Abstract
Proteins of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family are activated by various cellular stresses, including DNA damage, premature termination codon and nutritional status, and induce appropriate cellular responses. The importance of PIKK functions in the maintenance of genome integrity, accurate gene expression and the proper control of cell growth/proliferation is established. Recently, ATPase associated diverse cellular activities (AAA+) proteins RUVBL1 and RUVBL2 (RUVBL1/2) have been shown to be common regulators of PIKKs. The RUVBL1/2 complex regulates PIKK-mediated stress responses through physical interactions with PIKKs and by controlling PIKK mRNA levels. In this review, the functions of PIKKs in stress responses are outlined and the physiological significance of the integrated regulation of PIKKs by the RUVBL1/2 complex is presented. We also discuss a putative “PIKK regulatory chaperone complex” including other PIKK regulators, Hsp90 and the Tel2 complex.
Introduction
Genome maintenance and precise gene expression are critically important issues for all organisms. Cells have evolved defense systems from gene mutations, errors in gene expression and various environmental stresses. Phosphatidylinositol 3-kinase-related protein kinase (PIKK) family proteins engage with these defense systems at each level of gene expression. Six PIKKs, ATM (ataxia telangiectasia mutated), ATR (ATM- and Rad3-related), DNA-PKcs (DNA-dependent protein kinase catalytic subunit), SMG-1 (suppressor with morphogenetic effect on genitalia-1), TOR (target of rapamycin) and TRRAP (transformation/transcription domain associated protein), have been identified in vertebrates. All PIKKs, except for TRRAP, function as protein kinases and transduce cellular stresses as phosphorylation signals to downstream effectors and induce proper stress responses. In addition to the importance of each PIKK function, recent studies have suggested an interplay among PIKKs. In this review, we provide an overview of the functions of PIKKs and present recent findings of common regulators of PIKKs. We also discuss a possible role of common regulators of PIKKs in the coordination of PIKKs in cellular stress responses.
PIKK-Mediated Defense Systems Against Various Cellular Stresses
PIKK family
The phosphatidylinositol 3-kinase-related protein kinase (PIKK) family is known as an atypical Ser/Thr protein kinase family that has sequence homology to the catalytic domain of lipid PI3-kinases.Citation1 These kinases are characterized as large proteins (270−470 kDa) with shared domain structures: a highly conserved catalytic domain, FAT-C (FRAP, ATM and TRRAP C-terminal) and successive α-helical repeats in the N-terminal region that provides protein-protein interaction surfaces (). Among the six PIKKs reported, ATM, ATR, TRRAP and TOR are evolutionally conserved from Saccharomyces cerevisiae to Homo sapiens, whereas DNA-PKcs and SMG-1 appeared during metazoan evolution. ATM, ATR, DNA-PKcs and SMG-1 preferentially phosphorylate Ser or Thr followed by Glu; therefore, these proteins are called S/TQ directed kinases.Citation2 Every PIKK forms a protein complex with specific binding partners that can regulate the recruitment of PIKK to the activation site, substrate binding and kinase activity.Citation3 The PIKK complexes play central roles in cellular responses to various stresses, including DNA damage, aberrant mRNAs and nutrient availability ().
Figure 1. The domain structures of human PIKK family members. PIKKs share the highly conserved catalytic PIKK domain and the FAT-C (FRAP, ATM, and TRRAP C-terminal) domain. Although the PIKK domain has sequence homology to the catalytic domain of PI3-kinases, PIKKs act as Ser/Thr protein kinases except for TRRAP. The FAT-C domain located near the PIKK domain is thought to modulate the kinase activity. The N-terminal region of PIKK is composed of α-helical repeats, which contribute to protein-protein interactions.
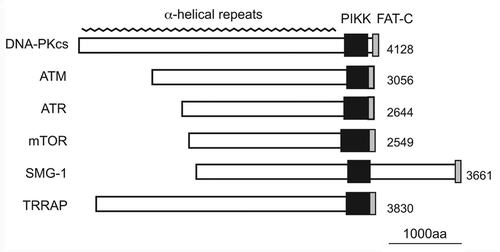
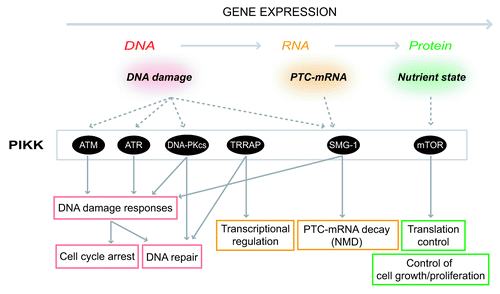
Figure 2. Summary of PIKK-mediated stress responses. PIKKs are activated various cellular stresses and induce proper cellular responses at various steps of gene expression. ATM and ATR are activated by DNA damages including DSBs to arrest cell cycle and activate DNA repair pathways. DNA-PKcs engages in a DSB repair process called NHEJ. TRRAP regulates transcription as a HAT complex component. SMG-1 recognizes PTC-mRNAs and leads to PTC-mRNA degradation. mTOR controls cellular translation activity and cell growth in response to nutrient status. Except for TRRAP, each PIKK induces proper stress responses through phosphorylations of downstream effector proteins.
ATM (reviewed in refs. Citation4 and Citation5)
ATM functions in responses to DNA double-strand breaks (DSBs), which are formed by ionizing radiation (IR) and DNA damaging agents. When DSBs appear, ATM is recruited to the adjacent region of the DSBs and is partially activated by autophosphorylation that transforms an inactive dimer to active monomers. In this early stage, ATM phosphorylates substrates including histone H2AX and p53. Phosphorylated histone H2AX becomes an initial signal for DNA damage and recruits DNA damage recognition/repair factors. Phosphorylated p53 induces the G1 checkpoint. Monomer ATM is recruited to DSBs by the Mre11-Rad50-Nbs1 (MRN) complex and is fully activated. Active ATM phosphorylates diverse downstream effectors and DNA break associated proteins, including Chk2, Nbs1, MDC1, BRCA1 and induces cell-cycle checkpoint, DNA repair and stress-induced transcription.
Besides DNA damage responses, ATM is involved in vesicle transport in the cytoplasm. For example, ATM associates with β-adaptin, one of the components of the clathrin-mediated endocytosis adaptor complex.Citation6 Cytoplasmic vesicular localization of ATM, including peroxisome, is also observed and ATM deficient cells show increased lysosomal accumulation and reduced oxidative metabolism.Citation7,Citation8 The cytoplasmic localization of ATM is especially appreciable in neural cells and ATM forms a complex with VAMP2 and Synapsin-I, two synaptic vesicle proteins, and modulates synaptic function through the regulation of the synaptic vesicular release cooperatively with ATR.Citation9 ATM also participates in insulin signaling by phosphorylating 4EBP1, a cap dependent negative translation regulator, collaborating with mTOR.Citation10
Mutations of the ATM gene are responsible for ataxia telangiectasia (A-T), an autosomal recessive disorder characterized by cerebellar ataxia telangiectasia, immunodeficiency, radiosensitivity and cancer susceptibility.Citation11,Citation12 ATM deficient mice show growth retardation, immunedefects, infertility, neurological defects and the majority of the mice develop thymic lymphomas.Citation7,Citation13 ATM depletion also impairs stem cell maintenance and causes aged phenotypes.Citation14,Citation15
ATR (reviewed in ref. Citation16)
ATR was originally discovered as a gene with sequence homology to ATM and is biochemically similar to ATM. In contrast to ATM, ATR is activated by a stalled replication fork during S phase and many types of DNA damage that give rise to single strand DNA (ssDNA) structures, including DSBs, base-crosslinks and agents, which cause DNA replication stress and DNA damage. ATR is recruited to the ssDNA coated with replication protein A (RPA) through the interaction with ATRIP. RPA also localizes the RAD9-RAD1-Hus1 (9–1-1) complex to the RPA-ssDNA sites. The 9–1-1 complex recruits TopBP1, an ATR activator, to ATR and induces ATR activation. Although ATR phosphorylates numerous substrates and regulates DNA replication, the cell cycle checkpoint and DNA repair, the best studied ATR substrate is Chk1. Activated Chk1 phospho-inactivates Ccd25 proteins, Cdk activators, thereby preventing the cell cycle transition. ATR-mediated Chk1 signaling is also critical for regulating DNA replication. ATR also phosphorylates replication related proteins, including PCNA, Polε, RPA, MCM proteins and DNA repair related proteins, including BRCA1, WRN and BLM. However, the physiological significance of these phosphorylation events is poorly understood. The kinase activity of ATR is also involved in replication-dependent histone mRNA degradation together with Upf1, a NMD transacting factor.Citation17
As expected in the critical regulation of replication stress, ATR is essential for viability across many organisms ranging from yeast to mammals.Citation18,Citation19 Moreover, deletion of the ATR gene in adult mice causes aged phenotypes and stem cell loss, in a similar manner to the ATM gene deletion.Citation20 Mutations of the ATR gene have been found in a few patients of the Seckel syndrome, an autosomal recessive disorder characterized by intrauterine growth retardation, microcephaly and mental retardation.Citation21
DNA-PKcs (reviewed in refs. Citation22 and Citation23)
DNA-PKcs is the catalytic subunit of the DNA-PK holoenzyme, which is composed of DNA-PKcs and the Ku70/80 heterodimer. DNA-PK (the DNA-PKcs and Ku70/80 complex) and its kinase activity are essential for non-homologous end-joining (NHEJ), a major DSB repair pathway in mammals. In NHEJ, Ku70/80 recognizes DNA ends and recruits DNA-PKcs to DSBs; thereby two DNA-PK molecules interact to connect the DNA ends. This interaction leads to the autophosphorylation of DNA-PK in trans, which induces conformational changes of DNA-PKcs and the release of the DNA-PKcs from the DNA ends, allowing the NHEJ- and repair factors to access the DSB and subsequent end-processing.Citation23 Besides autophoshorylation, a number of DNA-PK substrates including NHEJ factors have been identified in vitro. However, the physiological roles of these phosphorylation events in vivo have not been well defined. DNA-PK-mediated DNA end-processing is also required for the rejoining of DSBs generated by V(D)J recombination during T- and B-cell development, and a DNA-PKcs inactive mutation causes severe combined immunodeficiency (SCID).Citation24,Citation25 In addition to the role in NHEJ, recent studies have uncovered the involvement of DNA-PKcs in DNA damage signaling. For example, in response to IR, DNA-PKcs promotes cell survival through phosphorylation of Thr308 and Ser473 (this site is also phosphorylated by mTORC2, see below) residues of Akt (also called PKB) together with PDK1 and the subsequent transcriptional regulation of the p53-p21 pathway.Citation26 IR-dependent phospho-activation of nuclear caspase-2 by DNA-PKcs also contributes to NHEJ and the maintenance of the ATM-mediated G2/M checkpoint.Citation27 Parts of DNA-PK are localized to lipid rafts, microenvironments on cell membranes where signaling molecules accumulate, and such localization has been suggested to mediate DNA damage signals through phosphorylations in response to IR.Citation28 In addition, DNA-PKcs mediates exchange of UV-induced translation profiles, including the promotion of the synthesis of DNA-repair related proteins and the inhibition of global translation.Citation29 DNA-PKcs is also involved in replication stress induced histone mRNA destabilization together with Upf1, similarly to that observed for ATR.Citation30 Furthermore, DNA-PK associates with telomeres and DNA-PK defects induce telomere fusion and telomere aneuploidy without telomere shortening, suggesting DNA-PK’s critical role in telomere capping.Citation31
SMG-1 (reviewed in refs. Citation32 and Citation33)
SMG-1 forms an SMG-1 complex (SMG1C) with SMG-8 and SMG-9Citation34 and functions in an mRNA quality control mechanism called nonsense-mediated mRNA decay (NMD). NMD selectively degrades premature termination codon (PTC)-containing mRNAs, which can be generated by gene mutation, splicing and transcription errors. PTC-mRNAs also arise in a physiological process, the V(D)J recombination during T- and B-cell maturation.Citation35 Therefore NMD suppresses the production of potentially harmful or nonfunctional polypeptides and ensures the accuracy of gene expression. SMG-1 plays an essential role in NMD by phosphorylating Upf1, a central regulator of NMD. When a ribosome recognizes a PTC, SMG-1, Upf1 and eukaryotic releasing factors (eRF1 and eRF3) assemble to form the SMG-1-Upf1-eRF1-eRF3 (SURF) complex on the PTC-recognizing ribosome.Citation36 If an exon junction complex (EJC), a multiprotein complex deposited on an exon-junction in a splicing dependent manner, exists downstream of the PTC, the SURF associates with the EJC. The association between SURF and EJC establishes PTC recognition and induces SMG-1-mediated Upf1 phosphorylation.Citation36 Phosphorylated Upf1 recruits mRNA decay factors and phopho-Upf1 recognizing NMD factors,Citation37-Citation39 and advances subsequent decay processes. Therefore SMG-1-mediated Upf1 phosphorylation is an essential step in NMD. Although Upf1 is also identified as a substrate of other PIKKs (ATM, ATR, DNA-PKcs, see below), the function of SMG-1 in NMD cannot be compensated with other PIKKs.
