Abstract
Nuclear pore complexes (NPCs) are best known for their central role in controlling the molecular trafficking between the cytoplasm and the nucleus. NPCs are assembled from about 30 different proteins and a growing body of evidence suggests that these nucleoporins are not only acting in the context of NPCs, but also in the nucleoplasm and cytoplasm. In this context it is well accepted that a set of nucleoporins are important regulators of a variety of mitotic processes, including kinetochore assembly, spindle checkpoint control and cytokinesis, whereas others associate with chromatin and administer gene expression. However, the functional importance of nucleoporins go far beyond these roles and this review will provide an overview of the latest insights into the versatility of metazoan nucleoporins with an emphasis on their roles in cell migration, cellular signaling and tissue-specific activities.
Introduction
Nuclear pore complexes (NPCs) span the inner and outer nuclear membrane of the nuclear envelope (NE) and master all macromolecular exchange between the cytoplasm and the nucleus of interphase eukaryotic cells.Citation1,Citation2 NPCs are large multi-protein complexes that are composed of ~30 different proteins (nucleoporins or Nups), which are typically organized in repetitively arranged subcomplexes to form the NPC.Citation3-Citation5 NPCs exhibit 8-fold rotational symmetry and as a result nucleoporins are found in copy numbers of eight per NPC or multiple thereof. In total about 500 individual proteins form the NPC.Citation6 The principle structural organization of the NPC is evolutionary conserved and has been determined by distinct electron microscopy (EM) approaches mainly in Xenopus laevis oocyte nuclei, but also, for example, in yeast, amoebozoa, plants and human.Citation3,Citation6-Citation13 Overall, NPCs have a roughly tripartite architecture: a central framework (also called spoke complex, spoke-ring complex or scaffold-ring complex) that is decorated by the cytoplasmic filaments and the nuclear basket, an assembly formed of eight filaments that join into a distal ring (). The central framework consists of eight spokes (i.e., the scaffold) that are flanked by the cytoplasmic and nuclear ring moieties and it encloses a central pore through which nucleocytoplasmic transport occurs in a still controversially discussed manner.Citation1,Citation2 Despite this overall resemblance in the architecture, NPCs from different species show significant variance in protein density, most likely due to variations in nucleoporin sequence and position as well as NPC number between the species.Citation10
Figure 1. Nuclear pore complex (NPC) architecture and nucleoporin localization in the NPC. (A) Cross-sections along embedded nuclear envelopes of Xenopus oocyte nuclei allow side views of NPCs visualized by transmission electron microscopy. Shown are an NPC in a slightly worm's-eye view (left) and a schematic representation of the main structural components of the NPC (right). (B) Nucleoporins can be subdivided into different subgroups depending on their structural motifs and localization: transmembrane nucleoporins (white), scaffold nucleoporins (green), nucleoporins of the cytoplasmic filaments and the cytoplasmic ring (rose), central channel nucleoporins (blue) and nuclear basket and nuclear ring nucleoporins (yellow). Nup153 indicated at the nuclear ring and the distal ring refer to the respective anchoring sites of its N terminus and zinc-finger domain.
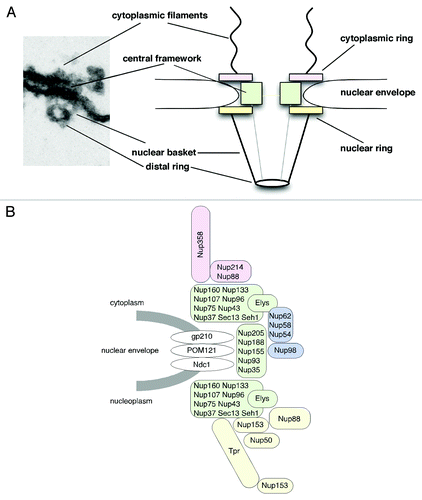
Molecular building blocks of NPCs are the nucleoporins that, according to their amino acid sequence and predicted structural motifs, fall into three groups.Citation14,Citation15 The first group comprises the transmembrane proteins that anchor the NPC to the NE and reside at the boundary between the central framework and the pore membrane. In metazoans, this group comprises gp210, Ndc1 and POM121 (). The second group of nucleoporins contains α-helical solenoid and β-propeller fold motifs and locates more toward the central channel of the NPC. This group of nucleoporins, which includes the Nup107-160 complex as well as the Nup93 complex, is crucial for the formation of the NPC scaffold. The third group comprises nucleoporins characterized by the presence of repetitive phenylalanine-glycine (FG) motifs (spaced by hydrophilic linkers) and/or by coiled-coil motifs, which are typically engaged in nucleocytoplasmic transport.Citation14,Citation15 This third group can be further subdivided into nucleoporins of the cytoplasmic filaments, such as Nup358 and Nup214, nucleoporins of the central channel, such as Nup98 and the Nup62 complex, and the nuclear basket nucleoporins Nup153, Nup50 and Tpr ().
