Abstract
Understanding the basis for multipotency, whereby stem cells and other progenitors can differentiate into certain tissues and not others, provides insights into the mechanism of cell programming in development, homeostasis, and disease. We recently reported a screen of diverse chromatin marks to obtain clues about chromatin states in the multipotent embryonic endoderm. Genetic and pharmacologic tests of certain marks’ function demonstrated that the relevant chromatin modifying factors modulate the fate choice for liver or pancreas induction in the endoderm. The information about chromatin states from embryonic studies can be used to predict lineage-specific developmental potential and chromatin modifiers to enhance particular cell fate transitions from stem cells.
Chromatin States in Pluripotent Cells
Epigenetic regulation, particularly through chromatin modification and DNA methylation, plays a critical role in controlling gene regulation and cell differentiation in the development of multicellular organism. Various studies revealed that chromatin alterations accompany cell lineage specification during mammalian development.Citation1,Citation2 In the mammalian embryo, early pluripotent cells initially develop into the endoderm, ectoderm, and mesoderm germ layers. Germ layer cells are multipotent and each layer further differentiates into certain tissues and organs, but not others. Fully understanding the changes of chromatin states in cell lineage specification, which is fundamental to the studies of organogenesis and regeneration medicine, will allow the efficient induction of stem cells into desired cell lineages.Citation3
Much has been learned about the chromatin states in pluripotent cells, specifically embryonic stem (ES) cells and how such states endow the competence to self-renew or initiate particular cell programs.Citation4,Citation5 Master transcription factors control self-renewal and pluripotency by extensive chromatin binding and autoregulatory loops,Citation6-Citation8 trimethylation of histone H3 on lysine residues K4 and K27 often marks silent genes with the potential to be activated in early development,Citation9 pluripotent chromatin is more loosely structured and accessible than differentiated cell chromatinCitation10 and P300, H3K27me3 and H3K4me1 are associated with poised enhancers at genes involved in early development.Citation11
While the combined pattern of transcription factors and histone modifications helps explain the competence for all tissue programs, it is not clear how such chromatin states will be relevant to multipotent cells, in which a restricted set of cell fates can be activated and other cell fates cannot. Furthermore, it is not known whether a common “open” chromatin state exists for silent genes in multipotent cells or if the silent genes for different fates are packaged into markedly different states. Superimposed upon these issues is the question of how chromatin states allow or deny the effect of inductive signals that promote cell fates. Lineage-restricted, multipotent progenitors exist at numerous stages of development, they are crucial for homeostatic cell replacement in adult tissues, and they are often the targets of human disease, such as cancer. Thus, it is important to understand how multipotent progenitors are made competent for their particular cell fate choices.
Chromatin States in Embryonic Endoderm Development
To address the above issues, we identified and tested the function of chromatin states in the embryonic endoderm. The ventral foregut endoderm of the mammalian embryo gives rise to liver and ventral pancreas progenitors and is an experimental model for investigating chromatin and inductive signaling.Citation12,Citation13 The multipotency of the ventral foregut endoderm has been shown by the ability of mouse embryo tissue to activate either the earliest liver or pancreas genes in explants,Citation14 by the ability of improperly positioned mouse endoderm to acquire a hepatic instead of a pancreatic fate in vivoCitation15 and by the ability of individually labeled zebrafish endoderm cells to give rise to hepatic and pancreatic descendants.Citation16 Endoderm cells contain occupied DNA binding sites for FoxA and GATA factors at an enhancer of the silent Alb1 gene;Citation17,Citation18 the latter being a marker of hepatic specification.Citation17 FoxA and GATA factors can bind and expose the local chromatin in compacted nucleosome arrays in vitroCitation19 and elicit chromatin opening in vivo.Citation20,Citation21 FoxA1 and FoxA2 are both required in the endoderm to activate the hepatic programCitation22 and GATA4 and GATA6 are necessary for liver bud development.Citation23-Citation25 The early developmental binding of the factors and their ability to open chromatin has led to the proposal that such factors function as competence or pioneer factors in the endoderm.Citation12,Citation26
Recently, we found that FoxA1 and FoxA2 can recruit the corepressor Grg3, causing compaction of the local chromatin domain.Citation27 Grg3 is co-expressed in the endoderm with FoxA factors, but then is sharply downregulated during hepatic induction.Citation28 Thus liver genes in endoderm could remain competent but silent by the apparent combined action of positive (FoxA and GATA) and negative (Grg3) factors. However, the role of chromatin modifications has been unknown.