In addition to NMD, SMG-1 is implicated in other stress responses, including DNA damage,Citation40 oxidative stress, hypoxiaCitation41,Citation42 and cytokine signaling.Citation43 In a similar fashion to ATM and ATR, SMG-1 activates by IR or UV and phosphorylates p53.Citation40 Moreover, SMG-1 depletion causes spontaneous DNA damage and sensitizes cells to IR.Citation40 SMG-1 also associates with the telomere and protects the telomere by inhibiting the association of telomeric repeat-containing RNA (TERRA) with telomeric DNA.Citation44
SMG-1 is essential for mouse embryogenesis.Citation45 SMG-1 null mutants in C. elegans and D. melanogaster are viable,Citation46,Citation47 and inactivation of SMG-1 shows oxidative stress resistance and longevity in analogy to TOR in C. elegans.Citation48 Since NMD suppresses the dominant phenotype of the heterozygote caused by a nonsense mutation and because NMD is not essential for viability in C. elegans, a temperature sensitive mutant of SMG-1 can be used for genetic screening to identify gene mutations in heterozygotes of C. elegans. Temperature sensitive mutants of SMG-1 have also been used for inducible expression of transgenes with long 3′UTRs, which are a NMD target.Citation49
mTOR (reviewed in ref. Citation50)
TOR was originally identified as the target protein of rapamycin, a macrolide produced by bacterium Streptomyces hygroscopicus.Citation51,Citation52 TOR regulates various cellular activities, including cell size control, cell proliferation, translation activity and cell metabolism in response to external stresses and nutritional status. From yeast to mammals, two distinct functional TOR complexes have been identified: TORC1 and TORC2. Mammalian TORC1 (mTORC1), which is directly inhibited by rapamycin, is composed of mTOR, Raptor and mLST8 (also called as GβL), whereas rapamycin insensitive mTORC2 is composed of mTOR, Rictor, mLST8, SIN1 and Protor.
mTORC1 functions as a sensor of external signals, such as growth factors, nutrients, redox stress and controls cellular translation activity.Citation53 The mTORC1 phosphorylates the p70 ribosomal S6 kinase (S6K) and eIF4E binding protein (4EBP), two key regulators of cap-dependent translation, thereby facilitating translation together with the regulation of ribosome biogenesis via transcriptional regulation.Citation54 mTORC1 also enhances the translation efficiency of newly synthesized spliced mRNAs through activation of S6K recruited to the spliced mRNAs by SKAR, an EJC component.Citation55 mTORC1-mediated S6K phosphorylation and translation enhancement are linked to cell size control.Citation56 mTORC1 also acts as a conserved negative regulator of autophagy in response to nutrient availability.Citation57
In contrast, mTORC2 regulates actin cytoskeletal organization by phosphorylating PKCαCitation58,Citation59; however, the upstream signals remain unclear. mTORC2 also phosphorylates Ser473 of Akt and controls cell growth, proliferation and cell migration.Citation60 Recently, another (m)TORC2 target, serum glucocorticoid–induced protein kinase 1 (SGK1) has been identified.Citation61 Through the phosphorylation of SGK1, TORC2 regulates fat metabolism, body size and development in C. elegans.Citation62,Citation63
Based on the critical role of mTOR in regulating cell proliferation and growth, mTOR knockout mice are embryonic lethal and mTOR-deficient embryonic stem (ES) cells fail to be maintained because of proliferation arrest.Citation64,Citation65 Moreover, (m)TORC1 signaling is linked to aging and the selective inhibition of TORC1 signaling commonly extends the life span of yeast, worms, flies and mice.Citation66
TRRAP (reviewed in ref. Citation67)
TRRAP, the only catalytically inactive PIKK member, was identified as an essential co-factor of c-Myc and E2F for the transcription/transformation activities of these oncogenic proteins and named “transformation/transcription domain–associated protein (TRRAP)”Citation68. Beside c-Myc and E2F, TRRAP associates with various transcription factors, including p53, E1A, ERα/β, β-catenin and regulates transcription. Importantly, TRRAP is a shared and essential component of distinct histone acetyltransferase (HAT) complexes, including PCAF (SAGA in S. cerevisiae), TIP60 (NuA4 in S. cerevisiae), TFTC and SILK HAT complexes from yeast to mammals. A HAT complex, which is composed of a catalytic subunit and its associated proteins, acetylates Lys residues of core histone tails to promote transcription. TRRAP appears to play a role in the recruitment of HAT complexes to the chromatin as a mediator between a HAT complex and various transcription factors. A genome-wide analysis revealed that TRRAP regulates various gene expressions involved in cell cycle progression, the cytoskeleton, cell adhesion, protein turnover, metabolism and signal transduction.Citation69 Consistent with this, TRRAP is essential for viability in S. cerevisiae and Mus musculus,Citation70,Citation71 and TRRAP-deficient cells show mitotic checkpoint failures and the severe suppression of cell proliferation.Citation70 TRRAP is also involved in DNA repair processes through the TIP60 HAT complexCitation72 and the NHEJ pathway.Citation73
RUVBL1 and RUVBL2 are Multifunctional ATPases (reviewed in ref. Citation74)
RUVBL1 and RUVBL2 are conserved AAA+ family proteins
RuvB-like (RUVBL) 1 and RUVBL2, also known as Pontin and Reptin (or TIP48 and TIP49), are evolutionally conserved ATPases that belong to the ATPase associated diverse cellular activities (AAA+) family. They have sequence homology to bacterial RuvB, a DNA helicase involved in homologs recombination and DNA repair.Citation75,Citation76 Both have been shown to have ATPase and DNA helicase activity in vitro.Citation77 RUVBL1 and RUVBL2 interact with each other and can form a double hexamer, probably consisting of two homo hexamers, which is a characteristic of AAA+ family proteins.Citation78,Citation79 In some circumstances, RUVBL1 and RUVBL2 act independently and have antagonistic effects.Citation80,Citation81 However, in most cases, they appear to form a complex and function together. The complex formation of RUVBL1 and RUVBL2 appears to be important in vivo, because the depletion of either protein causes co-depletion of the other protein.Citation82,Citation83 Based on their diverse functions (), both proteins are essential for viability/development of S. cerevisiae,Citation77,Citation85 D. melanogaster,Citation86 and C. elegans [wormbase (http://www.wormbase.org/)].
Figure 3. The RUVBL1/2 complex participates in diverse cellular processes. The RUVBL1/2 complex is composed of RUVBL1 and RUVBL2, and both proteins possess ATPase activity. The RUVBL1/2 complex is localized to nucleus and cytoplasm, and participates in diverse cellular processes together with specific interactors (shown below each box). The ATPase activity of the RUVBL1/2 complex is thought to essential for their functions in each process. The atomic structure of RUVBL1 is derived from reference Citation84.
