Abstract
In open mitosis the nuclear envelope (NE) reassembles at the end of each mitosis. This process involves the reformation of the nuclear pore complex (NPC), the inner and outer nuclear membranes, and the nuclear lamina. In human cells cell cycle-dependent NE subdomains exist, characterized as A-type lamin-rich/NPC-free or B-type lamin-rich/NPC-rich, which are initially formed as core or noncore regions on mitotic chromosomes, respectively. Although postmitotic NE formation has been extensively studied, little is known about the coordination of NPC and NE assembly. Here, we report that the nucleoporin ELYS/Mel28, which is crucial for postmitotic NPC formation, is essential for recruiting the lamin B receptor (LBR) to the chromosomal noncore region. Furthermore, ELYS/Mel28 is responsible for focusing of A-type lamin-binding proteins like emerin, Lap2α and the barrier-to-autointegration factor (BAF) at the chromosomal core region. ELYS/Mel28 biochemically interacts with the LBR in a phosphorylation-dependent manner. Recruitment of the LBR depends on the nucleoporin Nup107, which interacts with ELYS/Mel28 but not on nucleoporin Pom121, suggesting that the specific molecular interactions with ELYS/Mel28 are involved in the NE assembly at the noncore region. The depletion of the LBR affected neither the behavior of emerin nor Lap2α indicating that the recruitment of the LBR to mitotic chromosomes is not involved in formation of the core region. The depletion of ELYS/Mel28 also accelerates the entry into cytokinesis after recruitment of emerin to chromosomes. Our data show that ELYS/Mel28 plays a role in NE subdomain formation in late mitosis.
Introduction
In eukaryotes the nuclear envelope (NE) separates the nucleoplasm from the cytoplasm to coordinate the spatial and temporal regulation of genomic functions.Citation1,Citation2 Therefore, it is important to understand how the NE is formed, as this process is crucial to determine the overall architecture and function of the nucleus.
In metazoans the NE consists of the outer nuclear membrane (ONM), which fuses with the inner nuclear membrane (INM)Citation3 at places where the nuclear pore complexes (NPC) are embedded.Citation4,Citation5 The INM is attached to the nuclear lamina, a meshwork of A- and B-type lamins.Citation6 The NPC is a 125-MDa sophisticated assembly of ~30 nucleoporins (Nups),Citation7-Citation9 subdivided into scaffold nucleoporins that constitute inner and outer rings, peripheral nucleoporins that constitute cytoplasmic fibrils, the nuclear basket, the central channel forming a permeability barrier to mediate the selective nucleocytoplasmic exchange of moleculesCitation10-Citation12 and pore membrane proteins.Citation13-Citation15 While the ONM is continuous with the endoplasmic reticulum (ER), the INM has a subset of more than 60 INM proteins that interact with A- or B-type lamins.Citation16 The nuclear lamina, together with INM proteins, is important for determining nuclear shape and integrity, and is involved in chromatin organization. These properties of the nuclear lamina link the NE to various human diseases, collectively called laminopathies.Citation17,Citation18
Human cells possess two types of lamin: A-type and B-type. Lamins are type V intermediate filaments and contain an N-terminal globular head domain, a central α-helical rod domain, and a C-terminal tail domain.Citation6 Our group previously reported that in human cells, A-type lamins are enriched in the NPC-free regions of the NE, the so-called pore-free islands, while B-type lamins are enriched in the NPC-rich region of the NE.Citation19 Such NE subdomains appear and disappear periodically during the cell cycle. Different properties of A- and B-type lamins, which form separate, but interacting networks or subdomains at the NE have also been reported in different species and have been proposed to be important for the nuclear function.Citation20-Citation22
In eukaryotic cells, which undergo open mitosis, the NE, NPC and nuclear lamina disassemble at prophase to allow mitotic spindle formation and reassemble at the end of mitosis.Citation23 In human dividing cells the A- and B-type lamin-rich subdomains are formed at the end of mitosis, where they correspond to the core and noncore regions of mitotic chromosomes, respectively.Citation24,Citation25 The core region is a part of the mitotic chromosomes next to the spindle pole- and the central spindle areas in telophase.Citation25 The barrier-to-autointegration factor (BAF), an A-type lamin-binding protein, initiates formation of the core region by sequentially recruiting LEM domain-containing proteins, where L, E and M stand for the INM proteins Lap2α, emerin and MAN1, respectively.Citation26
The noncore region is characterized by NPCs, lamin B and the lamin B receptor (LBR). The LBR is one of the best-studied INM proteins and is reported to interact directly with DNACitation27 and many other proteins, such as lamin B,Citation28 heterochromatin-binding protein 1 (HP1),Citation29 HA95,Citation30 heterochromatic methyl-binding protein (MeCP1)Citation31 and importin βCitation32,Citation33 through its hydrophilic N-terminus. The LBR displays a C-14 sterol reductase activity at its C-terminus.Citation34 Furthermore, the LBR has been reported to play an important role in NE formation in Xenopus and sea urchin in vitro systems,Citation32,Citation35,Citation36 while the function of human LBR in NE formation has been described as redundant along with other INM proteins.Citation37 Moreover, human LBR is linked to the Pelger–Huët anomaly (PHA) through its function in granulocyte differentiation.Citation38
The nucleoporin ELYS/Mel28, initially identified as a putative transcription factor in mouse,Citation39 is essential in postmitotic NPC assembly.Citation40-Citation44 The binding of ELYS/Mel28 to mitotic chromosomes through its AT-hook domain was suggested to trigger NPC formation by recruitment of the Nup107-160 complex, followed by recruitment of transmembrane nucleoporin Pom121 and the Nup205–93 complex.Citation44-Citation46 These nucleoporins configure the NPC scaffold. The NPC matures as peripheral nucleoporins become incorporated to construct cytoplasmic fibrils, nuclear baskets and permeability barriers.Citation47 Recently, ELYS/Mel28 was shown to have a second function in mitosis beyond NPC assembly. ELYS/Mel28 localizes with the Nup107-160 complex to kinetochores in mitosis.Citation42 The Nup107-160 complex was suggested to play a crucial role for the mitotic spindle function,Citation48-Citation50 and its components such as Seh1 were shown to regulate the centromeric localization of the chromosomal passenger complex (CPC).Citation51 The CPC consists of INCENP, survivin, borealin and Aurora B kinase and is involved in the bipolar attachment of microtubule (MT) fibers at kinetochores, chromosome condensation, the spindle assembly checkpoint (SAC) and cytokinesis.Citation52,Citation53 It is unclear if ELYS/Mel28 has parallel functions to the Nup107-160 complex with which it interacts.Citation42
Several reports provide evidence that lamins, the INM proteins and NPC distribution are closely connected structurally and functionally during interphase.Citation19,Citation54-Citation60 However, how reformation of NPCs and the NE is coordinated on chromosomes during mitosis is not understood.
In this study, we examined the coordination of the postmitotic assembly of NPCs and the NE using HeLa cells, which display a clear divergence of A-type and B-type lamins and their interacting INM proteins on mitotic chromosomes. Through live cell imaging and the small interfering RNA (siRNA) technique, we found that recruitment of the LBR to the chromosomal noncore region and the focusing of emerin at the chromosomal core region were both dependent on ELYS/Mel28. The function of ELYS/Mel28 on NE assembly at the noncore region probably relies on a protein-protein interaction, besides its known interaction with the Nup107-160 complex, because immunoprecipitation (IP) experiments revealed that ELYS/Mel28 interacts specifically with the LBR in a phosphorylation-dependent manner. Behaviors of emerin or Lap2α were not affected upon depletion of the LBR, suggesting that the effect of ELYS on the recruitment of the LBR to the noncore region is different from its effects on INM proteins at the core region. Furthermore, the depletion of ELYS/Mel28 accelerated progression of late mitosis, particularly after the recruitment of emerin onto the chromosomes, which could be partially responsible for the improper formation of the core region. Our data show, that ELYS/Mel28 plays a role in events during late mitosis, including the NE subdomain formation.
