Abstract
Here, we review the sn- and sno-RNA transport pathways in S. cerevisiae and humans, aiming at understanding how they evolved and how common factors can have distinct functions depending on the RNA they bind. We give a particular emphasis on Tgs1, the cap hypermethylase that is conserved from yeast to humans and appears to play a central role in both sn- and sno-RNA biogenesis. In yeast, Tgs1 hypermethylates sn- and sno-RNAs in the nucleolus. In humans, Tgs1 occurs in two forms: a long isoform (Tgs1 LF), which locates in the cytoplasm and Cajal bodies, which is predominantly associated with snRNAs and a short isoform (Tgs1 SF), which is nuclear and mainly associates with snoRNAs. We show that Tgs1 LF is exported by CRM1 and that interaction with CRM1 competes for binding with the C-terminal domain of the core protein Nop58, which contains the Nucleolar localization signal of Box C/D snoRNPs (NoLS). Our data suggest a model where CRM1 removes Tgs1 LF from snoRNPs, thereby promoting nucleolar targeting via activation of their NoLS. In this review, we argue that CRM1, while first described as an export receptor, can also control the composition of nucleoplasmic complexes. Thus, it could coordinate the fate of these complexes with the general nucleo-cytoplasmic trafficking.
Keywords: :
Introduction
Eukaryotic cells contain many types of non-coding ribonucleoproteins (RNPs). Some of these RNPs reside in the nucleus and two such important groups are the small nuclear RNPs (snRNPs) that function in the maturation of mRNAs and the small nucleolar RNPs (snoRNPs) that localize to the nucleolus and are required for maturation of rRNAs. Progress has been made in understanding the mechanisms governing sn- and sno-RNP biogenesis, as well as sorting of these RNPs to their intra-cellular destinations.
Splicing snRNPs are comprised of a small nuclear RNA (snRNA), an heptad of Sm proteins and additional snRNP-specific proteins, all of which being well conserved from yeast to humans.Citation1 U1, U2, U4, U5 (but not U6 snRNA) are individually transcribed by RNA polymerase II and synthesized as m7G monomethylated capped precursors. Following assembly with Sm proteins, the cap is converted by trimethyl guanosine synthase 1 (Tgs1) into m2,2,7G.Citation2 In metazoans, this occurs in the cytoplasm and cap trimethylation is required to promote the nuclear uptake of snRNPs following an essential cytoplasmic phase of maturation (reviewed in ref. Citation3).
SnoRNAs are highly conserved from Archeabacteria to yeast and humans. In all organisms, the vast majority of these RNAs are known or predicted to function in the biogenesis of rRNAs.Citation4,Citation5 Based on sequence elements, snoRNAs were classified into two families, the box C/D and box H/ACA snoRNAs. Box C/D snoRNAs act as guides to direct 2′-O-methylation of rRNA and Box H/ACA to direct pseudo-uridylation. Similar to snRNAs, they can be transcribed by RNA polymerase II as independent units and, in this case, the m7G cap of the snoRNA precursor is also hypermethylated by Tgs1. However, they can also be transcribed as polycistronic units and processed by endonucleases, or located within introns and being released as uncapped RNA via a splicing-dependent pathway.Citation6 Every snoRNA associates with a small set of common proteins (reviewed in refs. Citation7 and Citation8): 15.5K (Snu13 in yeast), Nop56, Nop58 and fibrillarin (Nop1 in yeast) for C/D snoRNAs; Nhp2, Nop10, Gar1 and dyskerin (Cbf5 in yeast) for H/ACA snoRNAs. Biogenesis of both sn- and sno-RNP has common features that will be presented in this review. We will compare their trafficking pathways in yeast and humans, since these processes have changed during evolution.
SnRNP and snoRNP Biogenesis Involve Processing Factors that were Maintained from Yeast to Humans while their Trafficking Pathways have Diverged
In budding yeast, processing factors involved in the biogenesis of sn- and snoRNAs are well characterized (reviewed in refs. Citation9–Citation11). Assembly with core proteins is required for both RNA stability and intra-cellular targeting,Citation12 and both sn- and sno-RNAs transit through the nucleolus.Citation2 Indeed, Tgs1 appears to hypermethylate both sn- and snoRNAs in this compartment.Citation13 While snoRNA transcription occurs in the yeast nucleoplasm, their biogenesis involves a compartment inside the nucleolus called the nucleolar body (NB). Our data suggest that the components mediating snoRNP biogenesis have some ability to self-assemble and to form a distinct structure in the nucleolus. Growth on solid media led to formation of NB where Tgs1 can localize. Precursors of snoRNAs are also localized to this structure, consistent with its role in assembly of at least some snoRNPs ().Citation13 The fact that the yeast nucleolus is involved in the production of several small RNPs offers a way to coordinate major cellular metabolic activities, such as splicing and translation.