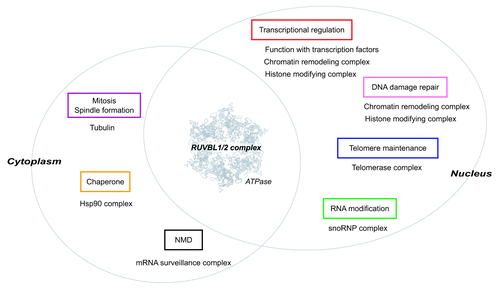
The RUVBL1/2 complex is involved in diverse cellular functions
While RUVBL1 and RUVBL2 participate in diverse nuclear processes, the best studied function of RUVBL1 and RUVBL2 is transcriptional control.Citation87 RUVBL1 and RUVBL2 are shared components of the Ino80 and SRCAP (Swr1 in S. cerevisiae) chromatin remodeling complex,Citation88,Citation89 and the TIP60 HAT complex.Citation90 The RUVBL1/2 complex is essential for the chromatin remodeling activity of the Ino80 complexCitation91 and the HAT activity of the TIP60 complex.Citation92 Both the Ino80 and TIP60 chromatin remodeling/modifying complexes are also implicated in DNA repair.Citation93,Citation94 Moreover, RUVBL1 and RUVBL2 interact with RNA polymerases and various transcription factors, including c-Myc, β-catenin, TATA binding protein and E2F, and regulate transcription of their target genes, thereby affecting cellular transformation, development and tumor metastasis.Citation87 Conditional depletion of Rvb1 or Rvb2 in S. cerevisiae was reported to influence transcription of > 5% of the genes of yeast.Citation95
The RUVBL1/2 complex is also involved in the maturation of small nucleolar RNPs (snoRNPs), which catalyze post-transcriptional modification of non-coding RNAs, such as rRNA (rRNA), tRNA (tRNA) and small nuclear RNA (snRNA).Citation96 The RUVBL1/2 complex associates with different snoRNPs and controls accumulation of snoRNAs, proper localization of the components to the nucleolus and snoRNP assembly.Citation97,Citation98
The RUVBL1/2 complex also participates in telomere maintenance. The RUVBL1/2 complex associates with telomerase reverse transcriptase (TERT) and regulates the accumulation of the telomerase RNA component (TERC) and the assembly of the functional telomerase complex with its ATPase activity. Therefore, depletion of RUVBL1 or RUVBL2 severely reduces telomerase activity.Citation83 The association of the RUVBL1/2 complex with TERT is observed to peak in the S phase,Citation83 suggesting that the RUVBL1/2 complex regulates telomerase activity in a cell cycle dependent manner.
In addition, the RUVBL1/2 complex was identified as one of interacting proteins of Hsp90, an essential molecular chaperone for cellular homeostasis, together with RPAP3 and NOP17 (Tah1 and Pih1 in S. cerevisiae, respectively), two conserved Hsp90 co-factorsCitation99,Citation100 (described later, see Section 4). The RUVBL1/2 complex was also found in another chaperone-like URI/prefoldin complex, which is involved in (m)Tor-dependent nutrient signalingCitation101 (described later, see Section 4).
RUVBL1 and RUVBL2 are also involved in mitosis. RUVBL1 interacts with α- and γ-tubulin, and regulates spindle assembly,Citation102 whereas RUVBL2 is localized to the midbody and suggested to function in cytokinesis.Citation103,Citation104
As mentioned above, the RUVBL1/2 complex participates in various cellular processes. In most cases, the RUVBL1/2 complex functions as a component of multiprotein complexes containing nucleic acids (DNA/RNA), and it commonly regulates the assembly of these functional complexes.Citation82,Citation83,Citation91 Inhibition of the RUVBL1/2 complex causes immature complex assembly and functional defects, suggesting a similar mode of action of the RUVBL1/2 complex on macromolecular complex formation/remodeling. Although the detailed mechanisms are unknown, their ATPase activity and nucleic binding properties may be important for these processes.
Integrated Regulation of the PIKK Family by the RUVBL1/2 Complex
The RUVBL1/2 complex regulates PIKK function through physical interaction and controls the levels of these kinases
Recently, we found an unexpected link between the RUVBL1/2 complex and the PIKK family. We had originally identified RUVBL1 and RUVBL2 as SMG-1 interacting proteins. Subsequent analyses revealed that the RUVBL1/2 complex associates not only with SMG-1 but also with any PIKK.Citation82 In addition to the physical interactions, the RUVBL1/2 complex regulates the levels of all PIKKs (). Either knockdown of RUVBL1 or RUVBL2 clearly decreased all PIKK proteins and suppressed PIKK signaling.Citation82 Thus, the RUVBL1/2 complex can modulate PIKK functions as a common interactor and regulator of their protein abundance.
Figure 4. The RUVBL1/2 complex can regulate PIKK functions through several ways. Three possible mechanisms for the RUVBL1/2 complex to regulate PIKK functions. (A) Control and balance the abundance of PIKK. The RUVBL1/2 complex and its ATPase activity is required for the maintenance of PIKK protein abundance. The RUVBL1/2 complex affects the mRNA level of some PIKKs. The character size of each PIKK shows the extent of the sensitivity. The RUVBL1/2 complex is also involved in the assembly and stabilization of newly synthesized PIKK protein complex probably together with Hsp90 and the Tel2 complex. (B) Functional control via physical interactions. The RUVBL1/2 complex physically interacts with PIKK and facilitates proper PIKK-mediated stress responses. Three mechanisms to control PIKK function; recruitment/localization of PIKK, activation of PIKK through posttranslational modification, and promotion of the functional complex assembly of PIKK during stress responses. (C) Function as a PIKK substrate. RUVBL2 is phosphorylated by ATM/ATR in response to DNA damage stress.Citation105 The RUVBL1/2 complex may have a role as a downstream effector protein of PIKKs. The atomic structure of RUVBL1in is derived from reference Citation84.
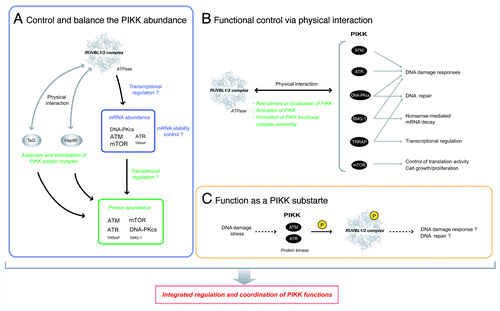
The detailed mechanism describing how the RUVBL1/2 complex controls the quantities of PIKKs is unknown; however, regulation appears to be at the mRNA level and the ATPase activities of both RUVBL1 and RUVBL2 are involved.Citation82 As one possibility, the RUVBL1/2 complex may regulate transcriptional activity of PIKKs together with E2F1 and c-Myc, because E2F1, one of RUVBL1 interacting transcription factors and regulated by c-Myc, can promote transcriptional activity of ATM and DNA-PKcs.Citation106,Citation107 E2F1 and c-Myc also facilitate translation activity of target mRNAs by inducing cap methylation;Citation108 therefore, the RUVBL1/2 complex may influence the translation activity of PIKK mRNAs. Actually, the effect of RUVBL1/2 knockdown on the PIKK protein levels is more severe than that on the PIKK mRNA levels,Citation82 indicating that an undefined mechanism at the protein level participates in the process. Given the association of the RUVBL1/2 complex with Hsp90 and the Tel2 complex, the RUVBL1/2 complex probably acts through the Hsp90 chaperone pathway for maturation and stabilization of PIKK proteins (, described later, see Putative “PIKK Regulatory Chaperone Complexes” Consisting of the RUVBL1/2 Complex, the Tel2 Complex and HSP90).
As described above, the RUVBL1/2 complex directly participates in the functions of at least two PIKKs, TRRAP and SMG-1. TRRAP and the RUVBL1/2 complex function together in transcriptional regulation and DNA repair processes as essential components of the TIP60 HAT complex.Citation72,Citation87,Citation90 On the other hand, the RUVBL1/2 complex associates with SMG-1 and facilitates rearrangement of the SMG-1-containing complex during NMD.Citation82 Since RUVBL1 and RUVBL2 interact with the N-terminal region of SMG-1,Citation82 the RUVBL1/2 complex is expected to interact with α-helical repeats of other PIKKs (). The α-helical region of PIKKs provides protein-protein interaction surfaces important for their functions, such as ATM-Nbs1, ATR-ATRIP, mTOR-Raptor and SMG-1-SMG-8/SMG-9;Citation109-Citation112 therefore the association of the RUVBL1/2 complex possibly influences the formation of PIKKs complexes.
In a different manner from TRRAP and SMG-1, a direct relationship between the RUVBL1/2 complex and other PIKKs has not been reported. However, previous studies suggest the involvement of the RUVBL1/2 complex in PIKK-mediated DNA damage response and repair. For example, the RUVBL1/2 complex-containing the TIP60 HAT complex acetylates the FAT-C domain of ATM, thereby activating ATM in response to DNA damage.Citation113 The requirement of the RUVBL1/2 complex for the TIP60 HAT activityCitation92 indicates a critical role of the RUVBL1/2 complex in ATM activation and the DNA damage response. The FAT-C domain is conserved among PIKKs and critical for kinase activity ();Citation114–Citation117 therefore other PIKKs may be activated by similar acetylation events.Citation118 The RUVBL1/2 complex may also be involved in ATR recruitment through physical interactions with RPA3,Citation85 a subunit of RPA, an ATR recruiter. Moreover, RUVBL2 is a DNA damage-induced ATM/ATR substrate.Citation105 These observations indicate that the RUVBL1/2 complex directly participates in the PIKK-mediated DNA damage response and repair process in addition to the quantity control of PIKKs ().
Although ATM, ATR and DNA-PKcs have been established as nuclear kinases, the RUVBL1/2 complex associates with PIKKs both in the nucleus and cytoplasm (unpublished data), suggesting that the RUVBL1/2 complex may also influence the nuclear localization of PIKKs or their cytoplasmic functions (see Section 1). For instance, a part of ATM, ATR and DNA-PKcs localizes to the centrosomeCitation119 and ATM/ATR activates the cell cycle checkpoint by inhibiting spindle assembly in response to DNA damage during mitosis.Citation120 As mentioned above, the RUVBL1/2 complex associates with α- and γ-tubulinCitation103,Citation121 and RUVBL1 regulates microtubule assembly during mitosis,Citation102 implying a relationship to the ATM/ATR-mediated DNA damage response during mitosis.