Results
The depletion of ELYS/Mel28 perturbs targeting of the LBR to chromosomes and focusing of emerin at the chromosomal core region in post-mitosis
To examine the function of ELYS/Mel28 in postmitotic NE reformation, we followed recruitment and localization of INM proteins to mitotic chromosomes using live imaging with HeLa cells stably expressing the yellow fluorescent protein (YFP) derivative Venus fused to the LBR and the super enhanced cyan fluorescent protein (SECFP) fused to emerin (). We compared control cells (control: either untreated or transfected with control oligo; see also Fig. S1) with cells depleted of ELYS/Mel28 (ELYS/Mel28 kd) by RNA interference (RNAi). The position of each cell observed with live imaging was marked and checked for an efficient depletion of ELYS/Mel28 by immediate immunofluorescence (IF) staining with ELYS/Mel28 antibody after live observation (see legend of ). Comparing the signal intensities of ELYS/Mel28 by IF staining () in control cells and cells depleted of ELYS/Mel28 showed an average depletion efficiency of 80%, which is in agreement with the results obtained by western blotting experiments (Fig. S2).
Figure 1. The targeting of the LBR depends on ELYS/Mel28 and the Nup107-160 complex but not on the nucleoporin Pom121. Immunofluorescence (IF) staining of mitotic cells depleted of (A) ELYS/Mel28 for 50 h, (B) Pom121 for 50 h, (C) or Nup107 after double transfection for 75–80 h. Two different siRNA oligos were used to for ELYS/Mel28 depletion (I and II). (D) The depletion efficiency of ELYS/Mel28, Pom121 and Nup107-Venus, observed in (A–C) respectively, was evaluated by comparing the IF signal intensities of the control cells and the depleted cells. (E) Endogenous Pom121, ELYS/Mel28, and LBR were detected by IF staining after LBR-depletion with RNAi (LBR kd). (F) Endogenous Lamin B, LBR, and Nup62 were detected by IF staining in ELYS/Mel28-depleted cells. Scale bars = 10 µm. The pictures are projections of image stacks (distance = 0.2 µm; three images).
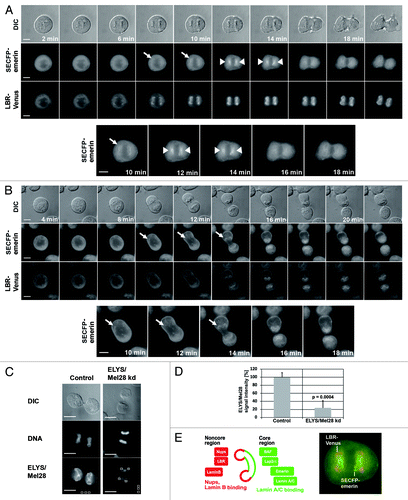
In the control cells, the LBR-Venus bound to the chromosomal noncore regions 8 min after anaphase onset (AO) (). From 8–14 min after AO, the LBR-Venus signal intensity increased. SECFP-emerin, like the LBR-Venus, was targeted to outer chromosome regions ~8 min after AO, but most of the chromosome signals of emerin focused at the core region of mitotic chromosomes ~12–14 min after AO, as reported previouslyCitation24 (, initial targeting of emerin is shown by arrows, focusing at the chromosomal core region shown by arrow heads, see also enlarged figures of emerin). About 20 min after AO, the SECFP-emerin and LBR-Venus signal slightly dispersed throughout the newly formed NE.
In ELYS/Mel28-depleted cells, chromosome recruitment of the LBR-Venus was strongly impaired (). Western blotting demonstrated that ELYS/Mel28 reduction did not affect protein levels of the LBR (Fig. S2A and B). On the other hand, in ELYS/Mel28-depleted cells, the SECFP-emerin signal appeared at the noncore region similarly as in control cells after AO, but unlike control cells, it did not focus to the core region and significant fluorescent signals remained all around the chromosomes (, see enlarged figures of SECFP-emerin). Loss of ELYS/Mel28 did not affect the timing of SECFP-emerin targeting to mitotic chromosomes after AO, but ELYS/Mel28-depleted cells completed mitosis faster than control cells (Fig. S1C). Quantified results clearly showed that in ELYS/Mel28-depleted cells, the time from the chromosome recruitment of emerin to cytokinesis (judged by DIC) was significantly reduced (Fig. S1C). These results show that the depletion of ELYS/Mel28 affects late mitotic events in NE-subdomain formation.
The recruitment of the LBR to mitotic chromosomes is dependent on ELYS/Mel28 and the Nup107-160 complex
To understand if ELYS/Mel28 specifically affects the recruitment of the LBR to mitotic chromosomes rather than binding of the LBR to other nucleoporins that interact with ELYS/Mel28, we tested the dependency of the recruitment of the LBR on different nucleoporins.
Transfection with two different siRNA oligos for ELYS/Mel28, which depleted ELYS/Mel28 by about 70–80% (, and Fig. S2B) severely impaired the recruitment of the endogenous LBR to mitotic chromosomes. As postmitotic NPC formation was inhibited, the Pom121 signal was absent from mitotic chromosomes and accumulated in the cytoplasm in ELYS/Mel28-depleted cells, as reported previously.Citation42 The depletion of Pom121 by siRNA did not affect the recruitment and localization of the LBR to chromosomes (; Fig. S2C), indicating that ELYS/Mel28 had a primary effect on LBR recruitment.
Figure 2. The targeting of the LBR depends on ELYS/Mel28 and the Nup107–160 complex but not on the nucleoporin Pom121. Immunofluorescence (IF) staining of mitotic cells depleted of (A) ELYS/Mel28 for 50 h, (B) Pom121 for 50 h, (C) or Nup107 after double transfection for 75–80 h. Two different siRNA oligos were used to for ELYS/Mel28 depletion (I and II). (D) The depletion efficiency of ELYS/Mel28, Pom121 and Nup107-Venus, observed in (A–C) respectively, was evaluated by comparing the IF signal intensities of the control cells and the depleted cells. (E) Endogenous Pom121, ELYS/Mel28, and LBR were detected by IF staining after LBR-depletion with RNAi (LBR kd). (F) Endogenous Lamin B, LBR, and Nup62 were detected by IF staining in ELYS/Mel28-depleted cells. Scale bars = 10 µm. The pictures are projections of image stacks (distance = 0.2 µm; three images).
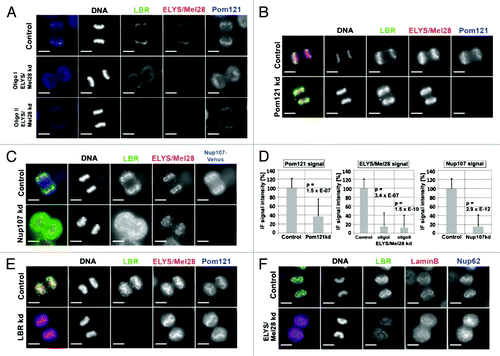
Since the Nup107-160 complex interacts with ELYS/Mel28, we examined if its depletion affected localization of the LBR. We depleted Nup107, as a representative for the Nup107-160 complex, by RNAi in cells stably expressing Nup107-Venus. IF staining showed that the binding of the LBR to mitotic chromosomes was reduced when Nup107 was depleted about 80% by RNAi (; Fig. S2D). Notably, the localization of ELYS/Mel28 was also affected by the Nup107-depletion; its binding to chromosomes was decreased and ELYS/Mel28 was partially dispersed into the cytoplasm. Conversely, the depletion of the LBR did not change the recruitment of ELYS/Mel28, Pom121 or other nucleoporins such as Nup153, Nup62 and Nup214 to mitotic chromosomes (; Figs. S3A and S3C). Furthermore, the functionality of interphase NPCs in the LBR-depleted cells was not changed when examined in an in vitro transport assay.Citation61 These results show that the LBR is not involved in postmitotic NPC formation (Fig. S3D).
