hnRNP G protein from repeated Northwestern assays. Note that while hnRNP G is found to bind two of the control RNAs, these interactions are substantially weaker than the one observed with WEC RNA.
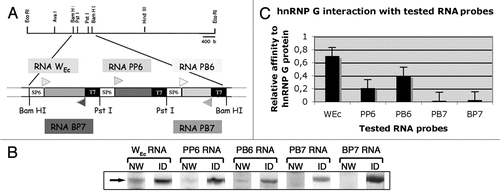
Figure 6 HnRNP G recognizes a short motif within the WEC RNA. (A) Schematic representation of WEC RNA structure and the eight small RNAs covering its sequence. (B) Mono-dimensional Northwestern assay on heLa cell nuclear proteins, using the eight small 32P-labeled RNas as probes. WEC RNA is used as a positive control. (C) Quantification of the RNA/hnRNP G interaction signals from (B). (D) Recognition of RNAB (WEC hairpin) by the major pI isoforms of hnRNP G on bi-dimensional Northwestern assay. NW, Northwestern assay; ID, immunodetection using an anti-hnRNP G antibody; pI, isoelectric point; kDa, kiloDalton. Isoelectric points are indicated on the top. Molecular masses are indicated to the left. (E) secondary structure of RNAB predicted by the ‘mfold web service’.Citation45,Citation46 VARNA applicationCitation47 was used to draw RNA. Arrows indicate the GGAAA pentaloop.
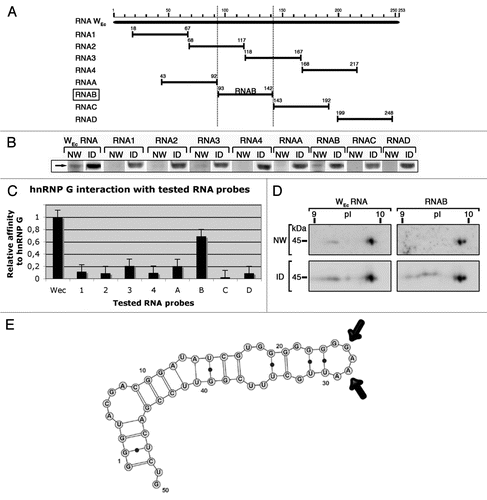
Figure 7 The C-ter region of ha-hnRNP G is required to interact with RNAB (WEC hairpin). (A) Schematic representation of the full-length protein and the deletion mutants used. HA, HA tag; NLS, SV40 nuclear localization signal, GST, Glutathione-s-transferase. (B) HA-tagged proteins were expressed in Xenopus oocytes by microinjection of their respective transcripts. Nuclear extracts were prepared and tested by Northwestern using a 32P-labeled RNAB as a probe. Newly expressed proteins were also detected by western blotting using the anti-HA antibody 16B12. Each lane corresponds to 10 nuclei. NW, Northwestern assay; ID, immunodetection. arrowheads indicate the positive RNA interaction signals with the newly made proteins. Note that a mutated hnRNP G that lacks its C-ter region does not have affinity for the RNA probe. (C) Proteins in fusion with GST were bacterially expressed and purified by affinity chromatography. Purified polypeptides were tested on Northwestern assay using a 32P-labeled RNAB as a probe. They were also detected by western blotting using an anti-GST antibody. PS, Ponceau S temporary protein staining; NW, Northwestern assay; ID, immunodetection. arrowhead indicates the positive RNA interaction signal shown by the polypeptide Fb (residues 334–391).