Figure 1. Schematic representation of sn- and sno-RNA biogenesis is S. cerevisiae and in human cells. (A) In S. cerevisiae, sn- and sno-RNA biogenesis occur in the nucleus and involves the NB. In particular, Tgs1, which hypermethylates sn- and snoRNAs, is localized to NB under certain growth conditions. (B) In human cells, sn- and sno-RNA use divergent trafficking pathways. While snoRNP biogenesis is exclusively nuclear, snRNP biogenesis involves a cytoplasmic phase. Hypermethylation of snoRNAs occurs in CB, while hypermethylation of snRNAs occur in the cytoplasm. PHAX and CRM1 are required for export of precursors of snRNAs but they also function sequentially inside the nucleus for capped snoRNP targeting. PHAX is first required to transport snoRNA precursors from the transcription site to CB and CRM1 is further required to transport mature snoRNP from CB to the nucleolus (capped and uncapped).
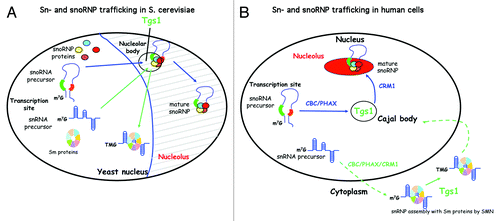
In contrast to budding yeast, sn- and sno-RNP biogenesis in metazoan cells involves distinct trafficking pathways. This evolution was accompanied by the appearance of new transport factors that are well characterized in vertebrates. While some of them are common to several pathways, the mechanisms for discrimination between the different pathways are not yet clearly understood. In metazoan, Sm protein assembly on snRNAs does not occur in the nucleoplasm but in the cytoplasm (). SnRNA precursors must be exported via CRM1 and its adaptor PHAX that recognizes the CBC complex on the 5′end of pre-snRNA.Citation14 In the cytoplasm, the SMN complex facilitates assembly of Sm proteins (reviewed in ref. Citation15) and recruitment of Tgs1 for hypermethylation of the cap.Citation16 Both events allow the formation of a bipartite signal for nuclear import of the snRNP particle. They finally transit through CB, where they bind to specific proteins to form mature snRNPs and are subjected to nucleotide modifications via scaRNAs (specific Cajal body RNA).Citation17
In contrast to snRNA, snoRNA biogenesis occurs entirely in the nucleus. Recently, we confirmed the lack of snoRNA export using heterocaryons from which only one nucleus was expressing an exogenous snoRNA.Citation18 The latter was not observed in the second nucleus, ruling out cytoplasmic transit during their biogenesis. This was shown for both Box C/D and Box H/ACA snoRNAs as well as for telomerase RNA, which share similarities with scaRNAs.Citation19 SnoRNAs follow a pathway from their transcription site via CB to the nucleolus.Citation20-Citation22 Using Box C/D snoRNA U3 as a model, we showed that CB transit is required for maturation and assembly of snoRNP, as well as hypermethylation of the cap.Citation13
In vertebrate cells, PHAX was first described as an adaptor for snRNA export.Citation14 However, we found PHAX was also associated with capped Box C/D snoRNA precursors.Citation23 Moreover, nuclear microinjection experiments using Cy3 labeled U3 snoRNA showed that PHAX is required for its localization in CB, while CRM1 is required for its nucleolar localization but not for CB localization (). Thus, PHAX and CRM1 act sequentially during snoRNP transport. For snoRNPs, CRM1 is recruited via a different mechanism than for snRNPs (e.g., independently of the cap, CBC and PHAX), as CRM1 is only found on the mature form of snoRNPs, and not on m7G capped precursors.Citation23 In agreement, microinjection experiments showed that CRM1 is required for nucleolar transport of uncapped intronic snoRNA.Citation18 Blocking nuclear pores with wheat germ agglutinin had the same effect as Leptomycin B (LMB)-driven inhibition of CRM1. Thus, CRM1 activity and functional nuclear pores are required for nucleolar targeting of capped and non-capped Box C/D snoRNAs. This led us to further investigate the role of CRM1 in snoRNP biogenesis and trafficking.