By 8.5 d gestation (~7–8 somite pairs; 7–8S), the liver genes Alb1, Afp, and Ttr are induced by BMP and FGF signals, while Pdx1, a pancreatic determination gene,Citation29-Citation31 is suppressed in the liver domain and activated in the adjacent caudal domain.Citation16,Citation32-Citation35 The BMP and FGF signals also differentially regulate Prox1, Hnf1b, and Hnf6;Citation35 which are necessary for both liver and pancreatic development. All three of these genes are expressed in endoderm spanning both the hepatic and pancreatic domains.Citation36-Citation40 By contrast, the induction of Alb1, Afp, Ttr and of Pdx1 are specific to the nascent hepatic and pancreatic domains, respectively, and thus the chromatin states of their promoters and enhancers in the endoderm should provide insight into the regulation of the liver or pancreas fate choice.
Due to the small cell numbers, 400–800 per embryo, the chromatin states in the endoderm, as well as in the mesoderm and ectoderm germ layers, have been elusive. To address this, we developed protocols for fluorescence activated cell sorting (FACS) of ventral foregut endoderm cells and for low cell number chromatin immunoprecipitation (ChIP).Citation41 We performed a screen for the presence of 15 different chromatin marks at regulatory elements for silent liver and pancreas specific genes in the endoderm. We also investigated such marks in nascent hepatoblasts and tested the functionality of the marks by genetic studies in mice.
Surprisingly, the results reveal a markedly different “pre-pattern” of chromatin states at the liver vs. pancreas regulatory elements, where the genes are poised but not active, in undifferentiated endoderm.Citation41 In contrast to the liver regulatory elements in undifferentiated endoderm, which were devoid of the positive (H3K9acK14ac) and negative (H3K27me3) marks, the pancreas regulatory elements, except area IV of the Pdx1 gene, contained both marks. Despite the diversity of chromatin marks tested, only H3K9acK14ac showed a consistent, significant increase at the liver elements when the undifferentiated endoderm cells differentiated into hepatoblasts. The co-existence of H3K9K14 hyperacetylation and H3K27me3 at the Pdx1 elements is retained in hepatoblasts. Currently, the low cell numbers preclude a sequential ChIP assay on the native embryonic cells. However, the persistence of both H3K9acK14ac and H3K27me3 marks on the silent Pdx1 gene in sorted Liv2+ hepatoblasts is consistent with their co-existence on individual genes. Such co-existence has been seen in embryonic stem cells.Citation42 The co-existence of the positive/negative marks of H3K9acK14ac and H3K27me3 may constitute a new kind of “bivalent” mark.