Functional relationships between the RUVBL1/2 complex and TOR have also been suggested. The (m)TORC1 acts as a positive regulator of transcription of rRNAs and ribosomal proteins.Citation54 In addition, TORC1 controls rRNA maturation through snoRNP localization/accumulation in the nucleolus like RUVBL1 in C. elegans,Citation122 suggesting that TOR and RUVBL1 function in the same pathway. A further study indicated that the RUVBL1/2 complex participates in (m)TOR signaling as components of the unconventional prefoldin URI complex together with RPB5Citation101 (described later, see Putative “PIKK Regulatory Chaperone Complexes” Consisting of the RUVBL1/2 Complex, the Tel2 Complex and HSP90).
Taken together, the RUVBL1/2 complex can regulate PIKK functions thorough several ways: (1) control of PIKKs levels (); (2) activation of PIKKs via post translational modifications (); (3) recruitment or localization of PIKKs; (4) promote assembly/rearrangement of PIKK complexes (); and (5) function as a downstream effector of PIKK signaling ().
Functional links among PIKKs and a possible role of the RUVBL1/2 complex in the coordination of PIKK-mediated stress responses
What is the significance of common PIKK regulators? Based on previous observations suggesting functional links among PIKKs, the RUVBL1/2 complex may participate in coordinating and regulating PIKK signaling for the appropriate stress responses (Possible models are illustrated in ).
Figure 5. Crosstalk and regulation among PIKKs. (A) Possible models of the regulation of PIKK signaling by the RUVBL1/2 complex. (a) The RUVBL1/2 complex integrates each PIKK signaling as an upstream regulator and induces proper stress responses. (b) When multiple PIKKs cooperatively function in response to stress signals, the RUVBL1/2 complex assists this process and coordinates multiple PIKK signals (the left model). The RUVBL1/2 complex coordinates the cross-regulation among PIKKs [see also (B)] thereby induce proper stress responses (the right model). The atomic structure of RUVBL1 is derived from reference.Citation84 (B) Cross-regulation among PIKKs. Several regulatory mechanisms among PIKKs have been observed. (a) Interdependent activation of ATM and ATR in response to DNA damage. (b) Regulation of other PIKK by direct phosphorylation: DNA-PKcs is phosphorylated by ATM and ATR in response to DNA damage stress to regulate cellular radio-resistance and NHEJ. (c) Regulation of other PIKK levels: DNA-PKcs and mTOR are required for the maintenance of ATM abundance. DNA-PKcs is also involved in the maintenance of SMG-1 abundance. (d) Regulation of other PIKK signals by indirect phosphorylations: Both upstream and downstream factors of mTORC1 signal are ATM/ATR substrates and mTORC1 signal is downregulated by DNA damage stresses. (C) Shared substrates among PIKKs. Histone H2Ax, p53, and Upf1 are shared substrates of DNA-PKcs, ATM, ATR and SMG-1. 4EBP and Akt, two well known mTOR substrates, are also phosphorylated by ATM and DNA-PKcs respectively.
![Figure 5. Crosstalk and regulation among PIKKs. (A) Possible models of the regulation of PIKK signaling by the RUVBL1/2 complex. (a) The RUVBL1/2 complex integrates each PIKK signaling as an upstream regulator and induces proper stress responses. (b) When multiple PIKKs cooperatively function in response to stress signals, the RUVBL1/2 complex assists this process and coordinates multiple PIKK signals (the left model). The RUVBL1/2 complex coordinates the cross-regulation among PIKKs [see also (B)] thereby induce proper stress responses (the right model). The atomic structure of RUVBL1 is derived from reference.Citation84 (B) Cross-regulation among PIKKs. Several regulatory mechanisms among PIKKs have been observed. (a) Interdependent activation of ATM and ATR in response to DNA damage. (b) Regulation of other PIKK by direct phosphorylation: DNA-PKcs is phosphorylated by ATM and ATR in response to DNA damage stress to regulate cellular radio-resistance and NHEJ. (c) Regulation of other PIKK levels: DNA-PKcs and mTOR are required for the maintenance of ATM abundance. DNA-PKcs is also involved in the maintenance of SMG-1 abundance. (d) Regulation of other PIKK signals by indirect phosphorylations: Both upstream and downstream factors of mTORC1 signal are ATM/ATR substrates and mTORC1 signal is downregulated by DNA damage stresses. (C) Shared substrates among PIKKs. Histone H2Ax, p53, and Upf1 are shared substrates of DNA-PKcs, ATM, ATR and SMG-1. 4EBP and Akt, two well known mTOR substrates, are also phosphorylated by ATM and DNA-PKcs respectively.](/cms/asset/7075ac80-295b-4694-9a5c-f01e2d921c38/kncl_a_10918926_f0005.gif)
Previous studies have often suggested functional relationships among PIKKs in DNA damage responses. For example, ATM and ATR are postulated to be activated by separate signals and act independently. However, interdependent activation between ATM and ATR is also observed [-(a)].Citation123-Citation125 In addition, ATM or ATR phosphorylates DNA-PKcs in response to IR or UV/replication stress and the former is important for cellular radio-resistance and NHEJ [-(b)].Citation126,Citation127 Conversely, DNA-PKcs regulates the abundance of ATM and SMG-1 [-(c)].Citation128,Citation129 SMG-1 also activates in response to IR and UV, and phosphorylates p53 together with ATM/ATR.Citation40 In DNA repair processes, TRRAP contributes to efficiency/fidelity of NHEJ in a HAT activity independent manner, in addition to DNA-PKcs.Citation73 ATM/ATR-mediated DNA damage signals link to various signal pathways. Upstream and downstream factors of mTORC1, Akt, TSC1, 4EBP and S6K have been identified as possible ATM/ATR substrates induced by IRCitation105 and downregulation of mTORC1 signaling by DNA damage stress has been reported [-(d)].Citation130 In contrast, mTOR regulates ATM levels [-(c)].Citation128 In addition, (m)TORC1 inhibition and tor1 (one of tor genes in S. pombe and forms TORC2) mutants show high sensitivity to DNA-damaging agents,Citation131-Citation133 suggesting a link between ATM/ATR-mediated DNA damage responses and (m)TOR signaling. We speculate that PIKKs function in DNA damage response and DNA repair in collaboration with each other at multiple levels, and this is important for proper DNA damage responses. In this context, the RUVBL1/2 complex can serve as a mediator among PIKKs and organize DNA damage responses.
Another possible functional link among PIKKs is telomere maintenance. The telomere is a protective end structure of chromosomes in eukaryotes and is essential for genome stability.Citation134 The telomere is maintained by telomerase, an RNP complex containing the telomerase reverse transcriptase catalytic subunit (TERT), and protected by multiple telomeric DNA binding proteins. Telomere maintenance closely links to DNA damage repair processesCitation135 and at least four of the six PIKKs are involved in telomere maintenance. For example, Tel1 and Rad3 (ATM and ATM orthologs in S. pombe) promote the recruitment of telomere protective proteins and telomerase.Citation136 ATM and ATR also cooperate with other repair machinery to form the proper telomeric structure on telomere replication.Citation137 DNA-PKcs and Ku70/80 associate with telomeres and are suggested to function in telomere capping.Citation31 SMG-1 also associates with telomeres and inhibits accumulation of TERRA around the telomere and SMG-1 depletion causes telomere loss and fusion.Citation44 In most somatic cells, telomerase expression is low, while progenitor germ/stem cells and putative cancer stem cells possess high activity of telomerase. When a silent TERT gene reactivates, c-Myc, TRRAP and its associating HAT activities are required.Citation138 TRRAP-containing SAGA HAT complex also regulates the turnover of critical telomere binding protein, TRF1.Citation139 Several reports also suggest the involvement of mTOR in telomere regulation. For example, mTORC1 inhibition causes downregulation of TERT mRNA expression and reduced telomerase activity.Citation140 On the other hand, Akt, a downstream effector of mTORC2, negatively regulates telomere length by phosphorylating TRF1,Citation141 which is consistent with another study showing the elongation of the telomere in a tor1 mutant in S. pombe.Citation131 As mentioned above, the RUVBL1/2 complex is critical for telomerase activity as this complex promotes the assembly of the telomerase complex.Citation83 Although the direct relationship among PIKKs and the RUVBL1/2 complex in telomere maintenance has not been defined, their cooperative actions and the coordination of PIKKs by the RUVBL1/2 complex may be important for telomere maintenance.
In addition to the above mentioned cases, several PIKK substrates, including p53, histone H2AX, Upf1, 4EBP and Akt are shared by multiple PIKKs (). Thus, the RUVBL1/2 complex may be involved in the selection of PIKKs through a cellular stress dependent mechanism.
Putative “PIKK Regulatory Chaperone Complexes” Consisting of the RUVBL1/2 Complex, the Tel2 Complex and Hsp90
Two other PIKK regulators, the Tel2 complex and Hsp90
In addition to the RUVBL1/2 complex, at least two common regulators of PIKK, the Tel2 complex and Hsp90, have been reported.
