Abstract
Inner nuclear membrane (INM) proteins can be important for positioning chromosomes within the nucleus. Little is known about INM proteins in the fission yeast Schizossacharomayces pombe. Telomeres are the most obvious chromosomal sites that are anchored to the nuclear envelope in this organism. A group of proteins that tether telomeres to the spindle-pole body (SPB) during meiotic prophase, such as Bqt1, Bqt2, and Sad1, has been identified previously, but proteins for anchoring telomeres to the nuclear envelope in vegetative cells have not been identified until recently. A recent report demonstrates that Bqt3 and Bqt4 are INM proteins that affect nuclear positioning of telomeres in vegetative cells, and consequently affect the telomere clustering in meiotic prophase. Interestingly, in the absence of Bqt4, telomeres are separated from the nuclear envelope but telomere silencing and telomere length are properly regulated. An important implication of these results is that the functional integrity of telomeres is maintained independently of their connection to the nuclear envelope.
Bouquet Formation of Chromosomes in Meiotic Prophase
Telomeres are dispersed along the nuclear envelope during the mitotic cell cycle. However, during a period of meiotic prophase, they form a cluster adjacent to the nuclear envelope to produce a characteristic bundled configuration of chromosomes, which is called the “bouquet” arrangement (reviewed in refs. Citation1–Citation4). Bouquet formation has been observed in a wide range of organisms and is important for pairing and recombination of homologous chromosomes, which in turn are essential for correct segregation of chromosomes during meiotic divisions. A striking example of chromosomal bouquet formation has been demonstrated in the fission yeast Schizosaccharomyces pombe. In this organism, the nucleus elongates and oscillates between the cell poles during meiotic prophase (appearing as what is known as the “horsetail” nucleus) and telomeres remain clustered to the SPB located at the leading edge of the moving nucleus.Citation4–Citation7
The SUN-KASH Protein Complex Links Chromosomes to the Cytoskeleton
Studies in S. pombe have provided the first clear demonstration that the conserved SUN-KASH protein complex mediates the intranuclear movements of telomeres gathering to the SPB.Citation4,Citation8,Citation9 In S. pombe, Sad1 and Kms1 are members of the SUN domain and KASH domain protein families, respectively (reviewed in ref. Citation9); Sad1 resides in the INM and Kms1 in the outer nuclear membrane (ONM) (). Sad1 and Kms1 are exclusively localized to the SPB in vegetative cells:Citation10,Citation11 during telomere clustering, Sad1-Kms1 complexes form several foci on the nuclear envelope where Bqt1 and Bqt2 connect telomeres to the nuclear envelope by linking Rap1 and Sad1 on the nucleoplasmic side. On the cytoplasmic side, Kms1 interacts with the dynein motor complex to tether multiple foci of telomeres to the SPB ().Citation4,Citation12,Citation13 In the kms1 mutant, Sad1 foci remain dispersed and telomeres fail to form a cluster at the SPB.Citation14
Bqt1 and Bqt2 are Meiosis-specific Telomere Binding Proteins
We previously identified meiosis-specific proteins, Bqt1 and Bqt2, that are required for telomere clustering during meiotic prophase.Citation8 Bqt1 and Bqt2 are not expressed in vegetative cells, but are expressed in the mating pheromone response during meiosis. Bqt1 and Bqt2 bind to the telomere through interaction with the telomere protein Rap1 (). Interestingly, Bqt1 or Bqt2 alone does not bind to Rap1, but together they form a complex which is able to bind to Rap1.