We next investigated histone modifiers for H3K9acK14ac and H3K27me3, to assess their roles in the liver vs. pancreas fate choice. Of the genes for mammalian lysine acetyltransferases (KATs), mice null for Gcn5l2, CBP or P300 die near gastrulation.Citation43,Citation44 By contrast, the KAT P/CAF is expressed at low levels in embryos and is dispensable, though it is expressed ubiquitously in adult cells.Citation44,Citation45 Mice heterozygous for Gcn5l2 or P300 are viable, though the latter can be lethal in certain backgrounds.Citation43,Citation44 Our genetics studies revealed that P300, but not Gcn5l2 or P/CAF, plays key roles in adding acetylation at the liver-specific regulatory elements, in liver-specific gene activation, and in modulating the liver over pancreas fate choice during embryogenesis.Citation41
Since H3K27me3 plays an important role in maintaining the silence of developmental regulatory genes,Citation46 we investigated whether diminishing this modification in the endoderm would be sufficient to alter the initiation of the pancreatic program. EZH2, a member of the PRC2 Polycomb complex, is a key histone methyltransferase for H3K27me3.Citation47 In embryos that we made deficient in Ezh2 in the endoderm, we discovered enhanced ventral pancreas specification with multiple bud-like structures, and the liver bud was smaller.Citation41 H3K9K14 acetylated chromatin and/or other locally acetylated substrates could play a role in allowing activation when the repressive H3K27me3 methyltransferase is lost. Thus, the chromatin of the pancreas gene in endoderm is set up so that simple loss of the repressive mark facilitates rapid induction. Indeed, the H3K27me3 mark is not seen at Pdx1 later in pancreas development.Citation48
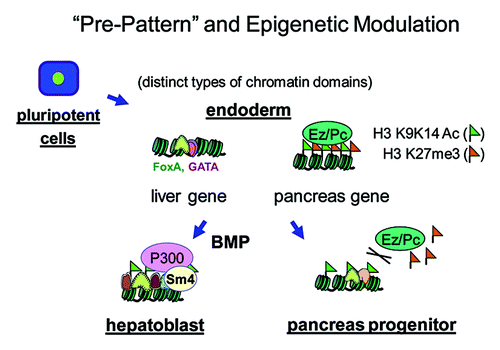
SMAD proteins are downstream effectors of BMP signalingCitation49 and can interact with P300.Citation50,Citation51 To directly test the role of SMAD4 in regulating P300 and histone acetylation, we conditionally disrupted Smad4 in the foregut endoderm.Citation35,Citation52 The study showed that the liver-inducing BMP signal is mediated by SMAD4 and P300 and results in histone acetylation at liver target elements and liver gene activation (). Our studies reveal chromatin “pre-programming” for different lineages in multipotent progenitor cells, they provide approaches that can be applied to other systems, and they provide new landmarks and molecular targets to track and modulate liver and pancreas specification from stem cells.
Figure 1. Chromatin “pre-pattern” and epigenetic modulation during liver and pancreas specification. In undifferentiated endoderm cells, the poised liver and pancreas regulatory elements have distinct chromatin “pre-patterns.” The pioneer factors of FoxA and GATA4/6 occupy liver regulatory elements, which have low or undetectable levels of the positive (H3K9acK14ac) and negative (H3K27me3) chromatin marks. By contrast, the pancreas regulatory elements of the Pdx1 gene, except for area IV, contain both marks. BMP signaling induces SMAD4 to recruit P300 to the liver elements, stimulating histone acetylation there and promoting liver specification. During this period, EZH2 restricts the specification of pancreas progenitors.