Human Tgs1 Functions both in the Cytoplasm and in the Nucleus at the Level of CB
We wanted to understand the mechanism by which CRM1 is involved in snoRNP trafficking and we focused our attention on Tgs1. Human Tgs1 occurs in two isoforms. The long isoform Tgs1 LF locates in the cytoplasm and CB and is predominantly associated with snRNAs. Cytoplasmic localization of Tgs1 LF is lost after LMB treatment, suggesting that it is exported in a CRM1-dependent manner. In addition to the full-length protein, a short, N-terminally truncated isoform of Tgs1 (Tgs1 SF) has been described as a proteasomal maturation product ().Citation24 In vivo, Tgs1 SF was shown to bind Box C/D snoRNP proteins and much less to Sm proteins. In addition, it localized to CB and nucleoplasm while absent from the cytoplasm. This suggests that snoRNPs are hypermethylated in the nucleus predominantly by Tgs1 SF, whereas snRNPs would be hypermethylated in the cytoplasm by Tgs1 LF. Co-immunoprecipitation and RNase protection assays showed that Box C/D snoRNA U3 associates with Tgs1 SF but also to a small extent with Tgs1 LF. Therefore, we cannot exclude that hypermethylation of capped Box C/D snoRNAs is sometimes mediated by the nuclear pool of Tgs1 LF.
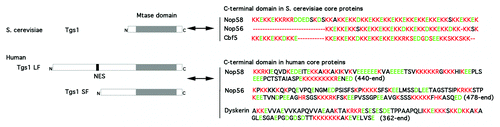
Figure 2. Conserved interaction of Tgs1 with the C-terminal domain of snoRNP core proteins from yeast to human. Tgs1 is the m7G cap hypermethylase conserved from yeast to human. In S. cerevisiae, it is essentially composed of the methylase domain (Mtase domain). Yeast Tgs1 interacts with KKE/D repeats found in the C-terminal domains of Nop58, Nop56 and Cbf5.2 In humans, Tgs1 has a conserved Mtase domain and an extended N-terminal domain that contains a Nuclear Export Sequence (NES). The full length protein (Tgs1 LF) is localized to the cytoplasm and nuclear CB in human cells. A shorter isoform (Tgs1 SF) is produced by proteosomal degradation and is restricted to CB.24 Both forms can interact with the highly charged C-terminal domain of Nop58, Nop56 and dyskerin. Amino acids sequences are shown, basic amino acids are in red and acidic are in green.
Interestingly, Tgs1 LF association with Box C/D snoRNAs appeared to be regulated by CRM1. This is not limited to capped snoRNAs, as intronic snoRNAs were also associated with Tgs1 LF, and LMB treatment increased association of both intronic and non-intronic Box C/D snoRNAs with Tgs1 LF. We wondered if we could extend this result to other RNA families. Therefore, we used microarrays to analyze the entire pool of RNAs that were immunoprecipitated with Tgs1 LF in the presence or absence of LMB.Citation18 In non-treated cells, Tgs1 LF associates preferentially with snRNAs; in the presence of LMB, Tgs1 LF association switched to intronic and non-intronic Box C/D snoRNAs, with a median increase in binding of 2-fold compared with non-treated cells (20-fold for the largest increase). Box H/ACA snoRNAs and scaRNAs were also enriched in Tgs1 LF IP after LMB treatment, but to a lesser extent.Citation18 Thus, CRM1 appears to control the binding of Tgs1 LF to snoRNAs and scaRNAs. Because most of the snoRNAs are uncapped, this suggests a new role for Tgs1 LF in snoRNP biogenesis that is unrelated to cap hypermethylation and that is regulated by CRM1.