Consistent with the previous data, the LBR targets mitotic chromosomes earlier than B-type lamin in anaphase.Citation62,Citation63 The depletion of ELYS/Mel28 or LBR did not affect the chromosome binding of B-type lamin (; Fig. S3B), indicating that ELYS/Mel28 specifically targets the LBR to mitotic chromosomes.
The LBR and ELYS/Mel28 interact in a phosphorylation-dependent manner
Next, we examined the interaction between the LBR and ELYS/Mel28 and the Nup107-160 complex. The LBR C-terminally fused to EGFP (LBR-EGFP) and stably expressed in HeLa cells () was immunoprecipitated with anti GFP-antibodies, and coprecipitation of ELYS/Mel28 and Nup107 was examined. Due to the insoluble properties of NPC and NE proteins, solubilization and extraction of these components were only successful with high-salt and detergent conditions (see Materials and Methods). Immunoprecipitation of LBR–EGFP from unsynchronized extracts did not yield coprecipitation of ELYS/Mel28 or other nucleoporins (, left blot). IP experiments using mitotic cell extracts, prepared either from cells synchronized to prometaphase with nocodazole (, middle blot) or released from nocodazole for 60 min to accumulate cells between metaphase and telophase (, right blot), showed that LBR-EGFP coprecipitates with ELYS/Mel28, but with decreased efficiency as mitosis progressed to telophase. No coprecipitation was observed with unsynchronized extracts (, left blot). Under the same conditions, LBR-EGFP neither coprecipitated Nup107, other nucleoporins, nor lamin B (, left, middle and right blot). The endogenous LBR coprecipitated with LBR-EGFP from all cell extracts, but with less efficiency in mitotic extracts.
Figure 3. The interaction between ELYS/Mel28 and the LBR is phosphorylation-dependent. (A) Coprecipitation of LBR–EGFP and ELYS/Mel28 was examined from extracts of unsynchronized cells (Unsynchronized), cells synchronized to prometaphase with nocodazole treatment (Prometaphase), and cells released from nocodazole treatment for 60 min (Meta/Telophase). The extracts were incubated with Protein G beads bound with (Anti-GFP-AB) or without (control) anti-GFP antibody. Starting materials (Input), bound fraction to beads (IP), and unbound fractions (unbound) were subjected to SDS-PAGE, followed by western blotting to detect LBR, ELYS/Mel28, Nup107, and lamin B. The upper membranes were probed with Mab414 antibody to detect FG-nucleoporins. (B) Phosphorylation-dependency of the interaction between the LBR and ELYS/Mel28 was tested by addition of either λ-protein phosphatase (λ-PPase) or phosphatase inhibitors to cell extract prepared from prometaphase synchronized cells (see Materials and Methods). HeLa cells (HeLa), or HeLa cells stably expressing LBR-EGFP (HeLa LBR-EGFP) were used. Asterisks mark nonspecific signals. (C) A still image of HeLa cells stably expressing LBR-EGFP used in A and B. Scale bar = 10 µm. (D) LBR contains eight transmembrane (TM) regions. The GST-LBR211aa-GFP fusion construct contains the soluble, nucleoplasmic N-terminus of LBR, including a bipartite nuclear localization sequence (NLS). (E) Pull down experiments performed with GST-LBR211aa-GFP from unsynchronized HeLa cell extract. (F) Same as E, except that pull down was performed from prometaphase synchronized extract with nocodazole treatment.
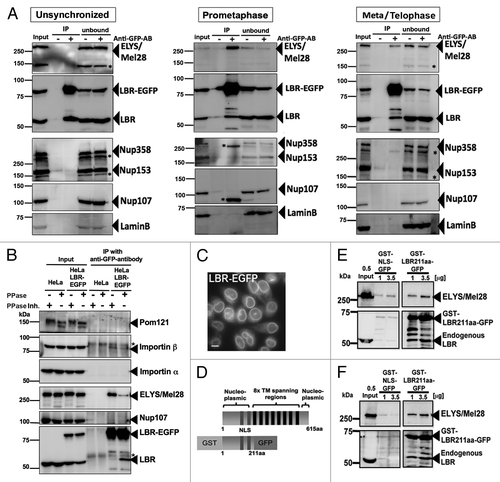
The LBR is phosphorylated by CDK1/cyclin B kinase and SRPK1 kinase in mitosis.Citation64,Citation65 To test the effect of phosphorylation in the interaction between the LBR and ELYS/Mel28, IP experiments were performed on mitotic extracts treated with either phosphatase or phosphatase inhibitors, for which the Pom121 signal served as an internal phosphorylation control (). ELYS/Mel28 coprecipitated with the LBR when a phosphatase inhibitor was added, whereas the addition of phosphatase hampered the coprecipitation. In contrast, the addition of phosphatase enhanced the coprecipitation of the endogenous LBR while addition of phosphatase inhibitor decreased the coprecipitation. In agreement with a previously reported interaction between the LBR and importin β,Citation32,Citation33 importin β co-precipitated with LBR-EGFP, while importin α did not.
We further performed pull-down experiments with a recombinant protein, consisting of the soluble N-terminal fragment of the LBR (amino acids 1–211) fused to GST and GFP (GST-LBR211aa-GFP; ) from mitotic or unsynchronized extracts. Since GST-LBR211aa-GFP contains a nuclear localization sequence (NLS),Citation66 we used a control fragment containing the SV40 T antigen NLS sequence (GST-NLS-GFP). ELYS/Mel28 bound to GST-LBR211aa-GFP from mitotic extracts () and unsynchronized extracts (). These pull-down experiments show that the LBR interacts with ELYS through its N-terminus (amino acids 1–211).
The biochemical interaction between the LBR and ELYS, which is phosphorylation-dependent, could be involved in the recruitment of the LBR to mitotic chromosomes as shown in and (see Discussion).
ELYS/Mel28 is involved in proper localization of BAF, Lap2α, and lamin A/C to the chromosomal core region
The depletion of ELYS/Mel28 altered the recruitment of emerin to the chromosomal core region ( and ). We next examined whether the depletion affects other INM components known to concentrate at the chromosomal core region in postmitosis. For this, we examined the localization of endogenous Lap2α (; Fig. S4D), lamin A/C (), and BAF (; Fig. S4C and S4D) with IF staining in control cells or cells depleted of ELYS/Mel28, using two RNAi oligos as described above. In control cells, Lap2α and BAF often accumulated first at both sides of the core region next to the central spindle- and spindle pole area, and then focused at the core region next to the central spindle area (Fig. S4A and S4B). In ELYS/Mel28-depleted cells, Lap2α and BAF either bound all around the chromosomes instead of accumulating mainly at the core region, or only at the core region next to the spindle pole area, which is the opposite to what was observed in control cells (, ; Fig. S4D: see arrows for localization at the spindle pole side). Lamin A/C, like Lap2α and BAF, was also affected by ELYS/Mel28 depletion, but differently, as it bound all around mitotic chromosomes in telophase and did not concentrate to a specific side or region (). The protein expression levels of lamin A/C were unchanged (Fig. S2B). More than 90% of ELYS/Mel28-depleted cells displayed aberrant localization of A-type lamin and Lap2α, as described above ().