Distinct Types of Positive-Negative Components at Poised Genes
We found the co-existence of the positive and negative marks of H3K9acK14ac and H3K27me3, respectively, at the silent Pdx1 elements in the endoderm, where the Pdx1 gene is competent or poised, to be activated. As described above, bivalent domains of H3K4me3/H3K27me3 often mark poised early developmental genes.Citation9 Also, the positive and negative effectors FoxA/GATA and Groucho/Grg3, respectively, appear to co-exist at the silent liver regulatory elements in the endoderm, where the genes are poised to be activated. Other examples of positive and negative components at poised genes include transcription factors bound to enhancers or promoters of silent genes in ES and iPS cells that are activated late in differentiation. In ES cells, the enhancer of the hypomethylated, silent Alb1 gene is occupied by FoxD3, which represses transcription.Citation53 During endoderm induction, FoxD3 is replaced by FoxA1, all prior to Alb1 activation.Citation54 Similarly, in nascent endoderm, the induced FoxA1 binds within the methylation-free Afp distal promoter, which is necessary for Afp activation during stem cell differentiation.Citation55 The positive factors are bound at unmethylated CpG residues amidst many methylated CpGs, which are typically considered repressive; thus together constituting a distinct kind of positive/negative mark. In ES cells, an intergenic enhancer of λ5-VpreB1 genes,Citation56 which are expressed in pro- and pre-B cells, is occupied with Sox2 and FoxD3. Sox2 acts as a positive factor, contributing to the establishment of the H3K4me2 mark, whereas FoxD3 is a negative factor, repressing intergenic transcription from the enhancer.Citation57 Another example is the pre-loading of RNA polymerase II, but its pausing, at silent genes with the potential to be activated in development.Citation58-Citation61 Since the poising of genes by simultaneous positive and negative chromatin components may be general, but involving diverse mechanisms, we suggest broadening the definition of “bivalency” in stem and progenitor cell chromatin to refer to the simultaneous presence of positive and negative components at poised genes, and not solely to the original H3K4me3/H3K27me3.Citation9 By having the poised state result from a dynamic equilibrium between positive and negative effectors, it may facilitate the synchronous activation of genes in the rapidly developing embryo.
Future Directions
While in principle it should be possible to characterize chromatin states in germ layer cells derived from embryonic stem cells in vitro, early in this study we found that the chromatin states in ES-derived endoderm can be different from those we have characterized in vivo (C.-R. Xu, P. Gadue, C. Nostro, G. Keller and K.Z. unpublished data). Knowing the chromatin states in native embryonic tissue can be used to define benchmarks of proper progenitor cell programming in stem cell studies and molecular targets for enzymatic modifiers that function in the cell fate transitions. It will be informative to perform genome wide analysis for H3K9/K14 acetylation and H3K27me3 in the undifferentiated endoderm, as the marks were found to be different between in vitro and in vivo populations. There may be key genes that demonstrate altered chromatin marks in ESC-derived endoderm compared with in vivo-derived endoderm that could limit in the in vitro differentiation process. The initial analysis of the genome wide ChIP data will focus on genes known to be involved in pancreatic and liver.
The mechanistic studies to date have focused on the chromatin regulation of hepatic specification. We don’t yet know how the repressive chromatin mark of H3K27me3 is lost from Pdx1 regulatory elements, which appears to be required for initiation of the pancreatic program. We hypothesize that cell signaling regulates the recruitment of a histone demethylase to Pdx1 elements upon pancreas specification. Such information could provide insight into ways to enhance pancreas specification from stem cells.
Our work has provided a paradigm for using antagonists of histone modifiers at specific developmental stages to alter cell lineage specification. This was enabled by a careful analysis of chromatin prepatterns of native progenitor cells in vivo. Such approaches can be taken with any stem or progenitor cell type to predict developmental potential and enhance differentiation of stem cells.
Acknowledgments
We thank the members of the Zaret group for advice and comments on the manuscript and ongoing projects, and Eileen Pytko for help in preparing the manuscript. This work was supported by NIH grants R37GM36477 and U01DK072503 to K.S.Z, and K01DK093886 to C.-R.X.