Tel2 (also called CLK2) is the mammalian homolog of S. cerevisiae telomere maintenance 2 (Tel2); however, the involvement of Tel2 in telomere maintenance has not been reported in mammals.Citation142 Tel2 forms a complex with the Tel2-interacting protein 1 (Tti1) (also called SMG-10) and Tel2-interacting protein 2 (Tti2)Citation128,Citation143-Citation145. Tel2, Tti1 and Tti2 are closely related and each protein level depends on one another.Citation128,Citation143,Citation144 The Tel2 complex associates with newly synthesized PIKK proteins and is required for the formation of PIKK-functional complexes, such as mTORC1, mTORC2 and ATR-ATRIP complexes.Citation144,Citation146 Inhibition of the Tel2 complex causes destabilization of PIKK proteins without affecting their mRNA levels.Citation142-Citation144 Moreover, Tel2 is required for the recruitment of Tel1 (ATM ortholog in S. pombe) to DNA damage sites and the activation of the Rad3 (ATR ortholog in S. pombe) and Rad3 mediated replication checkpoint.Citation147-Citation150 Tti1 was also isolated as one of the genes required for IR-resistance and is involved in PIKK-mediated DNA damage responses.Citation143
Hsp90 is an ATP-dependent molecular chaperone to specific proteins including various protein kinases. Hsp90 forms multiple complexes with its co-factors and facilitates structural maturation and complex assembly of its client proteins.Citation151 Although physical interactions of Hsp90 with PIKKs have been observed only in DNA-PKcs and SMG-1,Citation82,Citation152 Hsp90 inhibition leads to a reduction in all PIKK proteins and their downstream phosphorylation signals.Citation82,Citation128,Citation142 Moreover, Hsp90 inhibition impairs DSB repair and the cell cycle checkpoint to IR because of the attenuation of IR-induced ATM and DNA-PKcs activation without downregulation of ATM and DNA-PKcs.Citation153 Hsp90 was also identified as a Raptor interacting protein and the reduction in phosphorylation of mTOR and its substrates, 4EBP and S6K, were observed under Hsp90 inhibition without affecting the levels of mTOR.Citation154
The putative PIKK regulatory chaperone complex and its components
A recent report revealed that the Tel2 complex interacts with Hsp90 and that the associations between the Tel2 complex and PIKKs depends on the chaperone activity of Hsp90.Citation144 In addition, the RUVBL1/2 complex associates with the Tel2 complex and Hsp90.Citation82,Citation128,Citation144,Citation166 Both the RUVBL1/2 complex and the Tel2 complex also interact with two evolutionarily conserved Hsp90 co-factors, RPAP3 and NOP17,Citation82,Citation128,Citation144 which can affect ATPase activity of Hsp90.Citation155,Citation156 These observations strongly suggest that the RUVBL1/2 complex, the Tel2 complex and Hsp90 form a complex and function together to regulate PIKKs.
If it was the case, how do these molecules function together? One possibility is that the RUVBL1/2 complex and the Tel2 complex act as Hsp90 co-factors. Hsp90 cofactors play important roles in controlling Hsp90 chaperone activity through mediating client association and/or regulating the ATPase cycle of Hsp90.Citation151,Citation157 Since Tel2 directly interacts with ATM and mTOR,Citation142 the Tel2 complex may promote the association of Hsp90 with PIKKs. Alternatively, the RUVBL1/2 complex and the Tel2 complex may affect the Hsp90 ATPase cycle, thereby regulating its chaperone activity. Considering that RUVBL1 and RUVBL2 are AAA+ family ATPases, which are widely involved in molecular remodeling events via ATP hydrolysis,Citation158 it is also possible that the RUVBL1/2 complex may act as another molecular chaperone in PIKK regulation. Indeed, the ability of the RUVBL1/2 complex to maintain PIKK abundance requires its ATPase activity.Citation82 In this context, the associations between the RUVBL1/2 complex and Hsp90 cofactors (RPAP3, NOP17) may link two molecular chaperones like Hop, which mediates and coordinates two chaperones, Hsp70 and Hsp90.Citation159
Interestingly, the RUVBL1/2 complex interacts with another chaperone-related prefoldin complex containing URI, RPB5 and Monad.Citation101,Citation160 URI (also called RMP), an unconventional prefoldin, controls a part of nutrient sensitive gene expression and cell survival signaling downstream of (m)TOR,Citation101,Citation161 and its deficiency causes DNA breaks and cell cycle arrest in C. elegans.Citation162 URI interacts with all PIKK proteins, the Tel2 complex, and Hsp90.Citation128 RPB5, a shared subunit of RNA polymerases and a known URI interactor,Citation163,Citation164 associates with at least one PIKK, SMG-1, and is involved in NMD.Citation82 Monad (also called WDR92) interacts with at least the RUVBL1/2 complex, Tti1, RPAP3, NOP17, URI and RPB5.Citation82,Citation128,Citation160,Citation165 Based on the above mentioned observations, multiple chaperone-containing complexes are expected to collaboratively function to regulate PIKKs (). Together with the previous analyses, the putative PIKK regulatory chaperone complex may not only assist the maturation of PIKK complexes when PIKK proteins are synthesized, but also facilitate the remodeling of PIKK complexes when PIKKs activate in response to stress signals. Interestingly, some molecules including RUVBL2 have putative phosphorylation sites by PIKK (see ), suggesting that they can also function as PIKK downstream effectors and provide an additional intricate regulatory mechanism of PIKKs.
Figure 6. The putative “PIKK regulatory complex.” Three common PIKK regulators, the RUVBL1/2 complex, Hsp90 and the Tel2 complex interact with one another. Other factors (RPAP3, NOP17, RPB5, URI and Monad) are shared interactors of the RUVBL1/2 complex, Hsp90 and the Tel2 complex. They are possible PIKK regulators (see ). The interaction between the RUVBL1/2-URI-prefoldin complex and the Tel2 complex is mediated by NOP17 in a Tel2 phosphorylation dependent manner.Citation166
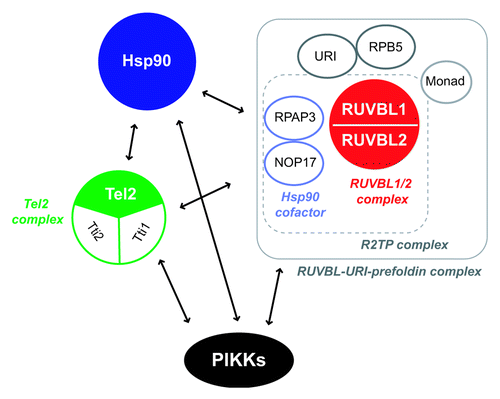
Table 1. List of common and possible PIKK regulators in mammals
Given that the majority of the putative PIKK regulatory chaperone complex components also physically and functionally associate with transcriptional machineryCitation167,Citation168 and RNP biogenesis,Citation169,Citation170 similar complexes probably function in other cellular processes.
On the other hand, the inhibition of the RUVBL1/2 complex or the Tel2 complex has been observed to have a different effect on the PIKK mRNA levels.Citation82,Citation142,Citation143 Concerning the regulation of the PIKK abundance, the mutual regulation among PIKKs is also exist [-(c)]. The regulatory mechanisms of the PIKK family appear to be involved in multiple unknown mechanisms. Further studies are required to understand the detailed molecular mechanisms of PIKK regulation by the putative PIKK regulatory chaperone complex.
Relationship of the RUVBL1/2 Complex to Cancer Biology
From a clinical point of view, PIKKs have been suggested to be potential therapeutic targets for cancer therapy. For example, the constitutive activation of mTOR signaling has been observed in multiple types of tumors, and rapamycin analogs, which inhibit mTORC1 and cause growth reduction of cancer cells, are under clinical trials as anti-cancer agents.Citation171 Further, ATM, ATR or DNA-PKcs-mediated DNA damage responses and DNA repair pathways are potential targets for cancer therapy in combination with irradiation and DNA-damaging chemical agents.Citation172 NMD inhibition is also attractive as a new therapeutic approach to cancer by inducing tumor immunity.Citation173 In addition to the regulation of all PIKKs, the RUVBL1/2 complex is implicated in telomerase activity and the Hsp90 pathway,Citation83,Citation99 both of which are promising targets of cancer therapy and the inhibitors of which are under clinical trials.Citation174,Citation175 RUVBL1 and RUVBL2 are also involved in c-Myc-mediated cellular transformation and cancer metastasis through the transcriptional regulation with β-catenin and the TIP60 HAT complex.Citation80,Citation176 Thus, the RUVBL1/2 complex represents a molecular target for cancer therapy through the simultaneous suppression of the above mentioned multiple pathways. In support of this idea, suppression of the RUVBL1/2 complex induces growth arrest and increased apoptosis of tumor cells in vitro and in vivo.Citation177
Conclusions and Perspectives
While much is known about the critical importance of PIKKs in cellular stress responses, their overall regulatory mechanisms and the interplay among PIKKs are not well defined. The finding that all PIKKs are regulated by common regulators provides important insights into these issues.
A common PIKK regulator, the RUVBL1/2 complex, can regulate each PIKK function by controlling PIKK levels and through physical interaction with each PIKK. This suggests that the RUVBL1/2 complex mediates PIKK signaling and coordinates each PIKK-mediated stress response as a common PIKK regulator. Based on its diverse cellular functions,Citation74 the RUVBL1/2 complex possibly links the PIKK-mediated stress responses to other cellular processes, thereby facilitating correct stress responses. Although the molecular mechanisms of the RUVBL1/2 complex-mediated PIKK regulation remain to be fully elucidated, recent studies have indicated the existence of putative PIKK regulatory chaperone complexes, including the RUVBL1/2 complex, the Tel2 complex and Hsp90. Future studies will clarify the PIKK-regulatory roles of the RUVBL1/2 complex as an ATPase and the function of the Tel2 complex in the chaperone activity of Hsp90. The putative PIKK regulatory chaperone complex contains additional factors, and their function and PIKK preference should also be examined. In addition, global analyses to evaluate the interplay among PIKKs and the linkage of PIKK signals to other cellular processes are important. Further analyses will reveal the physiological significance of the common regulators of PIKKs and help our understanding of the basic mechanisms underlying proper stress responses in living organisms.
| Abbreviations: | ||
| PIKK | = | Phosphatidylinositol 3-kinase-related protein kinase |
| ATM | = | ataxia telangiectasia mutated |
| ATR | = | ATM- and Rad3-related |
| DNA-PKcs | = | DNA-dependent protein kinase catalytic subunit |
| SMG-1 | = | suppressor with morphogenetic effect on genitalia-1 |
| mTOR | = | mammalian target of rapamycin |
| TRRAP | = | transformation/ transcription domain associated protein |
| AAA+ | = | ATPase associated diverse cellular activities |
| RUVBL1/2 | = | RuvB-like 1 and RuvB-like 2 |
| FAT-C | = | FRAP, ATM, and TRRAP C-terminal |
| DSBs | = | DNA double strand breaks |
| IR | = | ionizing radiation |
| UV | = | ultraviolet |
| NHEJ | = | non-homologous end-joining |
| NMD | = | nonsense-mediated mRNA decay |
| EJC | = | exon junction complex |
| PTC | = | premature termination codon |
| SURF | = | SMG-1-Upf1-eRF1-eRF3 |
| TERT | = | telomerase reverse transcriptase |
| TERRA | = | telomeric repeat-containing RNA |
| HAT | = | histone acetyltransferase |
| snoRNP | = | small nucleolar RNP |
| MRN | = | Mre11-Rad50-Nbs1 |
Acknowledgments
This work was supported, in part, by grants from the Japan Society for the Promotion of Science (to S.O. and N.I.), from the Ministry of Education, Culture, Sports, Science and Technology of Japan [a Grant-in-Aid for Scientific Research (A) (to S.O.), Young Scientists (A) (to A.Y.), Scientific Research on Innovative Areas “RNA regulation” (to A.Y.) and Scientific Research on Innovative Areas “Functional machinery for noncoding RNAs” (to A.Y.)], from the Japan Science and Technology Corporation (to A.Y. and S.O.), Takeda Science Foundation (to S.O.), Mitsubishi foundation (to S.O.), Uehara memorial foundation (to S.O.), and the Yokohama Foundation for Advancement of Medical Science (to A.Y.). N.I. is a Research Fellow of the Japan Society for the Promotion of Sciences.