Figure 5. The depletion of the LBR does not inhibit focusing of emerin and Lap2α at the core region. Live imaging was performed with cells stably expressing LBR-Venus/SECFP-emerin, like described in . (A) Untreated cells (control). (B) Cells depleted of LBR by RNAi for 48 h (LBR kd). Arrowheads show SECFP-emerin accumulating at the core region, and arrows show initial targeting of SECFP-emerin to the chromosomes. Scale bars = 10 µm. (C) The duration time of SECFP-emerin at the core region as observed in live imaging in , (n = 24 for the control, n = 15 for the ELYS/Mel28 kd, n = 10 for the LBR kd). (D) Endogenous Lap2α was examined by IF staining in HeLa cells stably expressing LBR-Venus/SECFP-emerin depleted of ELYS/Mel28 as in , or depleted of LBR as in . Scale bars = 10 µm. (E) Quantification results of D are shown. Control cells (n = 40), ELYS/Mel28 kd (n = 39) and LBR kd cells (n = 25).
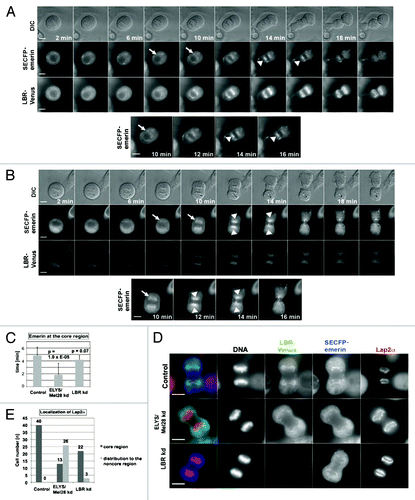
Figure 4. ELYS/Mel28 is involved in proper localization of BAF, Lap2α, and lamin A/C to the chromosomal core region. (A and B) Endogenous Lap2α, Pom121, Lamin A/C, Nup62, or ELYS/Mel28 was examined in HeLa cells (Control) or HeLa cells depleted of ELYS/Mel28 for 60 h (ELYS kd) by IF staining. Pictures are projections of image stacks (distance = 0.2 µm; three images). Scale bars = 10 µm. (C) Quantification results of (A and B) are shown. Localization at the core region of lamin A/C (n = 33 for the control, n = 29 for the ELYS/Mel28 kd) and Lap2α (n = 49 for the control, n = 38 for the ELYS/Mel28 kd). (D) Endogenous LBR and BAF were examined in HeLa cells (control) or HeLa cells depleted of ELYS/Mel28 for 50 h (ELYS kd) by IF staining. The pictures were deconvoluted using Softworx. Scale bars = 10 µm. White arrows in (A, B and D) indicate the localization of the A-type lamin-binding proteins to the chromosomal region adjacent to the spindle pole area.
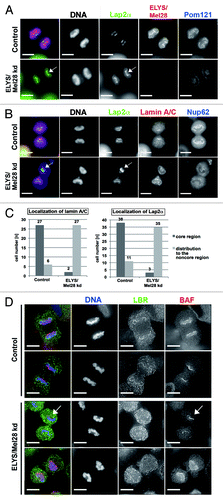
Observation of BAF in control cells in telophase (; Fig. S4A) as judged by the post-anaphase progression (PA) indexCitation67 (Fig. S4C) revealed that BAF localized to a clearly broad chromosomal region in cells (Fig. S4A and 4D, upper panel of control sample) with low PA indices (Fig. S4C), before it became focused to the core region (, lower panel of control sample) in cells with high PA indices (Fig. S4C). A high PA index indicates later mitotic stages (see figure legend of Fig. S4). Conversely, in ELYS/Mel28-depleted cells, BAF did not focus at the core region but stayed localized all around chromosomes (, upper panel of ELYS/Mel28 kd sample) in cells with high PA indices (Fig. S4C). In some cases, however, BAF bound at the spindle pole side of chromosomes, which is opposite from the control cells, where BAF focuses at the chromosomal region next to the central spindle area (, lower panel of ELYS/Mel28 kd sample; Fig. S4C).
Our results demonstrate that the depletion of ELYS/Mel28 affected the behavior of multiple INM proteins that concentrate to the chromosomal core region.
The depletion of ELYS/Mel28 affects NE formation of the chromosomal noncore region and core region differently
Depletion of ELYS/Mel28 specifically inhibited recruitment of the LBR to mitotic chromosomes, whereas lamin B was unaffected. Additionally, multiple INM proteins like BAF, emerin, Lap2α and A-type lamin, which bind to the core region, spread to the noncore region, when ELYS/Mel28 was reduced. Moreover, the depletion of ELYS/Mel28 clearly shortened the time from the chromosome recruitment of emerin to cytokinesis. We tried to address whether these effects of ELYS/Mel28-depletion were due to a failed recruitment of the LBR or caused via a specific function of ELYS/Mel28.
Therefore, we examined the effect of LBR-depletion using live imaging with LBR-Venus and SECFP-emerin stably expressing HeLa cells as in . The depletion efficiency of the LBR in all cells, examined by live imaging was confirmed by the reduction of Venus signals (average depletion efficiency was about 80%; see Fig. S5A). Different from the ELYS/Mel28-depletion, the depletion of the LBR did not affect the timing of SECFP–emerin to target mitotic chromosomes after AO (Fig. S5B). The depletion of the LBR slightly affected the duration of mitosis after emerin recruitment until cytokinesis, however, the effect could be negligible compared with the effect of the ELYS/Mel28-depletion (for comparison, see Fig. S5B). Therefore, such acceleration could be caused by a function of ELYS/Mel28 unrelated to the recruitment of the LBR.
We further examined whether the altered behavior of emerin or Lap2α upon ELYS/Mel28-depletion was a consequence of the absence of the LBR at mitotic chromosomes. Live imaging analysis showed that the LBR-depletion did not influence the accumulation of emerin at the chromosomal core region (). When examined by IF staining, depletion of the LBR did not change the behavior of Lap2α as it accumulated onto both chromosomal regions next to the spindle pole- and the central spindle area (). We thus concluded that the effects caused by ELYS/Mel28-depletion on INM proteins at the core region are different from those on the LBR at the noncore region, indicating that ELYS/Mel28 has specific, but distinct roles during late mitosis in NE subdomain formation at the chromosomal noncore and core regions.
Discussion
ELYS/Mel28 and the Nup107-160 complex recruit the LBR to the mitotic chromosomal noncore region
In this study, by using RNAi, we found that the recruitment of the LBR to the mitotic chromosomal noncore region relied on the nucleoporins ELYS/Mel28 and the Nup107-160 complex (, ). Biochemical data supported an interaction between ELYS/Mel28 and the LBR (), which was phosphorylation-dependent. Pull down experiments showed that ELYS interacts with the N-terminal region of the LBR ().
