Abstract
The mammalian interphase chromatin responds to DNA damages by altering the compactness of its architecture, thereby permitting local access of DNA repair machineries. Adding to the cellular strategies of chromatin remodeling following DNA damage, our recent work identified the 53BP1-EXPAND1 module in promoting chromatin dynamics in response to DNA double-strand breaks. Endowed with a nucleosome-binding PWWP domain, EXPAND1 tethers to the chromatin where it is involved in maintaining basal chromatin accessibility in unperturbed cells. Interestingly, through its direct interaction with the DNA damage mediator protein 53BP1, EXPAND1 accumulates at the damage-modified chromatin and triggers its further decondensation. These observations, together with the fact that EXPAND1 promotes cell survival following DNA damage, suggest that the chromatin-bound factor may facilitate DNA repair by regulating the organization of chromatin structure.
The mammalian chromatin, viewed as a static entity in the past, has revealed itself as a highly mobile structure that plays an active role in numerous fundamental processes, including DNA replication, transcription and DNA repair.Citation1 The dynamics of chromatin implicates sophisticated regulatory mechanisms that enable these processes to take place with temporal and spatial specificities. Indeed, recent research has uncovered numerous strategies via which the cell has evolved to coordinate DNA transaction events that operate on this architecture. These include chromatin bound proteins that are intrinsic to chromatin architecture and enzymatic activities that read and respond to cues to effect chromatin changes.
From the perspective of executing a DNA repair reaction, it is conceivable that DNA repair machinery must be able to locate perhaps as little as a single DNA break amongst the 6 billion base pairs of DNA in a human nucleus. Not only does this require exquisite signaling pathways to propagate the initial DNA damage signal, but the chromatin structure surrounding the DNA break must be altered such that the site of DNA damage becomes accessible to DNA repair enzymes.Citation2 Indeed, recent work has elucidated how a number of chromatin remodeling complexes are specifically recruited to a DNA break.Citation3,Citation4 In addition, a number of chromatin-decorating proteins were also found to respond to DNA damage, and that their damage-induced mobilization, which results in chromatin changes, have been associated with optimal DNA damage responses.Citation5,Citation6
More recently, the DNA damage mediator protein 53BP1 has been implicated, via promoting chromatin dynamics, to facilitate non-homologous end joining DNA repair.Citation7,Citation8 With the aim to better understand how 53BP1 may regulate chromatin dynamics, we undertook a tandem affinity purification (TAP) approach to identify proteins that interact with 53BP1.Citation9 We first tagged 53BP1 with our TAP tag (streptavidin binding peptide—S-protein tag), and overexpressed the fusion protein in 293T cells. Following two rounds of purification, we isolated and identified a few previously reported 53BP1-interacting proteins, including TOPBP1 and RIF1.Citation10,Citation11 In addition, EXPAND1/MUM1, an uncharacterized protein with a PWWP domain,Citation12 also copurified with the 53BP1 protein-complex. The PWWP domain is a member of the Tudor domain Royal Family, which are present in many chromatin-associated proteins. 13 The identification of EXPAND1 as a possible regulator of 53BP1 chromatin function, therefore, prompted us to further characterise EXPAND1 in more detail.
Identification of EXPAND1 as a DNA Damage Response Protein
If EXPAND1 interacts with 53BP1 to promote its function in DNA damage responses, one would expect that EXPAND1 might similarly localise to DNA damage sites. Indeed, like many other 53BP1-interacting proteins, EXPAND1 rapidly accumulated into microscopically visible foci in response to ionising radiation (IR), which perfectly overlapped with those of 53BP1 and the DNA damage marker γH2AX.Citation14 The observation that EXPAND1 concentrates at DNA breaks immediately raises the possibility that its recruitment may be coordinated by the defined DNA damage signaling pathway involving histone H2AX. DNA damage triggers the ATM/ATR-dependent phosphorylation of the histone variant H2AX (γH2AX), which constitutes one of the earliest events that initiates a signaling cascade to promote productive assembly of DNA damage checkpoint and repair proteins to the vicinity of DNA breaks.Citation15 Using a panel of human and mouse cells that are deficient in components of this signaling pathway, we ectopically expressed EXPAND1 and determined its subcellular localization after DNA damage. Interestingly, we uncovered genetic requirements not only for the H2AX-MDC1-RNF8 pathway,Citation16 but in the absence of 53BP1, EXPAND1 proteins also failed to concentrate to damage-induced foci. Conversely, EXPAND1 depletion did not noticeably affect 53BP1 foci formation. In addition, we found that EXPAND1 relocalisation to DNA breaks requires its 53BP1-binding domain, strongly supporting that the 53BP1-EXPAND1 interaction is responsible for loading EXPAND1 to the damage-modified chromatin.
Functional Roles of EXPAND1 at DNA Damage Sites
Having established a hierarchical relationship between 53BP1 and EXPAND1, we were interested in looking at how EXPAND1 may regulate 53BP1-dependent DNA damage responses. Although we were able to document a requirement of EXPAND1 in protecting cells from IR, we did not detect significant change in EXPAND1-depleted cells in parameters associated with checkpoint activation, suggesting that EXPAND1 may only regulate a subset of 53BP1 functions.Citation14 Since 53BP1 is also required for DNA repair, we next tested whether EXPAND1 may facilitate repair of DNA double-strand breaks (DSBs) by following the status of γH2AX in cells treated with a recoverable dose of IR. Indeed, we found that EXPAND1 was required for timely repair of IR-induced DNA breaks, and that this EXPAND1 attribute required its interaction with 53BP1. So how exactly does EXPAND1 promote DNA repair?