References
- Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell 1995; 83:1 - 4; http://dx.doi.org/10.1016/0092-8674(95)90225-2; PMID: 7553860
- Abraham RT. PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair (Amst) 2004; 3:883 - 7; http://dx.doi.org/10.1016/j.dnarep.2004.04.002; PMID: 15279773
- Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009; http://dx.doi.org/10.1016/j.dnarep.2009.04.006; PMID: 19464237
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008; 9:759 - 69; http://dx.doi.org/10.1038/nrm2514; PMID: 18813293
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003; 3:155 - 68; http://dx.doi.org/10.1038/nrc1011; PMID: 12612651
- Lim DS, Kirsch DG, Canman CE, Ahn JH, Ziv Y, Newman LS, et al. ATM binds to beta-adaptin in cytoplasmic vesicles. Proc Natl Acad Sci USA 1998; 95:10146 - 51; http://dx.doi.org/10.1073/pnas.95.17.10146; PMID: 9707615
- Barlow C, Ribaut-Barassin C, Zwingman TA, Pope AJ, Brown KD, Owens JW, et al. ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc Natl Acad Sci USA 2000; 97:871 - 6; http://dx.doi.org/10.1073/pnas.97.2.871; PMID: 10639172
- Watters D, Kedar P, Spring K, Bjorkman J, Chen P, Gatei M, et al. Localization of a portion of extranuclear ATM to peroxisomes. J Biol Chem 1999; 274:34277 - 82; http://dx.doi.org/10.1074/jbc.274.48.34277; PMID: 10567403
- Li J, Han YR, Plummer MR, Herrup K. Cytoplasmic ATM in Neurons Modulates Synaptic Function. Curr Biol 2009; http://dx.doi.org/10.1016/j.cub.2009.10.039; PMID: 19962314
- Yang DQ, Kastan MB. Participation of ATM in insulin signalling through phosphorylation of eIF-4E-binding protein 1. Nat Cell Biol 2000; 2:893 - 8; http://dx.doi.org/10.1038/35046542; PMID: 11146653
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995; 268:1749 - 53; http://dx.doi.org/10.1126/science.7792600; PMID: 7792600
- Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol 1997; 15:177 - 202; http://dx.doi.org/10.1146/annurev.immunol.15.1.177; PMID: 9143686
- Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev 1996; 10:2411 - 22; http://dx.doi.org/10.1101/gad.10.19.2411; PMID: 8843194
- Hishiya A, Ito M, Aburatani H, Motoyama N, Ikeda K, Watanabe K. Ataxia telangiectasia mutated (Atm) knockout mice as a model of osteopenia due to impaired bone formation. Bone 2005; 37:497 - 503; http://dx.doi.org/10.1016/j.bone.2005.05.012; PMID: 16027059
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell 2009; 137:1088 - 99; http://dx.doi.org/10.1016/j.cell.2009.03.037; PMID: 19524511
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616 - 27; http://dx.doi.org/10.1038/nrm2450; PMID: 18594563
- Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 2005; 12:794 - 800; http://dx.doi.org/10.1038/nsmb972; PMID: 16086026
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 2000; 10:479 - 82; http://dx.doi.org/10.1016/S0960-9822(00)00447-4; PMID: 10801416
- Naumovski L, Friedberg EC. A DNA repair gene required for the incision of damaged DNA is essential for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 1983; 80:4818 - 21; http://dx.doi.org/10.1073/pnas.80.15.4818; PMID: 6308653
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 2007; 1:113 - 26; http://dx.doi.org/10.1016/j.stem.2007.03.002; PMID: 18371340
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 2003; 33:497 - 501; http://dx.doi.org/10.1038/ng1129; PMID: 12640452
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene 2005; 24:949 - 61; http://dx.doi.org/10.1038/sj.onc.1208332; PMID: 15592499
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 2009; 417:639 - 50; http://dx.doi.org/10.1042/BJ20080413; PMID: 19133841
- Kirchgessner CU, Patil CK, Evans JW, Cuomo CA, Fried LM, Carter T, et al. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science 1995; 267:1178 - 83; http://dx.doi.org/10.1126/science.7855601; PMID: 7855601
- Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 1995; 80:813 - 23; http://dx.doi.org/10.1016/0092-8674(95)90360-7; PMID: 7889575
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell 2008; 30:203 - 13; http://dx.doi.org/10.1016/j.molcel.2008.02.024; PMID: 18439899
- Shi M, Vivian CJ, Lee KJ, Ge C, Morotomi-Yano K, Manzl C, et al. DNA-PKcs-PIDDosome: a nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell 2009; 136:508 - 20; http://dx.doi.org/10.1016/j.cell.2008.12.021; PMID: 19203584
- Lucero H, Gae D, Taccioli GE. Novel localization of the DNA-PK complex in lipid rafts: a putative role in the signal transduction pathway of the ionizing radiation response. J Biol Chem 2003; 278:22136 - 43; http://dx.doi.org/10.1074/jbc.M301579200; PMID: 12672807
- Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev 2009; 23:1207 - 20; http://dx.doi.org/10.1101/gad.516509; PMID: 19451221
- Müller B, Blackburn J, Feijoo C, Zhao X, Smythe C. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J Cell Biol 2007; 179:1385 - 98; http://dx.doi.org/10.1083/jcb.200708106; PMID: 18158334
- Gilley D, Tanaka H, Hande MP, Kurimasa A, Li GC, Oshimura M, et al. DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci USA 2001; 98:15084 - 8; http://dx.doi.org/10.1073/pnas.261574698; PMID: 11742099
- Yamashita A, Kashima I, Ohno S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta 2005; 1754:305 - 15; PMID: 16289965
- Abraham RT. The ATM-related kinase, hSMG-1, bridges genome and RNA surveillance pathways. DNA Repair (Amst) 2004; 3:919 - 25; http://dx.doi.org/10.1016/j.dnarep.2004.04.003; PMID: 15279777
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 2009; 23:1091 - 105; http://dx.doi.org/10.1101/gad.1767209; PMID: 19417104
- Li S, Wilkinson MF. Nonsense surveillance in lymphocytes?. Immunity 1998; 8:135 - 41; http://dx.doi.org/10.1016/S1074-7613(00)80466-5; PMID: 9491995
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 2006; 20:355 - 67; http://dx.doi.org/10.1101/gad.1389006; PMID: 16452507
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 2003; 12:1187 - 200; http://dx.doi.org/10.1016/S1097-2765(03)00443-X; PMID: 14636577
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 2008; 133:314 - 27; http://dx.doi.org/10.1016/j.cell.2008.02.030; PMID: 18423202
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 2011; http://dx.doi.org/10.1093/nar/gkr791; PMID: 21965535
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell 2004; 14:585 - 98; http://dx.doi.org/10.1016/j.molcel.2004.05.005; PMID: 15175154
- Chen RQ, Yang QK, Chen YL, Oliveira VA, Dalton WS, Fearns C, et al. Kinome siRNA screen identifies SMG-1 as a negative regulator of hypoxia-inducible factor-1alpha in hypoxia. J Biol Chem 2009; 284:16752 - 8; http://dx.doi.org/10.1074/jbc.M109.014316; PMID: 19406746
- Brown JA, Roberts TL, Richards R, Woods R, Birrell G, Lim YC, et al. A Novel Role for hSMG-1 in Stress Granule Formation. Mol Cell Biol 2011; 31:4417 - 29; http://dx.doi.org/10.1128/MCB.05987-11; PMID: 21911475
- Oliveira V, Romanow WJ, Geisen C, Otterness DM, Mercurio F, Wang HG, et al. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J Biol Chem 2008; 283:13174 - 84; http://dx.doi.org/10.1074/jbc.M708008200; PMID: 18326048
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007; 318:798 - 801; http://dx.doi.org/10.1126/science.1147182; PMID: 17916692
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci USA 2010; 107:12186 - 91; http://dx.doi.org/10.1073/pnas.1007336107; PMID: 20566848
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 1989; 123:301 - 13; PMID: 2583479
- Chen Z, Smith KR, Batterham P, Robin C. Smg1 nonsense mutations do not abolish nonsense-mediated mRNA decay in Drosophila melanogaster. Genetics 2005; 171:403 - 6; http://dx.doi.org/10.1534/genetics.105.045674; PMID: 15965240
- Masse I, Molin L, Mouchiroud L, Vanhems P, Palladino F, Billaud M, et al. A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PLoS ONE 2008; 3:e3354; http://dx.doi.org/10.1371/journal.pone.0003354; PMID: 18836529
- Jorgensen EM, Mango SE. The art and design of genetic screens: caenorhabditis elegans. Nat Rev Genet 2002; 3:356 - 69; http://dx.doi.org/10.1038/nrg794; PMID: 11988761
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006; 124:471 - 84; http://dx.doi.org/10.1016/j.cell.2006.01.016; PMID: 16469695
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991; 253:905 - 9; http://dx.doi.org/10.1126/science.1715094; PMID: 1715094
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994; 78:35 - 43; http://dx.doi.org/10.1016/0092-8674(94)90570-3; PMID: 7518356
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009; 10:307 - 18; http://dx.doi.org/10.1038/nrm2672; PMID: 19339977
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006; 25:6384 - 91; http://dx.doi.org/10.1038/sj.onc.1209883; PMID: 17041624
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 2008; 133:303 - 13; http://dx.doi.org/10.1016/j.cell.2008.02.031; PMID: 18423201
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 2005; 19:2199 - 211; http://dx.doi.org/10.1101/gad.351605; PMID: 16166381
- Chang YY, Juhasz G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK, et al. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans 2009; 37:232 - 6; http://dx.doi.org/10.1042/BST0370232; PMID: 19143638
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004; 14:1296 - 302; http://dx.doi.org/10.1016/j.cub.2004.06.054; PMID: 15268862
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004; 6:1122 - 8; http://dx.doi.org/10.1038/ncb1183; PMID: 15467718
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307:1098 - 101; http://dx.doi.org/10.1126/science.1106148; PMID: 15718470
- García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J 2008; 416:375 - 85; http://dx.doi.org/10.1042/BJ20081668; PMID: 18925875
- Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol 2009; 7:e60; http://dx.doi.org/10.1371/journal.pbio.1000060; PMID: 19260765
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev 2009; 23:496 - 511; http://dx.doi.org/10.1101/gad.1775409; PMID: 19240135
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol 2004; 24:9508 - 16; http://dx.doi.org/10.1128/MCB.24.21.9508-9516.2004; PMID: 15485918
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol 2004; 24:6710 - 8; http://dx.doi.org/10.1128/MCB.24.15.6710-6718.2004; PMID: 15254238
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta 2009; 1790:1067 - 74; http://dx.doi.org/10.1016/j.bbagen.2009.06.007; PMID: 19539012
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene 2007; 26:5358 - 72; http://dx.doi.org/10.1038/sj.onc.1210605; PMID: 17694078
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998; 94:363 - 74; http://dx.doi.org/10.1016/S0092-8674(00)81479-8; PMID: 9708738
- Herceg Z, Li H, Cuenin C, Shukla V, Radolf M, Steinlein P, et al. Genome-wide analysis of gene expression regulated by the HAT cofactor Trrap in conditional knockout cells. Nucleic Acids Res 2003; 31:7011 - 23; http://dx.doi.org/10.1093/nar/gkg902; PMID: 14627834
- Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, et al. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet 2001; 29:206 - 11; http://dx.doi.org/10.1038/ng725; PMID: 11544477
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR 3rd, et al. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J Biol Chem 1998; 273:26559 - 65; http://dx.doi.org/10.1074/jbc.273.41.26559; PMID: 9756893
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 2006; 8:91 - 9; http://dx.doi.org/10.1038/ncb1343; PMID: 16341205
- Robert F, Hardy S, Nagy Z, Baldeyron C, Murr R, Dery U, et al. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol Cell Biol 2006; 26:402 - 12; http://dx.doi.org/10.1128/MCB.26.2.402-412.2006; PMID: 16382133
- Jha S, Dutta A. RVB1/RVB2: running rings around molecular biology. Mol Cell 2009; 34:521 - 33; http://dx.doi.org/10.1016/j.molcel.2009.05.016; PMID: 19524533
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet 1997; 31:213 - 44; http://dx.doi.org/10.1146/annurev.genet.31.1.213; PMID: 9442895
- Yamada K, Ariyoshi M, Morikawa K. Three-dimensional structural views of branch migration and resolution in DNA homologous recombination. Curr Opin Struct Biol 2004; 14:130 - 7; http://dx.doi.org/10.1016/j.sbi.2004.03.005; PMID: 15093826
- Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, et al. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem 1999; 274:22437 - 44; http://dx.doi.org/10.1074/jbc.274.32.22437; PMID: 10428817
- Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, et al. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure 2008; 16:1511 - 20; http://dx.doi.org/10.1016/j.str.2008.08.009; PMID: 18940606
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol 2007; 366:179 - 92; http://dx.doi.org/10.1016/j.jmb.2006.11.030; PMID: 17157868
- Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 2005; 434:921 - 6; http://dx.doi.org/10.1038/nature03452; PMID: 15829968
- Diop SB, Bertaux K, Vasanthi D, Sarkeshik A, Goirand B, Aragnol D, et al. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep 2008; 9:260 - 6; http://dx.doi.org/10.1038/embor.2008.8; PMID: 18259215
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, et al. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal 2010; 3:ra27; http://dx.doi.org/10.1126/scisignal.2000468; PMID: 20371770
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 2008; 132:945 - 57; http://dx.doi.org/10.1016/j.cell.2008.01.019; PMID: 18358808
- Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem 2006; 281:38918 - 29; http://dx.doi.org/10.1074/jbc.M605625200; PMID: 17060327
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, et al. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem 1998; 273:27786 - 93; http://dx.doi.org/10.1074/jbc.273.43.27786; PMID: 9774387
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, et al. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J 2000; 19:6121 - 30; http://dx.doi.org/10.1093/emboj/19.22.6121; PMID: 11080158
- Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol 2007; 17:187 - 92; http://dx.doi.org/10.1016/j.tcb.2007.02.005; PMID: 17320397
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 2000; 406:541 - 4; http://dx.doi.org/10.1038/35020123; PMID: 10952318
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004; 303:343 - 8; http://dx.doi.org/10.1126/science.1090701; PMID: 14645854
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 2000; 102:463 - 73; http://dx.doi.org/10.1016/S0092-8674(00)00051-9; PMID: 10966108
- Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell 2004; 16:465 - 77; http://dx.doi.org/10.1016/j.molcel.2004.09.033; PMID: 15525518
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol 2008; 28:2690 - 700; http://dx.doi.org/10.1128/MCB.01983-07; PMID: 18285460
- Morrison AJ, Shen X. DNA repair in the context of chromatin. Cell Cycle 2005; 4:568 - 71; http://dx.doi.org/10.4161/cc.4.4.1612; PMID: 15753656
- van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair?. Nat Rev Mol Cell Biol 2005; 6:757 - 65; http://dx.doi.org/10.1038/nrm1737; PMID: 16167054
- Jónsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, et al. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem 2001; 276:16279 - 88; http://dx.doi.org/10.1074/jbc.M011523200; PMID: 11278922
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 2007; 8:209 - 20; http://dx.doi.org/10.1038/nrm2124; PMID: 17318225
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol 2001; 21:7731 - 46; http://dx.doi.org/10.1128/MCB.21.22.7731-7746.2001; PMID: 11604509
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 2004; 16:789 - 98; http://dx.doi.org/10.1016/j.molcel.2004.11.012; PMID: 15574333
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 2005; 120:715 - 27; http://dx.doi.org/10.1016/j.cell.2004.12.024; PMID: 15766533
- Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res 2007; 6:1963 - 73; http://dx.doi.org/10.1021/pr060595i; PMID: 17348703
- Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 2003; 302:1208 - 12; http://dx.doi.org/10.1126/science.1088401; PMID: 14615539
- Ducat D, Kawaguchi S, Liu H, Yates JR 3rd, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol Biol Cell 2008; 19:3097 - 110; http://dx.doi.org/10.1091/mbc.E07-11-1202; PMID: 18463163
- Sigala B, Edwards M, Puri T, Tsaneva IR. Relocalization of human chromatin remodeling cofactor TIP48 in mitosis. Exp Cell Res 2005; 310:357 - 69; http://dx.doi.org/10.1016/j.yexcr.2005.07.030; PMID: 16157330
- Skop AR, Liu H, Yates J 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 2004; 305:61 - 6; http://dx.doi.org/10.1126/science.1097931; PMID: 15166316
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160 - 6; http://dx.doi.org/10.1126/science.1140321; PMID: 17525332
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 2002; 16:245 - 56; http://dx.doi.org/10.1101/gad.949802; PMID: 11799067
- Berkovich E, Ginsberg D. ATM is a target for positive regulation by E2F-1. Oncogene 2003; 22:161 - 7; http://dx.doi.org/10.1038/sj.onc.1206144; PMID: 12527885
- Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene 2009; 28:1169 - 75; http://dx.doi.org/10.1038/onc.2008.463; PMID: 19137018
- Arias-Palomo E, Yamashita A, Fernandez IS, Nunez-Ramirez R, Bamba Y, Izumi N, et al. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev 2011; 25:153 - 64; http://dx.doi.org/10.1101/gad.606911; PMID: 21245168
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163 - 75; http://dx.doi.org/10.1016/S0092-8674(02)00808-5; PMID: 12150925
- Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 2005; 16:2372 - 81; http://dx.doi.org/10.1091/mbc.E04-11-1006; PMID: 15743907
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 2005; 25:5363 - 79; http://dx.doi.org/10.1128/MCB.25.13.5363-5379.2005; PMID: 15964794
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 2005; 102:13182 - 7; http://dx.doi.org/10.1073/pnas.0504211102; PMID: 16141325
- Morita T, Yamashita A, Kashima I, Ogata K, Ishiura S, Ohno S. Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J Biol Chem 2007; 282:7799 - 808; http://dx.doi.org/10.1074/jbc.M610159200; PMID: 17229728
- Priestley A, Beamish HJ, Gell D, Amatucci AG, Muhlmann-Diaz MC, Singleton BK, et al. Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res 1998; 26:1965 - 73; http://dx.doi.org/10.1093/nar/26.8.1965; PMID: 9518490
- Beamish HJ, Jessberger R, Riballo E, Priestley A, Blunt T, Kysela B, et al. The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res 2000; 28:1506 - 13; http://dx.doi.org/10.1093/nar/28.7.1506; PMID: 10710416
- Takahashi T, Hara K, Inoue H, Kawa Y, Tokunaga C, Hidayat S, et al. Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro. Genes Cells 2000; 5:765 - 75; http://dx.doi.org/10.1046/j.1365-2443.2000.00365.x; PMID: 10971657
- Jiang X, Sun Y, Chen S, Roy K, Price BD. The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J Biol Chem 2006; 281:15741 - 6; http://dx.doi.org/10.1074/jbc.M513172200; PMID: 16603769
- Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J Cell Biochem 2007; 101:451 - 65; http://dx.doi.org/10.1002/jcb.21195; PMID: 17171639
- Smith E, Dejsuphong D, Balestrini A, Hampel M, Lenz C, Takeda S, et al. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat Cell Biol 2009; 11:278 - 85; http://dx.doi.org/10.1038/ncb1835; PMID: 19182792
- Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, et al. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskeleton 2003; 56:79 - 93; http://dx.doi.org/10.1002/cm.10136; PMID: 14506706