The LBR has been reported to bind lamin BCitation28 and interacts with truncated Pom121137–513aa.Citation57 However, we did not detect any interactions between endogenous lamin B or Pom121 with the LBR-EGFP under the present experimental conditions. Transient associations between the LBR and lamin B and Pom121 could have been destroyed during extraction. On the other hand, the LBR also interacted with importin β (), as reported.Citation32,Citation33 It was previously shown, that binding between the human LBR and importin β is not phosphorylation-regulated,Citation68 corroborating our result, that the precipitation of importin β with the LBR did not change upon addition of phosphatase or phosphatase inhibitor (). Our IP data indicated that dephosphorylation strengthens the homotypic interaction of the LBR, as it was suggested before for oligomerization and chromatin binding of the LBR.Citation68,Citation69 Phosphorylation of the LBR, on the other hand, enhanced its interaction with ELYS/Mel28. Although there was no detectable coprecipitation between the full-length LBR and ELYS/Mel28 from interphase extracts, GST-LBR211aa-GFP precipitated ELYS/Mel28 from the interphase and mitotic extracts. The N-terminal fragment of the LBR is likely more accessible for interaction with ELYS/Mel28.
The Nup107-160 complex and ELYS/Mel28 interact physically and functionally, although whether they bind as a stoichiometric tight complex is not known.Citation70,Citation71 Our results () indicate that a mutual binding of ELYS/Mel28 and the Nup107-160 complex is required for recruitment of the LBR to mitotic chromosomes in late anaphase/telophase. However, coprecipitation of Nup107 with the LBR-EGFP was not detected in IP experiments (), indicating Nup107 could interact with the LBR through ELYS/Mel28. The biochemical data showed that the phosphorylation-dependent interaction between ELYS/Mel28 and the LBR was strongest in prometaphase and decreased as mitosis progressed to telophase (). Live imaging demonstrated that the LBR normally localizes to chromatin in anaphase/telophase (), where the interaction with ELYS/Mel28 should be weaker than in prometaphase. However, LBR is unable to target to chromatin in the absence of ELYS/Mel28 ( and ). We therefore hypothesize that ELYS/Mel28 and the Nup107-160 complex together are required for targeting but not for anchoring the LBR to chromatin. The LBR itself is known to possess DNA-binding activity.Citation27,Citation28 Altogether, dephosphorylation of the LBR would decrease the interaction with ELYS/Mel28 but would promote oligomerization or a competitive interaction with other proteins to bind stably to chromatin. Furthermore, our data support that the recruitment of the LBR to chromosomes relies on an interaction with single or few nucleoporins, instead of the NPC structure, because later-recruiting Pom121 did not affect the localization of the LBR (). Thus, the targeting of the LBR and the first steps of NPC assembly take place concomitantly in an ELYS/Mel28-controlled fashion, leading to the assembly of the NE at the noncore region. Importantly, the LBR was unnecessary for postmitotic NPC formation (; Fig. S3), implying that ELYS/Mel28 contributes to NPC assembly independently of its role in recruitment of the LBR.
The ELYS/Mel28-depletion affects the behavior of INM proteins at the chromosomal core region
Our results demonstrate that ELYS/Mel28 affects the focusing of several INM proteins like BAF, emerin, Lap2α and A-type lamins to the core region (; Fig. S4). Emerin and Lap2α behaved normally upon depletion of the LBR (), indicating that ELYS/Mel28 regulates localization of INM proteins to the core and noncore region by different mechanisms.
Formation of the core region is initiated by localization of BAF and takes place by the subsequent recruitment of Lap2α, emerin, and other LEM-domain containing proteins.Citation26 Each protein localizes to broader chromatin regions at first in anaphase and gradually concentrates at the core region during telophase (Fig. S4A and B).Citation25,Citation72 Closer examination of the dynamic localization of Lap2α and BAF using the PA index (Fig. S4C), clearly indicated that these proteins did not accumulate at the core region when ELYS/Mel28 was reduced but bound to all over chromatin or to chromosomal regions adjacent to the spindle pole area. Since BAF is necessary for other LEM-containing proteins to bind chromosomes and for a stable formation of the core region,Citation73,Citation74 we cannot exclude the possibility that ELYS/Mel28 primarily affects BAF.
ELYS/Mel28-depleted cells entered cytokinesis after emerin recruitment to chromosomes earlier than control cells (Fig. S5B), which significantly shortened the time of emerin at the core region (). This implies that the stable formation of the core region and events that lead to cytokinesis might be connected. ELYS/Mel28-depletion was reported to increase the number of cells with a midbody structure, which supported a role of ELYS/Mel28 in cytokinesis.Citation42
Through what molecular mechanism does the depletion of ELYS/Mel28 hinder the accumulation of the INM proteins at the core region? Considering that ELYS/Mel28 interacts tightly with the Nup107-160 complex,Citation42 and that this complex regulates the localization of the CPC,Citation51 it is plausible to speculate that phenomena seen in ELYS/Mel28-depleted cells were the consequences of a disturbed function of the CPC. We actually observed the mislocalization of CPC components, survivin and Aurora B, from centromeres and in later stages at the midzone of the central spindle (Fig. S5C, D and E). Many processes are regulated by the CPC through the activity of Aurora B kinase during mitosis.Citation52 It is necessary to identify the relevant substrates of Aurora B kinase to understand the role of the CPC in localization of the INM proteins at the core region. In addition to spindle dynamics, as suggested previously,Citation75 the state of chromatin might be regulated by the CPC. Alternatively, INM proteins might be phosphorylated by the CPC.
In contrast to ELYS/Mel28-depleted cells, which entered cytokinesis faster, cells depleted of Seh1, another component of the Nup107-160 complex, often became binucleated as a result of cytokinesis failure.Citation51 It might be possible that the behavior of the Nup107-160 complex changes due to the presence or absence of ELYS/Mel28, as suggested recently,Citation70,Citation71 which in turn might affect the function of the CPC. Instead, we cannot exclude the possibility that ELYS/Mel28 could interact with other important factors for cytokinesis.
In summary, our study showed that the nucleoporin ELYS/Mel28 regulates postmitotic NE formation, revealing new aspects how the postmitotic reassembly of NPCs, and the NE is coordinated. ELYS/Mel28 governs the formation of the B-type lamin/NPC-rich subdomain through its role in NPC assemblyCitation40-Citation44 and recruitment of the LBR. ELYS/Mel28 also affects formation of the A-type lamin/NPC-free subdomain, which might be connected to the function of the CPC. These results underline the versatile character of the nucleoporin ELYS/Mel28, which has a structural role at the NPC and contributes to mitotic processes; however, further studies are required to understand the function of ELYS/Mel28 in the structural organization of NE subdomains throughout the cell cycle.
Materials and Methods
Cell lines and siRNA transfection
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, SH30070, Thermo Scientific) under 5% CO2 at 37°C.
To obtain stable expression clones, a HeLa cell line with FRT sites was used with the Flp-In™ System (Invitrogen) with the plasmids described below. For stable LBR-Venus/SECFP-emerin expression under the EF1α promoter, a tandem expression vector was used, separating LBR-Venus and SECFP-emerin with insulator sequences (IxCitation2) as described by Yahata et al.Citation76 For stable LBR-EGFP expression, pFRT/V5-EF1α-LBR-EGFP (a gift from Dr Imamoto, Osaka University) was applied.
Transfections were performed with Oligofectamine™ reagent (12252–011, Invitrogen) and OptiMEMI™ serum (31985–062, GIBCO) according to the instruction manual. For live imaging and immunofluorescence staining, cells were treated with dsRNA for 50 h and for 72 h, respectively, except Nup107 depletion. For Nup107 depletion, cells were transfected twice 24 h apart (in total: 75–80 h incubation with dsRNA). The following siRNA oligos were used in this study: siRNA against ELYS/Mel28 (HSS146608 and HSS146610; Invitrogen), LBR (HSS105977; Invitrogen), Nup107 according to Boehmer et al.Citation77 (Integrated DNA Technologies) and Pom121 according to Funakoshi et al.Citation78 (Integrated DNA Technologies). As control, cells were treated with transfection reagent only or with the siRNA oligo against firefly luciferase GL2 according to Elbashir et al.Citation79 (Qiagen).