Human Tgs1 Possesses a NES and can Directly Interact with CRM1
The dual localization of Tgs1 LF to both cytoplasm and nuclear CB suggests that Tgs1 shuttles between the two compartments. Heterocaryons analysis using cells expressing GFP-Tgs1 LF confirmed that Tgs1 LF shuttles in a CRM1-dependent manner. This suggested that the N-terminal domain of Tgs1 LF, which is absent in Tgs1 SF, might bind CRM1 via a nuclear export signal (NES). We used truncated versions of human Tgs1 to map the region responsible for export; this confirmed the role of the N-terminal domain. Tgs1 LF contains numerous potential NES, nevertheless only NES1 and NES2 were functional. Deletion mutants showed that only NES2 was required for Tgs1 LF export (shown in ).
We used in vitro binding assays with recombinant proteins to prove the direct interaction of CRM1 with Tgs1 LF via its NES. Bacterially-synthesized human Tgs1 LF interacted with CRM1 in a Ran-GTP-dependent manner,Citation18 unlike Tgs1 LF lacking NES2. This shows that CRM1 binds directly to Tgs1 LF, and that this binding depends on NES2. Both Tgs1 SF and LF have several NLS that permit their localization to nucleus (data not shown). The motifs that are necessary for their localization to CB remain to be identified.
Interaction of Tgs1 with the Highly Charged C-terminal Domain of snoRNP Core Proteins
The C-terminal domains of Nop58 and Nop56 are highly charged and conserved through evolution (). In yeast, they are formed by repetition of KKD/E sequences, while in human they contain successive stretches of acidic and basic amino acids.Citation25,Citation26 In yeast, these domains directly interact with Tgs1,Citation24 and in humans, a direct interaction has also been proved for Nop58, and remains likely for Nop56 and dyskerin. Most interestingly, in vitro experiments with the human recombinant proteins have shown that binding of CRM1 to Tgs1 LF disrupt its interaction with Nop58, thereby providing a mechanistic explanation for the requirement of CRM1 in snoRNP transport. The C-terminal domain of human Nop58 also undergoes post-translational modifications like phosphorylation and sumoylation. We showed that a Nop58 protein mutated in its sumoylation consensus site prevents de novo assembly of snoRNP.Citation27 The possibility that sumoylation of the Nop58 C-terminal domain affects Tgs1 binding will require further experiments.
In both species, these domains are sufficient for the nucleolar localization of the corresponding proteins. Interestingly, budding yeast strains lacking KKD/E repeats of snoRNP core proteins show disrupted nucleolar morphology and snoRNP localization. Similar defects were observed upon deletion of Tgs1, while a catalytically defective allele of Tgs1 restored normal nucleolar structure.Citation28 Thus, in yeast, Tgs1 plays a role in nucleolar integrity that is independent of its hypermethylation function but is dependent on interaction with Nop56 and Nop58. Thus, the binding of Tgs1 to the C-terminal domains of Nop58 and Nop56 has been conserved during evolution, playing a dual role in driving cap hypermethylation and controlling snoRNP transport.
Tgs1 LF Masks the NoLS of Nop58 on Box C/D snoRNPs until it is Released by CRM1
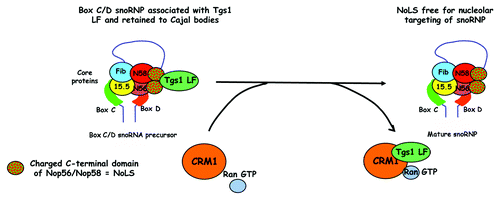
Our current understanding of how CRM1 might be involved in Box C/D snoRNP targeting to nucleolus comes from the following observations: (1) inhibition of CRM1 led to increased association of Tgs1 LF with both capped and non-capped snoRNP; (2) the retention of intronic Box C/D snoRNA in the nucleoplasm, together with Tgs1 LF, after CRM1 inhibition; (3) a Tgs1 mutant deleted for NES2, which cannot bind CRM1, also partially inhibits the transport of the snoRNA reporter to nucleoli; (4) in vitro, CRM1 inhibits the interaction of Tgs1 LF with the Nop58 C-terminal domain. Taken together, our data suggest a model where CRM1 removes Tgs1 LF from snoRNP, thereby activating the NoLS of Nop58. This would then lead to the nucleolar localization of Box C/D snoRNPs ().