References
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol 2010; 28:1079 - 88; http://dx.doi.org/10.1038/nbt.1684; PMID: 20944600
- Young RA. Control of the embryonic stem cell state. Cell 2011; 144:940 - 54; http://dx.doi.org/10.1016/j.cell.2011.01.032; PMID: 21414485
- Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev 2011; 21:140 - 6; http://dx.doi.org/10.1016/j.gde.2011.01.015; PMID: 21316216
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol 2009; 10:526 - 37; http://dx.doi.org/10.1038/nrm2727; PMID: 19603040
- Chambers D, Mason I. Expression of sprouty2 during early development of the chick embryo is coincident with known sites of FGF signalling. Mech Dev 2000; 91:361 - 4; http://dx.doi.org/10.1016/S0925-4773(99)00288-9; PMID: 10704865
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000; 24:372 - 6; http://dx.doi.org/10.1038/74199; PMID: 10742100
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005; 122:947 - 56; http://dx.doi.org/10.1016/j.cell.2005.08.020; PMID: 16153702
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008; 132:1049 - 61; http://dx.doi.org/10.1016/j.cell.2008.02.039; PMID: 18358816
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 26; http://dx.doi.org/10.1016/j.cell.2006.02.041; PMID: 16630819
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10:105 - 16; http://dx.doi.org/10.1016/j.devcel.2005.10.017; PMID: 16399082
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470:279 - 83; http://dx.doi.org/10.1038/nature09692; PMID: 21160473
- Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet 2002; 3:499 - 512; http://dx.doi.org/10.1038/nrg837; PMID: 12094228
- Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet 2008; 9:329 - 40; http://dx.doi.org/10.1038/nrg2318; PMID: 18398419
- Deutsch G, Jung J, Zheng M, Lóra J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development 2001; 128:871 - 81; PMID: 11222142
- Bort R, Martinez-Barbera JP, Beddington RS, Zaret KS. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 2004; 131:797 - 806; http://dx.doi.org/10.1242/dev.00965; PMID: 14736744
- Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell 2008; 15:738 - 48; http://dx.doi.org/10.1016/j.devcel.2008.08.019; PMID: 19000838
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 1996; 10:1670 - 82; http://dx.doi.org/10.1101/gad.10.13.1670; PMID: 8682297
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development 1998; 125:4909 - 17; PMID: 9811575
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 2002; 9:279 - 89; http://dx.doi.org/10.1016/S1097-2765(02)00459-8; PMID: 11864602
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005; 122:33 - 43; http://dx.doi.org/10.1016/j.cell.2005.05.008; PMID: 16009131
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008; 132:958 - 70; http://dx.doi.org/10.1016/j.cell.2008.01.018; PMID: 18358809
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature 2005; 435:944 - 7; http://dx.doi.org/10.1038/nature03649; PMID: 15959514
- Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development 2005; 132:4005 - 14; http://dx.doi.org/10.1242/dev.01978; PMID: 16079152
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol 2005; 25:2622 - 31; http://dx.doi.org/10.1128/MCB.25.7.2622-2631.2005; PMID: 15767668
- Watt AJ, Zhao R, Li J, Duncan SA. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol 2007; 7:37; http://dx.doi.org/10.1186/1471-213X-7-37; PMID: 17451603
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011; 25:2227 - 41; http://dx.doi.org/10.1101/gad.176826.111; PMID: 22056668
- Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 2007; 28:291 - 303; http://dx.doi.org/10.1016/j.molcel.2007.10.002; PMID: 17964267
- Santisteban P, Recacha P, Metzger DE, Zaret KS. Dynamic expression of Groucho-related genes Grg1 and Grg3 in foregut endoderm and antagonism of differentiation. Dev Dyn 2010; 239:980 - 6; http://dx.doi.org/10.1002/dvdy.22217; PMID: 20108349
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994; 371:606 - 9; http://dx.doi.org/10.1038/371606a0; PMID: 7935793
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 1995; 121:11 - 8; PMID: 7867492
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996; 122:983 - 95; PMID: 8631275
- Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science 1999; 284:1998 - 2003; http://dx.doi.org/10.1126/science.284.5422.1998; PMID: 10373120
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev 2001; 15:1998 - 2009; http://dx.doi.org/10.1101/gad.904601; PMID: 11485993