Bqt1 also binds to the INM protein Sad1, providing a link between telomeres and the nuclear envelope (). Although Sad1 is exclusively localized at the SPB, it is potentially an INM protein. In fact, when Sad1 is overexpressed it is localized to the nuclear envelope beside the SPB.Citation8,Citation10 Experiments monitoring fluorescence recovery after photobleaching show that Sad1 diffuses to the nuclear envelope in a few minutes (Yuji Chikashige, unpublished). These observations show that Sad1 shuttles between the SPB and the nuclear envelope, but apparently it remains at the SPB for most of the time due to the presence of robust interacting partners which are found exclusively at the SPB. On the other hand, Bqt1 accumulates at telomeres in meiotic prophase and can provide Sad1 with additional attachment sites on the nuclear envelope where telomeres locate. Indeed, multiple Sad1 foci can be seen on the nuclear envelope in addition to the SPB during the process of telomere clustering in meiotic prophase.Citation4,Citation8
It should be pointed out, however, that localization of Sad1 is restricted to the nuclear envelope by its transmembrane (TM) sequence, and thus telomeres must locate close to the nuclear envelope in order to interact with Sad1. As telomeres tend to reside near the nuclear envelope in vegetative cells, there might be proteins that anchor telomeres prior to expression of Bqt1 and Bqt2, but such proteins have not been found in S. pombe. We hypothesized that if telomeres were not anchored to the nuclear envelope when Bqt1 and Bqt2 appear, bouquet formation would be subsequently affected as telomeres would fail to encounter Sad1. Thus, we revisited the library of gene disruptions we used to identify Bqt1,Citation8 and identified new INM proteins Bqt3 and Bqt4 in S. pombe.Citation15
Bqt4 Anchors Telomeres to the Nuclear Envelope in Vegetative Cells
While loss of Bqt1 or Bqt2 resulted in complete loss of telomere clustering, we picked up one strain that showed partial loss of telomere clustering in meiotic prophase, and named the protein responsible Bqt3. However, Bqt3 did not show a direct interaction with Rap1. Thus, we searched for other proteins that interact with Bqt3 by yeast two-hybrid screening. Of the resulting candidate proteins, we found one protein that appears commonly in our collection of Rap1 interacting proteins (Yuji Chikashige, unpublished) and confirmed that it binds to both Bqt3 and Rap1. Loss of this protein also resulted in partial loss of meiotic telomere clustering, and thus we named it Bqt4.
Fluorescence microscopy studies have shown that Bqt3 and Bqt4 are localized uniformly at the nuclear envelope in vegetative cells as well as in meiotic cells. Electron microscopy experiments further determined their localization to the INM. Because Bqt4 is an INM protein that directly interacts with the Rap1 telomere protein, we concluded that Bqt4 anchors telomeres to the nuclear envelope. Although Bqt3 and Bqt4 were found as proteins required for telomere clustering in meiotic prophase, it should be emphasized that these proteins are necessary for anchoring telomeres to the nuclear envelope in vegetative cells and not only in meiotic cells.Citation15 These proteins provide an attachment surface for telomeres along the nucleoplasmic surface of the nuclear envelope ().
Bqt3 Protects Bqt4 from Protein Degradation
We have shown that Bqt4 is degraded in the absence of Bqt3 and this degradation requires localization of Bqt4 at the nuclear envelope.Citation15 Bqt4 lacking its TM sequence localizes to the nucleoplasm and escapes protein degradation. Thus, we speculate that a putative protease is bound to the nuclear envelope. Alternatively, the TM sequence could be a target of a putative protease. When we replaced the TM of Bqt4 with other TM sequences, these chimeric Bqt4 proteins were localized at the nuclear envelope and partially complemented a defect in telomere clustering in bqtΔ cells, but escaped protein degradation in the absence of Bqt3 (). In contrast to the native Bqt4, which disappears from the nuclear envelope in the absence of Bqt3, the chimeric Bqt4 variants retained their localization at the nuclear envelope as shown in . The TM of Bqt4 is necessary for its interaction with Bqt3, and these chimeric Bqt4 with foreign TMs did not interact with Bqt3 in a yeast two-hybrid assay (data not shown). Thus, the most likely explanation of these results is that the TM of Bqt4 is a target of a putative protease, and it is masked by its interaction with Bqt3.
Roles for the putative protease are unknown. One possible role is to regulate the amount of Bqt4 as part of protein quality control: excess Bqt4, which is unbound to Bqt3, might be degraded by this putative protease. To identify this putative protease, we are currently searching, by genetic screening and cytological screening, for mutants in which Bqt4 is stable in the absence of Bqt3.
Telomere Functions in bqt Mutants
Whereas Bqt1 and Bqt2 are expressed specifically upon mating pheromone signaling during meiotic prophase, Bqt3 and Bqt4 are also expressed in vegetative cells. Bqt3 is upregulated by mating pheromone signaling and Bqt4 by nitrogen starvation. Deletion of any bqt genes does not cause defects in mitotic growth. In sharp contrast with the conserved SUN-KASH proteins and telomere proteins, no obvious homologues of Bqt1, Bqt2 and Bqt3 have been found in other organisms except for another fission yeast Schizosaccharomyces japonicus (; for Bqt1 and Bqt2, reviewed in ref. Citation4). On the other hand, homologues of Bqt4 can be seen but only in some other fungi (). Bqt4 may play a similar role in these fungi.