Figure 3. A model to explain the role of CRM1 in the transport of snoRNPs to nucleoli. Tgs1 LF binds the highly charged C-terminal domains of Nop58 and Nop56 that act as a Nucleolar localization signal (NoLS). This interaction is prevented by the binding of Tgs1 LF to CRM1. Thus direct binding of CRM1 to Tgs1 LF in a Ran-GTP dependent manner would unmask the NoLS on Box C/D snoRNPs, promoting the targeting of mature Box C/D snoRNPs to the nucleolus.
We did not test whether CRM1 can function in a similar way with Box H/ACA snoRNAs. Dyskerin was shown to interact with Tgs1 SFCitation24 and it seems likely to interact with LF. Using microarrays, we found an enrichment of Tgs1 LF with Box H/ACA snoRNAs that was increased after LMB treatment. This suggests that the association of Tgs1 LF with Box H/ACA snoRNAs would be also regulated by CRM1. The C-terminal domain of dyskerin, which binds Tgs1, might be masked, thus preventing nucleolar targeting of Box H/ACA snoRNAs. This role could be tested as above, namely by localizing a Box H/ACA snoRNA reporter in human cells and by analyzing the effect of CRM1 on Tgs1 LF interaction with dyskerin in vitro. It will also be interesting to determine if CRM1 has a role in the localization of telomerase RNA to the nucleolus and/or telomeres.
CRM1 is an Important Regulator in RNP Trafficking Capable of Linking Nucleoplasmic Events to Nucleo-Cytoplasmic Trafficking
Our recent data indicate that CRM1 controls the composition of nucleoplasmic pre-snoRNPs. Previous studies showed that CRM1, independently of its function in nuclear export, regulates the composition of protein complex during mitosis. Indeed, a CRM1-containing complex is involved in the formation of mitotic spindles (reviewed in ref. Citation29). Nevertheless, CRM1 is best known as a nuclear export receptor (reviewed in ref. Citation30), and no such role was previously demonstrated in interphase nuclei. Our data suggest that CRM1 could control the composition of many nucleoplasmic complexes, thereby coupling nucleo-cytoplasmic trafficking with other nuclear functions. In agreement, CRM1 has been shown to favor retention of snRNPs to CB.Citation31
SnoRNP transport must be coordinated with ribosome biogenesis and CRM1 could play a central role in this process. For instance, blocking nuclear pores with wheat germ agglutinin inhibits snoRNP transport to nucleoliCitation18 and this could be explained by the titration of CRM1 by accumulating ribosomal subunits.
| Abbreviations: | ||
| RNP | = | ribonucleoprotein |
| snRNA | = | small nuclear RNA |
| snoRNA | = | small nucleolar RNA |
| scaRNA | = | specific Cajal body RNA |
| CBC | = | cap binding complex |
| CB | = | Cajal body |
| NB | = | nucleolar body |
| LMB | = | leptomycin B |
| NES | = | nuclear export signal |
| NLS | = | nuclear localization signal |
| NoLS | = | nucleolar localization signal |
| TMG | = | trimethyl guanosine |
| Tgs1 LF | = | Tgs1 long form |
| Tgs1 SF | = | Tgs1 short form |
| CRM1 | = | export receptor |
| PHAX | = | adaptor protein for RNA export |
| CBC | = | 5′cap binding |
| Tgs1 | = | 5′cap hypermethylation, SMN, survival motor neuron complex implicated in snRNP assembly |
Acknowledgments
We thank Remy Bordonné and Robert Hipkins for critical reading of the manuscript. This work was supported by the “Centre National de la Recherche Scientifique” and by grants from the “Association pour la Recherche sur le Cancer,” “La Ligue contre le Cancer” and “Agence National pour la Recherche.”