- Calmont A, Wandzioch E, Tremblay KD, Minowada G, Kaestner KH, Martin GR, et al. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell 2006; 11:339 - 48; http://dx.doi.org/10.1016/j.devcel.2006.06.015; PMID: 16950125
- Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science 2009; 324:1707 - 10; http://dx.doi.org/10.1126/science.1174497; PMID: 19556507
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet 2000; 25:254 - 5; http://dx.doi.org/10.1038/76996; PMID: 10888866
- Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev 2002; 118:147 - 55; http://dx.doi.org/10.1016/S0925-4773(02)00240-X; PMID: 12351178
- Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol 2003; 258:105 - 16; http://dx.doi.org/10.1016/S0012-1606(03)00115-5; PMID: 12781686
- Haumaitre C, Barbacci E, Jenny M, Ott MO, Gradwohl G, Cereghini S. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci U S A 2005; 102:1490 - 5; http://dx.doi.org/10.1073/pnas.0405776102; PMID: 15668393
- Lokmane L, Haumaitre C, Garcia-Villalba P, Anselme I, Schneider-Maunoury S, Cereghini S. Crucial role of vHNF1 in vertebrate hepatic specification. Development 2008; 135:2777 - 86; http://dx.doi.org/10.1242/dev.023010; PMID: 18635606
- Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science 2011; 332:963 - 6; http://dx.doi.org/10.1126/science.1202845; PMID: 21596989
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8:532 - 8; http://dx.doi.org/10.1038/ncb1403; PMID: 16570078
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 1998; 93:361 - 72; http://dx.doi.org/10.1016/S0092-8674(00)81165-4; PMID: 9590171
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 2000; 26:229 - 32; http://dx.doi.org/10.1038/79973; PMID: 11017084
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, et al. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A 2000; 97:11303 - 6; http://dx.doi.org/10.1073/pnas.97.21.11303; PMID: 11027331
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6:838 - 49; http://dx.doi.org/10.1038/nrm1761; PMID: 16261189
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039 - 43; http://dx.doi.org/10.1126/science.1076997; PMID: 12351676
- van Arensbergen J, García-Hurtado J, Moran I, Maestro MA, Xu X, Van de Casteele M, et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res 2010; 20:722 - 32; http://dx.doi.org/10.1101/gr.101709.109; PMID: 20395405
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003; 425:577 - 84; http://dx.doi.org/10.1038/nature02006; PMID: 14534577
- Pouponnot C, Jayaraman L, Massagué J. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem 1998; 273:22865 - 8; http://dx.doi.org/10.1074/jbc.273.36.22865; PMID: 9722503
- de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, et al. The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem 2000; 275:2115 - 22; http://dx.doi.org/10.1074/jbc.275.3.2115; PMID: 10636916
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development 2004; 131:3501 - 12; http://dx.doi.org/10.1242/dev.01248; PMID: 15215210
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, et al. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci U S A 2007; 104:12377 - 82; http://dx.doi.org/10.1073/pnas.0704579104; PMID: 17640912
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev 2009; 23:2824 - 38; http://dx.doi.org/10.1101/gad.1861209; PMID: 20008934
- Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem 2010; 285:16135 - 44; http://dx.doi.org/10.1074/jbc.M109.088096; PMID: 20348100
- Minaee S, Farmer D, Georgiou A, Sabbattini P, Webster Z, Chow CM, et al. Mapping and functional analysis of regulatory sequences in the mouse lambda5-VpreB1 domain. Mol Immunol 2005; 42:1283 - 92; http://dx.doi.org/10.1016/j.molimm.2005.01.003; PMID: 15950724
- Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, et al. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell 2010; 7:114 - 26; http://dx.doi.org/10.1016/j.stem.2010.05.020; PMID: 20621055
- Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature 2007; 450:198 - 202; http://dx.doi.org/10.1038/nature06324; PMID: 17994086
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A 2008; 105:7762 - 7; http://dx.doi.org/10.1073/pnas.0802406105; PMID: 18505835
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 2010; 327:335 - 8; http://dx.doi.org/10.1126/science.1181421; PMID: 20007866
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell 2010; 141:432 - 45; http://dx.doi.org/10.1016/j.cell.2010.03.030; PMID: 20434984