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol 2008; 18:1355 - 64; http://dx.doi.org/10.1016/j.cub.2008.07.097; PMID: 18804378
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 2006; 8:37 - 45; http://dx.doi.org/10.1038/ncb1337; PMID: 16327781
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 2006; 281:9346 - 50; http://dx.doi.org/10.1074/jbc.M513265200; PMID: 16431910
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, et al. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 2006; 25:5775 - 82; http://dx.doi.org/10.1038/sj.emboj.7601446; PMID: 17124492
- Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, et al. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem 2007; 282:6582 - 7; http://dx.doi.org/10.1074/jbc.M611605200; PMID: 17189255
- Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol 2006; 26:7520 - 8; http://dx.doi.org/10.1128/MCB.00048-06; PMID: 16908529
- Izumi N, Yamashita A, Hirano H, Ohno S. Hsp90 regulates PIKK family proteins together with the RUVBL1/2 and Tel2-containing co-factor complex. Cancer Sci 2012; 103:50 - 7; http://dx.doi.org/10.1111/j.1349-7006.2011.02112.x; PMID: 21951644
- Peng Y, Woods RG, Beamish H, Ye R, Lees-Miller SP, Lavin MF, et al. Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res 2005; 65:1670 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-04-3451; PMID: 15753361
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA 2005; 102:8204 - 9; http://dx.doi.org/10.1073/pnas.0502857102; PMID: 15928081
- Schonbrun M, Laor D, Lopez-Maury L, Bahler J, Kupiec M, Weisman R. TOR complex 2 controls gene silencing, telomere length maintenance, and survival under DNA-damaging conditions. Mol Cell Biol 2009; 29:4584 - 94; http://dx.doi.org/10.1128/MCB.01879-08; PMID: 19546237
- Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, et al. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell 2005; 120:747 - 59; http://dx.doi.org/10.1016/j.cell.2004.12.040; PMID: 15797377
- Shen C, Lancaster CS, Shi B, Guo H, Thimmaiah P, Bjornsti MA. TOR signaling is a determinant of cell survival in response to DNA damage. Mol Cell Biol 2007; 27:7007 - 17; http://dx.doi.org/10.1128/MCB.00290-07; PMID: 17698581
- O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 2010; 11:171 - 81; PMID: 20125188
- d'Adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev 2004; 18:1781 - 99; http://dx.doi.org/10.1101/gad.1214504; PMID: 15289453
- Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet 2009; 5:e1000622; http://dx.doi.org/10.1371/journal.pgen.1000622; PMID: 19714219
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 2006; 127:709 - 20; http://dx.doi.org/10.1016/j.cell.2006.09.034; PMID: 17110331
- Nikiforov MA, Chandriani S, Park J, Kotenko I, Matheos D, Johnsson A, et al. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol Cell Biol 2002; 22:5054 - 63; http://dx.doi.org/10.1128/MCB.22.14.5054-5063.2002; PMID: 12077335
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, et al. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 2009; 35:352 - 64; http://dx.doi.org/10.1016/j.molcel.2009.06.015; PMID: 19683498
- Zhou C, Gehrig PA, Whang YE, Boggess JF. Rapamycin inhibits telomerase activity by decreasing the hTERT mRNA level in endometrial cancer cells. Mol Cancer Ther 2003; 2:789 - 95; PMID: 12939469
- Chen YC, Teng SC, Wu KJ. Phosphorylation of telomeric repeat binding factor 1 (TRF1) by Akt causes telomere shortening. Cancer Invest 2009; 27:24 - 8; http://dx.doi.org/10.1080/07357900802027081; PMID: 19160102
- Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell 2007; 131:1248 - 59; http://dx.doi.org/10.1016/j.cell.2007.10.052; PMID: 18160036
- Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 2010; 24:1939 - 50; http://dx.doi.org/10.1101/gad.1934210; PMID: 20810650
- Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev 2010; http://dx.doi.org/10.1101/gad.1956410; PMID: 20801936
- Hayashi T, Hatanaka M, Nagao K, Nakaseko Y, Kanoh J, Kokubu A, et al. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 2007; 12:1357 - 70; http://dx.doi.org/10.1111/j.1365-2443.2007.01141.x; PMID: 18076573
- Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem 2010; 285:20109 - 16; http://dx.doi.org/10.1074/jbc.M110.121699; PMID: 20427287
- Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol 2007; 9:391 - 401; http://dx.doi.org/10.1038/ncb1555; PMID: 17384638
- Anderson CM, Korkin D, Smith DL, Makovets S, Seidel JJ, Sali A, et al. Tel2 mediates activation and localization of ATM/Tel1 kinase to a double-strand break. Genes Dev 2008; 22:854 - 9; http://dx.doi.org/10.1101/gad.1646208; PMID: 18334620
- Shikata M, Ishikawa F, Kanoh J. Tel2 is required for activation of the Mrc1-mediated replication checkpoint. J Biol Chem 2007; 282:5346 - 55; http://dx.doi.org/10.1074/jbc.M607432200; PMID: 17189249
- Rendtlew Danielsen JM, Larsen DH, Schou KB, Freire R, Falck J, Bartek J, et al. HCLK2 is required for activity of the DNA damage response kinase ATR. J Biol Chem 2009; 284:4140 - 7; http://dx.doi.org/10.1074/jbc.M808174200; PMID: 19097996
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 2010; 11:515 - 28; http://dx.doi.org/10.1038/nrm2918; PMID: 20531426
- Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett 2005; 579:6350 - 4; http://dx.doi.org/10.1016/j.febslet.2005.10.020; PMID: 16263121
- Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res 2006; 66:9211 - 20; http://dx.doi.org/10.1158/0008-5472.CAN-06-2181; PMID: 16982765
- Ohji G, Hidayat S, Nakashima A, Tokunaga C, Oshiro N, Yoshino K, et al. Suppression of the mTOR-raptor signaling pathway by the inhibitor of heat shock protein 90 geldanamycin. J Biochem 2006; 139:129 - 35; http://dx.doi.org/10.1093/jb/mvj008; PMID: 16428328
- Eckert K, Saliou JM, Monlezun L, Vigouroux A, Atmane N, Caillat C, et al. The Pih1-Tah1 cochaperone complex inhibits Hsp90 molecular chaperone ATPase activity. J Biol Chem 2010; http://dx.doi.org/10.1074/jbc.M110.138263; PMID: 20663878
- Millson SH, Vaughan CK, Zhai C, Ali MM, Panaretou B, Piper PW, et al. Chaperone ligand-discrimination by the TPR-domain protein Tah1. Biochem J 2008; 413:261 - 8; http://dx.doi.org/10.1042/BJ20080105; PMID: 18412542
- Hahn JS. The Hsp90 chaperone machinery: from structure to drug development. BMB Rep 2009; 42:623 - 30; http://dx.doi.org/10.5483/BMBRep.2009.42.10.623; PMID: 19874705
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 2005; 6:519 - 29; http://dx.doi.org/10.1038/nrm1684; PMID: 16072036
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 2004; 5:781 - 91; http://dx.doi.org/10.1038/nrm1492; PMID: 15459659
- Cloutier P, Al-Khoury R, Lavallee-Adam M, Faubert D, Jiang H, Poitras C, et al. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods 2009; 48:381 - 6; http://dx.doi.org/10.1016/j.ymeth.2009.05.005; PMID: 19450687
- Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell 2007; 28:28 - 40; http://dx.doi.org/10.1016/j.molcel.2007.08.010; PMID: 17936702
- Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. URI-1 is required for DNA stability in C. elegans. Development 2006; 133:621 - 9; http://dx.doi.org/10.1242/dev.02235; PMID: 16436622
- Woychik NA, Liao SM, Kolodziej PA, Young RA. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev 1990; 4:313 - 23; http://dx.doi.org/10.1101/gad.4.3.313; PMID: 2186966
- Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N, et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol 1998; 18:7546 - 55; PMID: 9819440
- Itsuki Y, Saeki M, Nakahara H, Egusa H, Irie Y, Terao Y, et al. Molecular cloning of novel Monad binding protein containing tetratricopeptide repeat domains. FEBS Lett 2008; 582:2365 - 70; http://dx.doi.org/10.1016/j.febslet.2008.05.041; PMID: 18538670
- Horejsí Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, et al. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol Cell 2010; 39:839 - 50; http://dx.doi.org/10.1016/j.molcel.2010.08.037; PMID: 20864032
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 2007; 27:262 - 74; http://dx.doi.org/10.1016/j.molcel.2007.06.027; PMID: 17643375
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 2010; 39:912 - 24; http://dx.doi.org/10.1016/j.molcel.2010.08.023; PMID: 20864038
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol 2008; 180:579 - 95; http://dx.doi.org/10.1083/jcb.200708110; PMID: 18268104
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol 2008; 180:563 - 78; http://dx.doi.org/10.1083/jcb.200709061; PMID: 18268103
- Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene 2006; 25:6436 - 46; http://dx.doi.org/10.1038/sj.onc.1209886; PMID: 17041628
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008; 8:193 - 204; http://dx.doi.org/10.1038/nrc2342; PMID: 18256616
- Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 2010; 465:227 - 30; http://dx.doi.org/10.1038/nature08999; PMID: 20463739
- Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer 2008; 8:167 - 79; http://dx.doi.org/10.1038/nrc2275; PMID: 18256617
- Taldone T, Gozman A, Maharaj R, Chiosis G. Targeting Hsp90: small-molecule inhibitors and their clinical development. Curr Opin Pharmacol 2008; 8:370 - 4; http://dx.doi.org/10.1016/j.coph.2008.06.015; PMID: 18644253
- Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell 2000; 5:321 - 30; http://dx.doi.org/10.1016/S1097-2765(00)80427-X; PMID: 10882073
- Huber O, Menard L, Haurie V, Nicou A, Taras D, Rosenbaum J. Pontin and reptin, two related ATPases with multiple roles in cancer. Cancer Res 2008; 68:6873 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-08-0547; PMID: 18757398