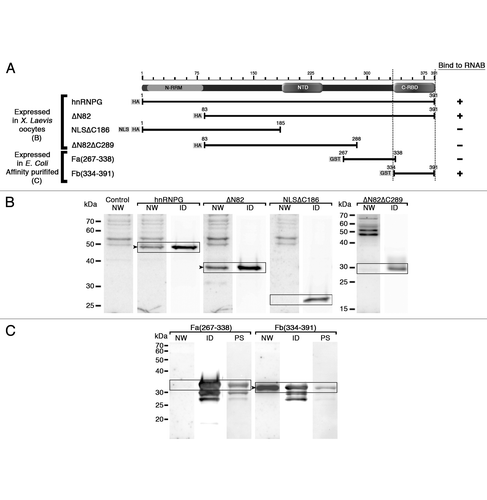
Figure 8 Sequence conservation of the NTD and C-ter RBD. Multiple sequence alignments of NTD domain (residues 186–236) (A and C) and C-ter RBD domain (residues 334–391) (B and D) of human hnRNP G with its orthologues from different species (A and B) and with other proteins members of RBMX/RBMY family (C and D). percent values of sequence similarities with the human domain are marked to the left. Accession numbers of hnRNP G sequences used in the alignment are: human, P38159; cow, XP_880704.2; dog, XP_852809; mouse, NP_035382; rat, NP_001020834; platypus (marsupial), XP_001510789; opossum (marsupial), XP_001367196; chicken, NP_001073196; zebra finch (bird), XP_002190045; Xenopus laevis, NM_001091512; zebrafish, NP_997763; Astatotilapia burtoni (fish), ABG02278; puffer fish, CAG09825. Xenopus tropicalis hnRNP G sequence was generated by blasting the X. tropicalis genome assembly 4.1 of the JGI's Xenopus Genome Sequencing project (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). Accession numbers of RBMX/RBMY protein family sequences used in the alignment are: RBMXL1, NP_062556; hnRNPG-T, NP_055284; RBMY, NP_005049. The clustal W programCitation48 was used to generate the alignments and the cLc sequence viewer 6 software (http://www.clcbio.com/index.php?id=28) to generate the graphical output.
Table 1 Schematic representation of the full-length human protein and the deletion mutants that were expressed in stage IV–V xenopus oocytes
Table 2 Stem structure capped by the pentaloop GGAAA is a potential RNA binding target for hnRNP G protein
Acknowledgements
We thank Natalya Novikova for expert technical assistance. We are very grateful to Dr. M. Soulard and Dr. L. Chabanne for the gift of the anti-hnRNP G sera, and Dr. R.G. Roeder for providing the anti-RPC15 and anti-RPC53 antibodies. We also thank Dr. V. Della Valle for providing the cDNA clones used for the production of the GST-tagged proteins. Finally, we would like to thank our guest Editor-in-Chief, Dr. T. Pederson and the anonymous reviewers for their very insightful and constructive critique of our manuscript. M.B. is supported by a CAREER award from the National Science Foundation.
References
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 2005; 17:251 - 256
- de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett 2008; 582:1971 - 1976
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 1993; 62:289 - 321
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 2002; 3:195 - 205
- Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol 2008; 28:6033 - 6043
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol 2007; 623:123 - 147
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582:1977 - 1986
- Nasim MT, Chernova TK, Chowdhury HM, Yue BG, Eperon IC. HnRNP G and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum Mol Genet 2003; 12:1337 - 1348
- Takemoto T, Nishio Y, Sekine O, Ikeuchi C, Nagai Y, Maeno Y, et al. RBMX is a novel hepatic transcriptional regulator of SREBP-1c gene response to high-fructose diet. FEBS Lett 2007; 581:218 - 222
- Zhao S, Korzan WJ, Chen CC, Fernald RD. Heterogeneous nuclear ribonucleoprotein A/B and G inhibits the transcription of gonadotropin-releasing-hormone 1. Mol Cell Neurosci 2008; 37:69 - 84
- Shin KH, Kim RH, Kang MK, Kim RH, Kim SG, Lim PK, et al. p53 promotes the fidelity of DNA end-joining activity by, in part, enhancing the expression of heterogeneous nuclear ribonucleoprotein G. DNA Repair (Amst) 2007; 6:830 - 840
- Shin KH, Kim RH, Kim RH, Kang MK, Park NH. hnRNP G elicits tumor-suppressive activity in part by upregulating the expression of Txnip. Biochem Biophys Res Commun 2008; 372:880 - 885
- Tsend-Ayush E, O'Sullivan LA, Grutzner FS, Onnebo SM, Lewis RS, Delbridge ML, et al. RBMX gene is essential for brain development in zebrafish. Dev Dyn 2005; 234:682 - 688
- Dichmann DS, Fletcher RB, Harland RM. Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev Dyn 2008; 237:1755 - 1766
- Delbridge ML, Lingenfelter PA, Disteche CM, Graves JA. The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome. Nat Genet 1999; 22:223 - 224
- Mazeyrat S, Saut N, Mattei MG, Mitchell MJ. RBMY evolved on the Y chromosome from a ubiquitously transcribed X–Y identical gene. Nat Genet 1999; 22:224 - 226