The EXPAND1 PWWP Link
The PWWP domain on the EXPAND1 polypeptide offered us the only hint to further dissect its function. Knowing that PWWP domains bind to histones and DNA, and that PWWP domain-containing proteins often associate with chromatin, we examined whether the EXPAND1 PWWP might be endowed with similar properties. Using a series of biochemical assays, we found that EXPAND1 PWWP interacted with nucleosomes in vitro, and was required for EXPAND1 chromatin association in vivo, suggesting that EXPAND1 promotes DNA damage repair in part through its chromatin-binding ability. In line with this idea, the EXPAND1 PWWP was indispensable for DNA repair as well as clonogenic survival upon IR treatment.
Previous work suggested that 53BP1, while stabilizing DNA ends, promotes NHEJ by enhancing chromatin dynamics.Citation7,Citation8 With this in mind, we employed the micrococcal nuclease sensitivity assay, which allows detection of damage-induced chromatin changes, to monitor chromatin status in the absence of EXPAND1. Strikingly, not only was EXPAND1 itself pivotal for damage-induced chromatin decondensation, but there was a strict requirement for its PWWP domain. Collectively, this data led us to propose that EXPAND1 may facilitate DNA repair by regulating chromatin decondensation.
From being a mere template of genetic information, the interphase chromatin is now being recognised as a dynamic platform that actively engages in preserving its integrity. As such, the cell utilises a series of protective protocols that, in response to DNA damage, coordinate chromatin changes with DNA repair. Significantly, these mechanisms not only restore the physical integrity of the damaged DNA, but also allow epigenetic marks to be reinstated. Our identification of the 53BP1-EXPAND1 module in regulating the early events on the damaged chromatin highlights the precise coordination of DNA damage signaling and chromatin changes.
On the other hand, it remains enigmatic why the cell has evolved so many different ways to influence chromatin reorganization. Are each of these pathways devoted for repair of a sub-class of DNA lesions? Or are they integral to enforcing to the fullest extent of chromatin change in response to DNA Damage?
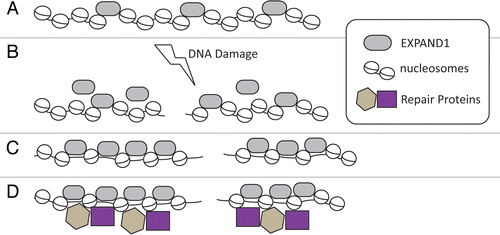
Likewise, although EXPAND1 is required for chromatin decondensation, as measured by the use of partial microccocal nuclease digestion assay, we do not know whether this effect is limited to chromatin domains surrounding DSBs or if this is a global effect. Although purely speculative, it is possible that EXPAND1 regulates nucleosome spacing, and thus promote basal access of transaction activities to chromatin. By the same token, we envision that the 53BP1-dependent concentration of EXPAND1 to the vicinity of DSBs might further open up chromatin domains flanking DSBs by spacing out local nucleosomal structures (). Whether these 53bp1-dependent events involve redistribution of chromatin-bound EXPAND1 or recruitment of a soluble fraction of EXPAND1 will require further studies. Nevertheless, given that EXPAND1 inactivation also triggered chromatin condensation in unperturbed cells, it seems likely that EXPAND1, as a chromatin-associated factor, may also regulate other cellular processes that require access to chromatin.
Finally, as PWWP domains were recently shown to specifically interact with methylated histones,Citation17,Citation18 it is tempting to speculate that EXPAND1, via its PWWP domain, may also preferentially associate with certain methylated histones in vivo. Given that histone methylation contributes to the epigenetic landscape and is intimately involved in numerous cellular processes, it will be of significant interest to identify this mark, which will offer further routes via which to dissect EXPAND1 functions, and to appreciate a fuller and a more comprehensive picture of the mammalian DNA damage response.
Figures and Tables
Figure 1 Proposed model of EXPAND1 in chromatin structure regulation. (A) EXPAND1 associates with chromatin through its ability to interact with nucleosomes. (B) DNA damage triggers the accumulation of EXPAND1 to sites of DNA breaks. (C) Binding of EXPAND1 to chromatin domains surrounding DNA break site opens up its structure. (D) Efficient loading of DNA repair proteins to the damage-modified chromatin. For simplicity, 53BP1 is omitted from illustration (see text).
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (CA089239, CA092312 and CA100109 to J.C.) and from Seed Funding for Basic Research (Project Code: 200908159008; HKU to M.S.Y.H.). M.S.Y.H. would like to thank J.C. for his continuous support. J.C. is a recipient of an Era of Hope Scholar award from the Department of Defence and a member of the Mayo Clinic Breast SPORE program (P50 CA116201).
References
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell 2007; 128:721 - 733
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009; 10:243 - 254
- Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev 2007; 17:126 - 131
- Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, et al. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 2009; 11:865 - 872
- Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, et al. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol 2009; 11:92 - 96
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 2008; 453:682 - 686
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 2008; 456:524 - 528
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 2008; 456:529 - 533
- Huen MS, Huang J, Leung JW, Sy SM, Leung KM, Ching YP, et al. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell 2010; 37:854 - 864
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol 2002; 22:555 - 566
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 2004; 18:2108 - 2119
- Coulie PG, Lehmann F, Lethe B, Herman J, Lurquin C, Andrawiss M, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA 1995; 92:7976 - 7980
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation?. FEBS Lett 2000; 473:1 - 5
- Huen MS, Huang J, Leung JW, Sy SM, Leung KM, Ching YP, et al. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell 2010; 37:854 - 864
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858 - 5868
- Huen MS, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 2010; 35:101 - 108
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol 2010; 17:617 - 619
- Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR 3rd, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell 2009; 33:428 - 437