References
- Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 2001; 13:290 - 301; http://dx.doi.org/10.1016/S0955-0674(00)00211-8; PMID: 11343899
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonné R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 2002; 9:891 - 901; http://dx.doi.org/10.1016/S1097-2765(02)00484-7; PMID: 11983179
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 1998; 67:265 - 306; http://dx.doi.org/10.1146/annurev.biochem.67.1.265; PMID: 9759490
- Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in Archaea. Science 2000; 288:517 - 22; http://dx.doi.org/10.1126/science.288.5465.517; PMID: 10775111
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O’Brien J, Lehrach H, Bachellerie JP, et al. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 2001; 20:2943 - 53; http://dx.doi.org/10.1093/emboj/20.11.2943; PMID: 11387227
- Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol Cell 2003; 12:113 - 23; http://dx.doi.org/10.1016/S1097-2765(03)00267-3; PMID: 12887897
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 2007; 8:209 - 20; http://dx.doi.org/10.1038/nrm2124; PMID: 17318225
- Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 2007; 35:1452 - 64; http://dx.doi.org/10.1093/nar/gkl1172; PMID: 17284456
- Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol 1999; 11:378 - 84; http://dx.doi.org/10.1016/S0955-0674(99)80053-2; PMID: 10395551
- Bertrand E, Bordonné R. Assembly and traffic of small nuclear RNPs. Prog Mol Subcell Biol 2004; 35:79 - 97; http://dx.doi.org/10.1007/978-3-540-74266-1_4; PMID: 15113080
- Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine DL, Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol Cell Biol 2000; 20:5415 - 24; http://dx.doi.org/10.1128/MCB.20.15.5415-5424.2000; PMID: 10891482
- Verheggen C, Mouaikel J, Thiry M, Blanchard JM, Tollervey D, Bordonné R, et al. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J 2001; 20:5480 - 90; http://dx.doi.org/10.1093/emboj/20.19.5480; PMID: 11574480
- Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonné R, et al. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 2002; 21:2736 - 45; http://dx.doi.org/10.1093/emboj/21.11.2736; PMID: 12032086
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000; 101:187 - 98; http://dx.doi.org/10.1016/S0092-8674(00)80829-6; PMID: 10786834
- Chari A, Paknia E, Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr Opin Cell Biol 2009; 21:387 - 93; http://dx.doi.org/10.1016/j.ceb.2009.02.004; PMID: 19286363
- Mouaikel J, Narayanan U, Verheggen C, Matera AG, Bertrand E, Tazi J, et al. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep 2003; 4:616 - 22; http://dx.doi.org/10.1038/sj.embor.embor863; PMID: 12776181
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 2003; 22:1878 - 88; http://dx.doi.org/10.1093/emboj/cdg187; PMID: 12682020
- Pradet-Balade B, Girard C, Boulon S, Paul C, Azzag K, Bordonné R, et al. CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J 2011; 30:2205 - 18; http://dx.doi.org/10.1038/emboj.2011.128; PMID: 21522132
- Jády BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 2004; 164:647 - 52; http://dx.doi.org/10.1083/jcb.200310138; PMID: 14981093
- Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell 1999; 10:2131 - 47; PMID: 10397754
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J 1998; 17:3747 - 57; http://dx.doi.org/10.1093/emboj/17.13.3747; PMID: 9649444
- Narayanan A, Lukowiak A, Jády BE, Dragon F, Kiss T, Terns RM, et al. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J 1999; 18:5120 - 30; http://dx.doi.org/10.1093/emboj/18.18.5120; PMID: 10487763
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777 - 87; http://dx.doi.org/10.1016/j.molcel.2004.11.013; PMID: 15574332
- Girard C, Verheggen C, Neel H, Cammas A, Vagner S, Soret J, et al. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J Biol Chem 2008; 283:2060 - 9; http://dx.doi.org/10.1074/jbc.M704209200; PMID: 18039666
- Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol 1997; 17:7088 - 98; PMID: 9372940
- Lyman SK, Gerace L, Baserga SJ. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 1999; 5:1597 - 604; http://dx.doi.org/10.1017/S1355838299991288; PMID: 10606270
- Westman BJ, Verheggen C, Hutten S, Lam YW, Bertrand E, Lamond AI. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol Cell 2010; 39:618 - 31; http://dx.doi.org/10.1016/j.molcel.2010.07.025; PMID: 20797632
- Colau G, Thiry M, Leduc V, Bordonné R, Lafontaine DL. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology. Mol Cell Biol 2004; 24:7976 - 86; http://dx.doi.org/10.1128/MCB.24.18.7976-7986.2004; PMID: 15340060
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 2005; 7:626 - 32; http://dx.doi.org/10.1038/ncb1263; PMID: 15908946
- Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 2007; 581:2783 - 93; http://dx.doi.org/10.1016/j.febslet.2007.05.013; PMID: 17509569
- Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci 2007; 120:1540 - 50; http://dx.doi.org/10.1242/jcs.001529; PMID: 17405816