Plasmid construction
The plasmid pGEX2T-NLS-GFP was a gift from Dr. Yoneda, which contains GST fused to the SV40 large T-antigen nuclear localization sequence (PPKKKRKVEDP) and GFP (S65T). The pGEX-GST-LBR211-GFP was constructed by exchanging the NLS region of pGEX2T-NLS-GFP to LBR1-633nt amplified from human LBR cDNA by PCR with the primers 5′-GGGAAGCTTATGCCAAGTAGGAAATTTGCC and 5′-AAAGGGCCCTCCTCCAAACTCCAAGTCC.
Cell synchronization
To examine mitotic cells in live imaging and IF staining, cells were synchronized with 2 mM thymidine (T1895, SIGMA) for between 12–16 h and released by washing for 10 h. For preparation of cell extracts for immunoprecipitation (), HeLa cells expressing LBR-EGFP were synchronized with 2 mM thymidine overnight, released for 8 h and incubated with 0.1 µg/ml nocodazole for 2 h and either harvested directly (Prometaphase) or released for 60 min into mitosis (Meta/Telophase).
Live imaging and indirect immunofluorescence
Immunofluorescence staining (IF) and live imaging were performed as described by Funakoshi et al.Citation57 with minor modifications. Cells were seeded onto a glass-bottom dish with a grid to mark the position of each cell. Before live imaging observations, the medium was changed to DMEM without phenolred (21063-029, Gibco) supplied with 10% FBS. Time-lapse images of metaphase cells were captured every 2 min by a DeltaVisionRT microscopy using PlanApo 60x/1.40 (Olympus), 10 h after releasing cells from the thymidine treatment. To evaluate the knockdown efficiency, cells were immunostained with anti-ELYS/Mel28 antibody immediately after live imaging. The position of each cell observed with live imaging on the grid was used to find cells after IF staining and to monitor the IF signal intensities of ELYS/Mel28 of control cells and cells depleted of ELYS/Mel28. IF Signal intensities were analyzed with Softworx software’s polygon tool by comparing the signal of the same area for ELYS/Mel28 at the nuclear rim subtracted from the background signal outside the cell. For BAF staining cells were fixed with 3.7% paraformaldehyde and 0.2% glutaraldehyde supplied in medium for 15 min, permeabilized with 0.1% Triton X-100 for 5 min and incubated with anti-BAF antibody at 4°C over night. In all IF stainings, DAPI (4',6-diamidino-2-phenylindole; 10236276001, Roche) in a final concentration of 1 μg/ml was used for chromatin staining. The following antibodies were used in this study: rabbit anti-BAF (H00008815-M01, Abnova), mouse anti-ELYS/Mel28 (BMR00513, Biomatrix Research), mouse anti-Lamin A/C (SC-7292, Santa Cruz), goat anti-Lamin B (SC-6217, Santa Cruz), rabbit anti-Lap2α (IQ175, ImmuQuest), rabbit anti-LBR (1398–1, Epitomics; IF staining), rat anti-Nup62 according to Maeshima et al.Citation19 and rat anti-Pom121 according to Funakoshi et al.Citation78 Secondary antibodies (Molecular Probes, Invitrogen) were goat anti rabbit Alexa 488 (A21206), goat anti rabbit Alexa 594 (A11037), goat anti mouse Alexa 594 (A11005), goat anti mouse Alexa 488 (A11001), goat anti rat Alexa 647 (A21235), donkey anti goat Alexa 594 (A11058), donkey anti-rat Alexa 488 (A21208), donkey anti-rabbit Alexa 647 (A31573), and donkey anti mouse Alexa 488 (A21202). The images were captured with a DeltavisionRT microscope (Applied Precision) using PlanApo 60x/1.40 (Olympus), acquired by Softworx (Applied Precision), processed by ImageJ (http://rsb.info.nih.gov/ij) and arranged in Photoshop CS (Adobe).
Immunoprecipitation
Unsynchronized LBR-EGFP expressing HeLa cells were directly used for extraction or synchronized to harvest cells in different mitotic stages as described above. Protein extraction was performed according to Hawryluk-Gara et al.Citation55 About 5–6 × 106 cells were resuspended in lysis buffer A (10 mM Tris, pH 7.4, 400 mM NaCl, 1% Triton X-100, 2 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail; complete mini, 04 0693 124001, Roche), sonicated, centrifuged at 14,800 rpm at 4°C for 15 min, and diluted 3.75× with dilution buffer (10 mM Tris, pH 7.4, 2 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail; complete mini, 04-0693-124001, Roche). The extracts were treated with 0.6 U/µl of lambda-protein phosphatase (λ-PPase; P0753, New England BioLabs) at 4°C for 30 min. Untreated samples contained phosphatase inhibitors (PhosSTOP; 04-906-837-001, Roche) from lysis onward. The samples were precleared with magnetic Dynabeads® Protein G (100.03D, Invitrogen). Finally, the precleared samples were incubated with 2 µg anti-GFP-antibody (11-814-460-001, Roche) bound to Dynabeads® Protein G for 1 h, washed with wash buffer (= lysis buffer 3.75x diluted with dilution buffer), and resuspended with sodium dodecyl sulfate-PAGE (SDS-PAGE) sample buffer. 1/2 of the precipitate and 1/30th of the input (extract before IP) was used for SDS-PAGE. The proteins were blotted to a polyvinylidene fluoride (PVDF) membrane and probed with an anti-LBR antibody and anti-ELYS/Mel28 antibody used for IF staining. To detect importins and nucleoporins, Nup153 and Nup358, following antibodies were used: rat anti-importin α (D168-3, MBL), rabbit anti-importin β (SC-11367, Santa Cruz) and mAb414 (MMS-120P, Covance).
Protein purification and pull down
GST-LBR211aa-GFP and GST-NLS-GFP were purified from Escherichia coli strain BL21 after induction with 0.3 mM IPTG at 20°C overnight. The bacteria were resuspended and incubated in lysis buffer B (50 mM Tris, pH 7.4, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 × protease inhibitor cocktail; complete mini, 04-0693-124001, Roche) with 0.3 mg/ml lysozyme (126-02671, Wako) for 20 min, sonicated, incubated with lysis buffer B containing 1% Triton X-100 for 30 min, and centrifuged at 12,000 g at 4°C for 30 min. The recombinant proteins were isolated from the final supernatants with Glutathione-Sepharose™ 4B beads (17-0765-05, GE Healthcare) and eluted with elution buffer (50 mM Tris, pH 8.0, 10 mM l-glutathione; G6529, Sigma). The eluates were dialyzed against dialysis buffer (10 mM Tris, 107 mM NaCl, 0.3% Triton X-100, pH 7.4, 2 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail; complete mini, 04-0693-124001, Roche). Then, 1.0–3.5 µg of either GST-NLS-GFP or GST-LBR211aa-GFP, immobilized on 10 µl Glutathione-Sepharose™ 4B beads, were incubated with mitotic or unsynchronized HeLa cell extracts, prepared as described for IP with phosphatase inhibitors (PhosStop; 04-906-837-001, Roche) under rotation at 4°C for 1 h. After repeated washing with IP washing buffer, beads were resuspended in sample buffer and the proteins were separated by SDS-PAGE (7% polyacrylamide gels), blotted, and probed with an anti-LBR antibody and anti-ELYS/Mel28 antibody used in IF staining.