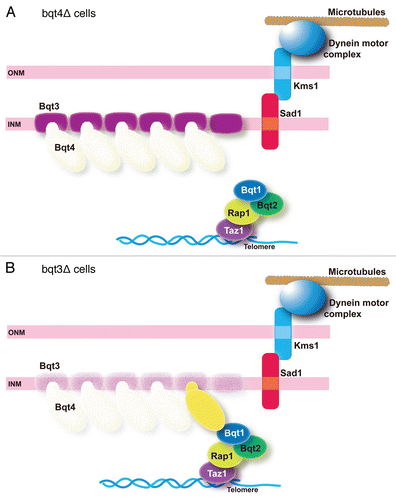
The level of defects in telomere clustering varies between the bqt mutants. In the absence of Bqt1 or Bqt2, telomere clustering is completely lost.Citation8 This is because these proteins have a direct role in connecting telomeres to the SUN-KASH protein complex, which transmits the cytoskeletal motive force (reviewed in refs. Citation4 and Citation9). Thus, telomeres have no chance of contacting Sad1 in the absence of Bqt1 or Bqt2 (as in ). On the other hand, in the absence of Bqt4, telomeres are not physically connected to the nuclear envelope but can happen to reside close to the nuclear envelope (). In the absence of Bqt3, Bqt4 is degraded generating phenotypes similar to those exhibited by mutants that lack Bqt4, but residual amounts of Bqt4 may result in even weaker phenotypes (). In those cells, Bqt1 and Bqt2 are expressed and accumulate to telomeres, thus providing telomeres with some chance of contacting Sad1. This explains why telomere clustering is only partially lost.Citation15
In vegetative cells, telomeres lose their attachment to the nuclear envelope in the absence of Bqt4. It should be pointed out that telomere silencing is maintained and telomere length is properly regulated in those cells.Citation15 This is likely because molecular components crucial for telomere functions are present at telomeres in the absence of Bqt4. Thus, the bqt4Δ mutation provides a unique opportunity for separating the effects of the nuclear positioning of telomeres from their functions. Many other telomere protein mutations, such as Taz1 and Rap1, affect both telomere position and function,Citation16–Citation20 making it difficult to separate these issues. An important implication of these results is that the functional integrity of telomeres is maintained even when they are separated from the nuclear envelope.
Figures and Tables
Figure 1 A group of bouquet proteins. (A) Bqt3 and Bqt4 form an attachment surface for telomeres on the nuclear envelope in vegetative cells. (B) Bqt1 and Bqt2 link telomeres to the SUN-KASH protein complex in meiotic prophase.
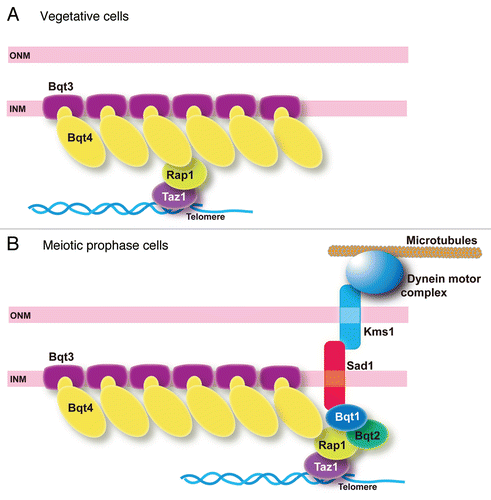
Figure 2 Localization of Bqt4 variants bearing chimeric TM regions. (A) Replacement of the TM sequence of Bqt4 by other TM sequences. The TM of Bqt4 (indicated by the filled box) was replaced by other TM sequences of Sad1 or Lem2 (SPAC18G6.10). Sad1 and Lem2 are INM proteins of S. pombe. Membrane geometry of these proteins is shown in this diagram. The TM of Sad1 and the first TM of Lem2 (indicated by the hatched box) were used to construct the chimeric Bqt4 variants. These TM sequences orient their N-terminus to the nucleoplasm as the TM of Bqt4 does. To construct the chimeric Bqt4 proteins, we replaced the TM of Bqt4 (the C-terminal 19 amino acid residues) with the following sequences: LL WFG IT L FST LLI IT L L (TM of Sad1; 171–188 aa) in Bqt4-Sad1TM, SSY LVH IFM ILL GVV AAI FLA LL (the first TM of Lem2; 316–338 aa) in Bqt4-Lem2TM. (B) Telomere clustering in bqt4Δ cells expressing the GFP-fused Bqt4 variant proteins. Percentages of the nuclei bearing a single cluster of telomeres are shown for each of the variants as indicated. Numbers of cells examined: 124 (GFP-Bqt4-wt), 56 (GFP-Bqt4-dTM), 64 and 85 (GFP-Bqt4-Sad1TM in bqt3+ and bqt3Δ, respectively), 49 and 101 (GFP-Bqt4-Lem2TM in bqt3+ and bqt3Δ respectively). (C) Localization of Bqt4 and its variants bearing chimeric TM fused with GFP in vegetatively growing cells. The bar indicates 10 µm. To express GFP-fused Bqt4 variants, the bqt4 promoter region (1,000 base pair region upstream the coding sequence plus the initial ATG), the sequence encoding GFP-S65T and the coding sequence of the bqt4+ gene, or its variant constructs, followed by the nmt1 terminator sequence were ligated in-frame into an integration vector pYC36;Citation21 the resulting plasmid was integrated into the chromosome at the lys1 gene locus in bqt4Δ cells.