- Venables JP, Elliott DJ, Makarova OV, Makarov EM, Cooke HJ, Eperon IC. RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum Mol Genet 2000; 9:685 - 694
- Lingenfelter PA, Delbridge ML, Thomas S, Hoekstra HE, Mitchell MJ, Graves JA, et al. Expression and conservation of processed copies of the RBMX gene. Mamm Genome 2001; 12:538 - 545
- Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl 2004; 27:328 - 334
- Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, et al. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep 2007; 8:372 - 379
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem 2009; 284:14303 - 14315
- Soulard M, Della Valle V, Siomi MC, Pinol-Roma S, Codogno P, Bauvy C, et al. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res 1993; 21:4210 - 4217
- Morgan GT. Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res 2002; 10:177 - 200
- Austin C, Novikova N, Guacci V, Bellini M. Lampbrush chromosomes enable study of cohesin dynamics. Chromosome Res 2009; 17:165 - 184
- Soulard M, Della Valle V, Larsen CJ. Autoimmune antibodies to hnRNPG protein in dogs with systemic lupus erythematosus: epitope mapping of the antigen. J Autoimmun 2002; 18:221 - 229
- Beenders B, Jones PL, Bellini M. The tripartite motif of nuclear factor 7 is required for its association with transcriptional units. Mol Cell Biol 2007; 27:2615 - 2624
- Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol 2003; 4:975 - 980
- Murphy C, Wang Z, Roeder RG, Gall JG. RNA polymerase III in Cajal bodies and lampbrush chromosomes of the Xenopus oocyte nucleus. Mol Biol Cell 2002; 13:3466 - 3476
- Penrad-Mobayed M, Moreau N, Angelier N. Evidence for specific RNA/protein interactions in the differential segment of the W sex chromosome in the amphibian Pleurodeles waltl. Dev Growth Differ 1998; 40:147 - 156
- Penrad-Mobayed M, Sourrouille P, Bonnanfant-Jais ML, N'Da E, Edstrom JE, Angelier N. Microdissection and cloning of DNA from landmark loops of amphibian lampbrush chromosomes. Chromosoma 1991; 101:180 - 188
- Lin TY, Chan LC, Fan YH, Lin CH, Chow KC, Lin SL, et al. Use of a recombinant protein containing major epitopes of hnRNP G to detect anti-hnRNP G antibodies in dogs with systemic lupus erythematosus. Res Vet Sci 2006; 81:335 - 339
- Sallacz NB, Jantsch MF. Chromosomal storage of the RNA-editing enzyme ADAR1 in Xenopus oocytes. Mol Biol Cell 2005; 16:3377 - 3386
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 2000; 25:381 - 388
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136:777 - 793
- Soulard M, Barque JP, Della Valle V, Hernandez-Verdun D, Masson C, Danon F, et al. A novel 43-kDa glycoprotein is detected in the nucleus of mammalian cells by autoantibodies from dogs with autoimmune disorders. Exp Cell Res 1991; 193:59 - 71
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol 1995; 129:551 - 560
- Iijima M, Suzuki M, Tanabe A, Nishimura A, Yamada M. Two motifs essential for nuclear import of the hnRNP A1 nucleocytoplasmic shuttling sequence M9 core. FEBS Lett 2006; 580:1365 - 1370
- Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B” and their cognate RNAs. Nature 1990; 345:502 - 506
- Hung CJ, Lee YJ, Chen DH, Li C. Proteomic analysis of methylarginine-containing proteins in HeLa cells by two-dimensional gel electrophoresis and immunoblotting with a methylarginine-specific antibody. Protein J 2009; 28:139 - 147
- Ginisty H, Amalric F, Bouvet P. Two different combinations of RNA-binding domains determine the RNA binding specificity of nucleolin. J Biol Chem 2001; 276:14338 - 14343
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 2005; 309:2054 - 2057
- Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet 2002; 11:2037 - 2049
- Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes 3. Optimum incubation conditions. J Exp Zool 1973; 184:321 - 333
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell 1998; 9:2987 - 3001
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 1999; 288:911 - 940
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406 - 3415
- Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009; 25:1974 - 1975
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947 - 2948