| Abbreviations: | ||
| AO | = | anaphase onset |
| CPC | = | chromosomal passenger complex |
| DAPI | = | 4',6-diamidino-2-phenylindole |
| DIC | = | differential interference contrast |
| GFP | = | green fluorescent protein |
| INM | = | inner nuclear membrane |
| IP | = | immunoprecipitation |
| LBR | = | lamin B receptor |
| NE | = | nuclear envelope |
| NPC | = | nuclear pore complex |
| nup | = | nucleoporin |
| ONM | = | outer nuclear membrane |
| SECFP | = | super enhanced cyan fluorescent protein |
| TM | = | transmembrane |
Additional material
Download Zip (18.5 MB)Acknowledgements
We thank Dr. Y. Yoneda for the gift of the pGEX vector and Dr F. Imamoto for the LBR-construct and are very grateful for comments and the kind present of the anti-BAF antibody by Dr T. Haraguchi and Dr T. Koujin. We also thank members of the Cellular Dynamics Lab for helpful comments and discussions. This work was supported by RIKEN Special Funding for Basic Science (Cellular System) and funding awarded to N.I. by the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for Next Generation World-Leading Researchers (NEXT Program),” initiated by the Council for Science and Technology Policy (CSTP). M.C. is supported by the RIKEN Joint Graduate School Program, “International Program Associate” (IPA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest have been disclosed.
References
- Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 2005; 21:347 - 80; http://dx.doi.org/10.1146/annurev.cellbio.21.090704.151152; PMID: 16212499
- D’Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cell Mol Life Sci 2006; 63:316 - 32; http://dx.doi.org/10.1007/s00018-005-5361-3; PMID: 16389459
- Watson ML. The nuclear envelope; its structure and relation to cytoplasmic membranes. J Biophys Biochem Cytol 1955; 1:257 - 70; http://dx.doi.org/10.1083/jcb.1.3.257; PMID: 13242591
- Foisner R. Cell cycle dynamics of the nuclear envelope. ScientificWorldJournal 2003; 3:1 - 20; http://dx.doi.org/10.1100/tsw.2003.06; PMID: 12806116
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000539; http://dx.doi.org/10.1101/cshperspect.a000539; PMID: 20300205
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev 2002; 16:533 - 47; http://dx.doi.org/10.1101/gad.960502; PMID: 11877373
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature 2007; 450:695 - 701; http://dx.doi.org/10.1038/nature06405; PMID: 18046406
- Beck M, Lucicá V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 2007; 449:611 - 5; http://dx.doi.org/10.1038/nature06170; PMID: 17851530
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem 2011; 80:613 - 43; http://dx.doi.org/10.1146/annurev-biochem-060109-151030; PMID: 21495847
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol 2001; 13:310 - 9; http://dx.doi.org/10.1016/S0955-0674(00)00213-1; PMID: 11343901
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 2000; 148:635 - 51; http://dx.doi.org/10.1083/jcb.148.4.635; PMID: 10684247
- Madrid AS, Weis K. Nuclear transport is becoming crystal clear. Chromosoma 2006; 115:98 - 109; http://dx.doi.org/10.1007/s00412-005-0043-3; PMID: 16421734
- Mansfeld J, Güttinger S, Hawryluk-Gara LA, Panté N, Mall M, Galy V, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell 2006; 22:93 - 103; http://dx.doi.org/10.1016/j.molcel.2006.02.015; PMID: 16600873
- Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol 1993; 122:513 - 21; http://dx.doi.org/10.1083/jcb.122.3.513; PMID: 8335683
- Gerace L, Ottaviano Y, Kondor-Koch C. Identification of a major polypeptide of the nuclear pore complex. J Cell Biol 1982; 95:826 - 37; http://dx.doi.org/10.1083/jcb.95.3.826; PMID: 7153248
- Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci 2010; 123:1973 - 8; http://dx.doi.org/10.1242/jcs.019042; PMID: 20519579
- Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp Cell Res 2007; 313:2167 - 79; http://dx.doi.org/10.1016/j.yexcr.2007.03.012; PMID: 17451680
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res 2007; 313:2121 - 33; http://dx.doi.org/10.1016/j.yexcr.2007.03.028; PMID: 17467691
- Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, et al. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci 2006; 119:4442 - 51; http://dx.doi.org/10.1242/jcs.03207; PMID: 17074834
- Goldberg MW, Fiserova J, Huttenlauch I, Stick R. A new model for nuclear lamina organization. Biochem Soc Trans 2008; 36:1339 - 43; http://dx.doi.org/10.1042/BST0361339; PMID: 19021552
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev 2008; 22:3409 - 21; http://dx.doi.org/10.1101/gad.1735208; PMID: 19141474
- Furukawa K, Ishida K, Tsunoyama TA, Toda S, Osoda S, Horigome T, et al. A-type and B-type lamins initiate layer assembly at distinct areas of the nuclear envelope in living cells. Exp Cell Res 2009; 315:1181 - 9; http://dx.doi.org/10.1016/j.yexcr.2008.12.024; PMID: 19210986
- Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol 2008; 20:669 - 77; http://dx.doi.org/10.1016/j.ceb.2008.09.010; PMID: 18938243
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, et al. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci 2000; 113:779 - 94; PMID: 10671368
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci 2008; 121:2540 - 54; http://dx.doi.org/10.1242/jcs.033597; PMID: 18628300
- Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol 2007; 261:1 - 46; http://dx.doi.org/10.1016/S0074-7696(07)61001-8; PMID: 17560279
- Duband-Goulet I, Courvalin JC. Inner nuclear membrane protein LBR preferentially interacts with DNA secondary structures and nucleosomal linker. Biochemistry 2000; 39:6483 - 8; http://dx.doi.org/10.1021/bi992908b; PMID: 10828963
- Ye Q, Worman HJ. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J Biol Chem 1994; 269:11306 - 11; PMID: 8157662
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ 3rd. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun 2005; 331:929 - 37; http://dx.doi.org/10.1016/j.bbrc.2005.04.016; PMID: 15882967
- Martins SB, Eide T, Steen RL, Jahnsen T, Skålhegg B S, Collas P. HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. J Cell Sci 2000; 113:3703 - 13; PMID: 11034899
- Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp Cell Res 2009; 315:1895 - 903; http://dx.doi.org/10.1016/j.yexcr.2009.01.019; PMID: 19331822
- Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen, et al. Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci 2007; 120:520 - 30; http://dx.doi.org/10.1242/jcs.03355; PMID: 17251381
- Lu X, Shi Y, Lu Q, Ma Y, Luo J, Wang Q, et al. Requirement for lamin B receptor and its regulation by importin beta and phosphorylation in nuclear envelope assembly during mitotic exit. J Biol Chem 2010; 285:33281 - 93; http://dx.doi.org/10.1074/jbc.M110.102368; PMID: 20576617
- Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta 1998; 1392:233 - 44; PMID: 9630650
- Collas P, Courvalin JC, Poccia D. Targeting of membranes to sea urchin sperm chromatin is mediated by a lamin B receptor-like integral membrane protein. J Cell Biol 1996; 135:1715 - 25; http://dx.doi.org/10.1083/jcb.135.6.1715; PMID: 8991085
- Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos SD. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J 1996; 15:7108 - 19; PMID: 9003786
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol 2009; 186:183 - 91; http://dx.doi.org/10.1083/jcb.200901106; PMID: 19620630
- Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma 2007; 116:227 - 35; http://dx.doi.org/10.1007/s00412-007-0094-8; PMID: 17245605
- Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, et al. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells 2002; 7:435 - 46; http://dx.doi.org/10.1046/j.1365-2443.2002.00529.x; PMID: 11952839
- Fernandez AG, Piano F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr Biol 2006; 16:1757 - 63; http://dx.doi.org/10.1016/j.cub.2006.07.071; PMID: 16950115
- Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol 2006; 16:1748 - 56; http://dx.doi.org/10.1016/j.cub.2006.06.067; PMID: 16950114
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A 2006; 103:17801 - 6; http://dx.doi.org/10.1073/pnas.0608484103; PMID: 17098863
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 2007; 17:1657 - 62; http://dx.doi.org/10.1016/j.cub.2007.08.041; PMID: 17825564
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 2008; 19:3982 - 96; http://dx.doi.org/10.1091/mbc.E08-01-0012; PMID: 18596237
- Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 2003; 113:195 - 206; http://dx.doi.org/10.1016/S0092-8674(03)00235-6; PMID: 12705868
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, et al. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol 2008; 180:857 - 65; http://dx.doi.org/10.1083/jcb.200707026; PMID: 18316408
- Fernandez-Martinez J, Rout MP. Nuclear pore complex biogenesis. Curr Opin Cell Biol 2009; 21:603 - 12; http://dx.doi.org/10.1016/j.ceb.2009.05.001; PMID: 19524430
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, et al. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell 2006; 17:3806 - 18; http://dx.doi.org/10.1091/mbc.E05-11-1061; PMID: 16807356
- Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J 2007; 26:1853 - 64; http://dx.doi.org/10.1038/sj.emboj.7601642; PMID: 17363900
- Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 2010; 12:164 - 9; http://dx.doi.org/10.1038/ncb2016; PMID: 20081840
- Platani M, Santarella-Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell 2009; 20:5260 - 75; http://dx.doi.org/10.1091/mbc.E09-05-0377; PMID: 19864462
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 2007; 8:798 - 812; http://dx.doi.org/10.1038/nrm2257; PMID: 17848966
- Meyer H, Drozdowska A, Dobrynin G. A role for Cdc48/p97 and Aurora B in controlling chromatin condensation during exit from mitosis. Biochem Cell Biol 2010; 88:23 - 8; http://dx.doi.org/10.1139/O09-119; PMID: 20130676
- Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J 2000; 19:3918 - 31; http://dx.doi.org/10.1093/emboj/19.15.3918; PMID: 10921874
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell 2005; 16:2382 - 94; http://dx.doi.org/10.1091/mbc.E04-10-0857; PMID: 15703211
- Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, et al. Functional association of Sun1 with nuclear pore complexes. J Cell Biol 2007; 178:785 - 98; http://dx.doi.org/10.1083/jcb.200704108; PMID: 17724119
- Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell 2011; 22:1058 - 69; http://dx.doi.org/10.1091/mbc.E10-07-0641; PMID: 21289085
- Lussi YC, Hügi I, Laurell E, Kutay U, Fahrenkrog B. The nucleoporin Nup88 is interacting with nuclear lamin A. Mol Biol Cell 2011; 22:1080 - 90; http://dx.doi.org/10.1091/mbc.E10-05-0463; PMID: 21289091
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol 2011; 194:27 - 37; http://dx.doi.org/10.1083/jcb.201012154; PMID: 21727197
- Yewdell WT, Colombi P, Makhnevych T, Lusk CP. Lumenal interactions in nuclear pore complex assembly and stability. Mol Biol Cell 2011; 22:1375 - 88; http://dx.doi.org/10.1091/mbc.E10-06-0554; PMID: 21346187
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol 1990; 111:807 - 16; http://dx.doi.org/10.1083/jcb.111.3.807; PMID: 2391365
- Chaudhary N, Courvalin JC. Stepwise reassembly of the nuclear envelope at the end of mitosis. J Cell Biol 1993; 122:295 - 306; http://dx.doi.org/10.1083/jcb.122.2.295; PMID: 8391536
- Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol 2000; 149:1179 - 92; http://dx.doi.org/10.1083/jcb.149.6.1179; PMID: 10851016
- Courvalin JC, Segil N, Blobel G, Worman HJ. The lamin B receptor of the inner nuclear membrane undergoes mitosis-specific phosphorylation and is a substrate for p34cdc2-type protein kinase. J Biol Chem 1992; 267:19035 - 8; PMID: 1326541
- Takano M, Koyama Y, Ito H, Hoshino S, Onogi H, Hagiwara M, et al. Regulation of binding of lamin B receptor to chromatin by SR protein kinase and cdc2 kinase in Xenopus egg extracts. J Biol Chem 2004; 279:13265 - 71; http://dx.doi.org/10.1074/jbc.M308854200; PMID: 14718546
- Smith S, Blobel G. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J Cell Biol 1993; 120:631 - 7; http://dx.doi.org/10.1083/jcb.120.3.631; PMID: 8381121
- Uehara R, Goshima G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J Cell Biol 2010; 191:259 - 67; http://dx.doi.org/10.1083/jcb.201004150; PMID: 20937700
- Tseng LC, Chen RH. Temporal control of nuclear envelope assembly by phosphorylation of lamin B receptor. Mol Biol Cell 2011; 22:3306 - 17; http://dx.doi.org/10.1091/mbc.E11-03-0199; PMID: 21795390
- Nikolakaki E, Drosou V, Sanidas I, Peidis P, Papamarcaki T, Iakoucheva LM, et al. RNA association or phosphorylation of the RS domain prevents aggregation of RS domain-containing proteins. Biochim Biophys Acta 2008; 1780:214 - 25; http://dx.doi.org/10.1016/j.bbagen.2007.10.014; PMID: 18022399
- Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell 2009; 20:4031 - 42; http://dx.doi.org/10.1091/mbc.E09-02-0150; PMID: 19625448
- Ródenas E, González-Aguilera C, Ayuso C, Askjaer P. Dissection of the NUP107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein MAD1 in C. elegans. Mol Biol Cell 2012; http://dx.doi.org/10.1091/mbc.E11-11-0927; PMID: 22238360
- Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, et al. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J Cell Sci 2004; 117:6117 - 28; http://dx.doi.org/10.1242/jcs.01529; PMID: 15546916
- Shumaker DK, Lee KK, Tanhehco YC, Craigie R, Wilson KL. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J 2001; 20:1754 - 64; http://dx.doi.org/10.1093/emboj/20.7.1754; PMID: 11285238
- Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci U S A 2005; 102:3290 - 5; http://dx.doi.org/10.1073/pnas.0408364102; PMID: 15728376
- Guse A, Mishima M, Glotzer M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr Biol 2005; 15:778 - 86; http://dx.doi.org/10.1016/j.cub.2005.03.041; PMID: 15854913
- Yahata K, Kishine H, Sone T, Sasaki Y, Hotta J, Chesnut JD, et al. Multi-gene gateway clone design for expression of multiple heterologous genes in living cells: conditional gene expression at near physiological levels. J Biotechnol 2005; 118:123 - 34; http://dx.doi.org/10.1016/j.jbiotec.2005.02.020; PMID: 15961178
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A 2003; 100:981 - 5; http://dx.doi.org/10.1073/pnas.252749899; PMID: 12552102
- Funakoshi T, Maeshima K, Yahata K, Sugano S, Imamoto F, Imamoto N. Two distinct human POM121 genes: requirement for the formation of nuclear pore complexes. FEBS Lett 2007; 581:4910 - 6; http://dx.doi.org/10.1016/j.febslet.2007.09.021; PMID: 17900573
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411:494 - 8; http://dx.doi.org/10.1038/35078107; PMID: 11373684