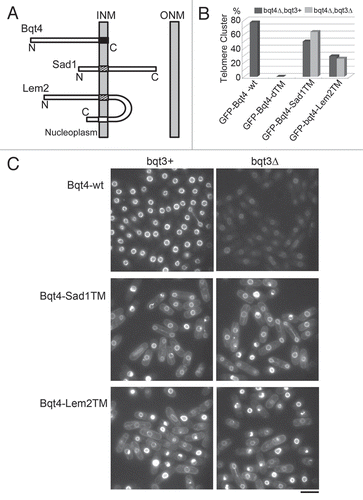
Figure 3 Homologues of Bqt3 and Bqt4 in fungi. Comparison of amino acid sequences of Bqt3 and Bqt4. Identical residues are dark shaded with white letters and conserved residues are light shaded. Species: Sp, Schizossacharomyces pombe; Sj, Schizossacharomyces japonicus; Nc, Neurospora crassa; An, Aspergillus nidulans. Gene ID: Sj bqt3 (SJAG_05226), Sj bqt4 (SJAG_01263), Nc bqt4 (NCU06560) and An bqt4 (AN0162.2).

Figure 4 Phenotypes of bouquet mutants. (A) In the absence of Bqt4 (transparent), telomeres are not anchored to the nuclear envelope. However, telomeres may have some chance of contact with Sad1 when telomeres locate near the nuclear envelope through interaction between Bqt1 and Sad1. (B) In the absence of Bqt3 (transparent), degradation of Bqt4 results in basically the same situation as in (A), but residual Bqt4 (yellow) may provide a chance for telomeres to contact with Sad1.
Acknowledgements
We thank Tomoko Kojidani, Kasumi Okamasa, Chihiro Tsutsumi and Miho Yamane for technical assistance. This work was supported by grants from the Japanese Ministry MEXT (Y.C., T.H. and Y.H.).
Extra View on: Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, et al. Membrane proteins Bqt3 and −4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol 2009; 187:413 - 427
References
- Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol 2001; 2:621 - 627
- Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci 2004; 117:4025 - 4032
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma 2006; 115:158 - 174
- Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma 2007; 116:497 - 505
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, et al. Telomere-led premeiotic chromosome movement in fission yeast Schizosaccharomyces pombe. Science 1994; 264:270 - 273
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci 1998; 111:701 - 712
- Hiraoka Y. Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells 1998; 3:405 - 413
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006; 125:59 - 69
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell 2009; 17:598 - 605
- Hagan I, Yanagida M. The product of the spindle formation gene sadl+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 1995; 129:1033 - 1047
- Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J 2000; 19:3831 - 3840
- Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O. The 14 kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell 2002; 13:930 - 946
- Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics 2004; 270:449 - 461
- Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, et al. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet 1997; 254:238 - 249
- Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, et al. Membrane proteins Bqt3 and −4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol 2009; 187:413 - 427
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 1997; 385:744 - 747
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 1998; 392:825 - 828
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 1998; 392:828 - 831
- Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 2001; 11:1618 - 1623
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 2001; 11:1624 - 1630
- Chikashige Y, Kurokawa R, Haraguchi T, Hiraoka Y. Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells 2004; 9:671 - 684