Besides being important structural elements of the NPC and fundamental for general nucleocytoplasmic transport, nucleoporins often exhibit an enormously dynamic character due to which they are either directly or indirectly engaged in a variety of other cellular processes, both in interphase and mitotic cells. Several recent review articles have addressed the role of nucleoporins in mitosis and gene expression control,Citation16-Citation20 and we will focus here on the dynamic character of nucleoporins and their functions in distinct cellular signaling pathways, cell migration and differentiation. Nucleoporin function in these processes occurs often in a tissue-specific manner and only became evident from studies using animal models.
Nucleoporin Dynamics, Dynamic Nucleoporins
NPCs have been assumed to be rather static of nature, in contrast to the highly mobile cargoes and transport receptors that traverse the NPC during nucleocytoplasmic transport events. In fact, in mammalian cells, the NPC as a whole appears stably anchored to the NE.Citation21,Citation22 Single NPCs only move in arrays in response to changes in nuclear shape during the cell cycle but show no independent movement within the plane of the NE.Citation22 In contrast to the whole NPC, individual nucleoporins can be very dynamic. The turnover of nucleoporins at NPCs range from seconds to days, both in dividing and non-dividing cells.Citation21,Citation23 In this context especially the so-called scaffold nucleoporins, such as the Nup107-160 complex (), are very stably associated with the NPC and long-lived with basically no turnover during the cell cycle, whereas, for example, the nuclear basket proteins Nup153 and Nup50 () are characterized by their highly dynamic behavior.Citation21,Citation23
Nup153 and Nup50 both play important roles in facilitated nucleocytoplasmic transport. Nup153 is known to be key for nuclear protein import as well as most RNA export pathways and to interact with virtually every nuclear transport receptor studied.Citation24 Nup153 dynamically associates with the NPC and its exchange at NPCs is inhibited when RNA polymerases I or II are blocked.Citation25 Fluorescence recovery after photobleaching (FRAP) experiments revealed a two-step recovery of GFP-Nup153 at NPCs in both HeLa and NRK cells with an overall recovery rate of t1/2 41 sec.Citation23,Citation25 Beyond Nup153 as a whole, its FG-repeat domain further increases Nup153’s dynamic nature.Citation26-Citation28 Nup153 is organized in three domains and immuno-EM studies have revealed their complex topology in the NPC, with the N-terminal and zinc-finger domain anchoring Nup153 to the NPC ().Citation28-Citation30 The C-terminal FG-repeat domain of Nup153 in contrast is highly flexible and dynamic within the NPC,Citation26,Citation28 a feature that was also observed for the FG-repeat domains of Nup214, Nup98 and Nup62.Citation31-Citation33 Consistent with their role in facilitated nucleocytoplasmic transport, the spatial distribution of FG-repeat domains alters in a transport- and energy-dependent manner.Citation31,Citation34,Citation35
Nup50 (also known as Npap60) enhances nucleocytoplasmic transport by acting as cofactor for importin α/β-mediated nuclear protein import.Citation36 Nup50 has binding sites for importin α, importin β and RanGTP, and shuttles between the nucleus and the cytoplasm together with nuclear import complexes.Citation36 A similar dynamic behavior has been described for Nup2p and Nup60p, the yeast orthologs of Nup50.Citation37 Analogous to the role of Nup50 in importin α/β-mediated nuclear import, Nup98 appears to act as mobile cofactor for CRM1-mediated nuclear export.Citation38 Nup98 is anchored to the center of the NPC and likely the major constituent of the NPC’s permeability barrier.Citation33,Citation39-Citation42 Nup98 dynamically associates with the NPC in a transcription-dependent manner,Citation25,Citation43 and it binds directly to CRM1 in a RanGTP-dependent manner. Exogenously expressed GFP-Nup98 sequesters endogenous CRM1 from the nucleoplasm and NE to Nup98-intranuclear dots (so-called GLFG bodiesCitation44), which results in the inhibition of nuclear export of the Ran-binding protein RanBP1.Citation38
Nucleoporins and Cell Migration
A dynamic character of nucleoporins appears not only important for nucleocytoplasmic transport, but also for cell migration. Three nucleoporins, Nup62, Nup153 and Nup358, appear of importance in this context (). Nup62 belongs to the group of FG nucleoporins and is part of the Nup62 complex (). The Nup62 complex further includes Nup58, its splice-variant Nup45 as well as Nup54Citation45,Citation46 and it is located toward the central pore of the NPC.Citation32 Nucleoporins of the Nup62 complex have medium turnover rates at the NPC.Citation23 In Jurkat cells infected by human immunodeficiency virus (HIV)-1, however, Nup62 translocates into the cytoplasm in association with the viral RNA and the HIV proteins Rev and Gag.Citation47 Hence, Nup62 apparently leaves the NPCs as part of the growing HIV-1 vRNA-RNP complex.Citation47 Moreover, in HeLa and activated human erythroleukemia cells, Nup62 was found to cycle between the plasma membrane and the perinuclear recycling compartment, which appears crucial for cell migration.Citation48,Citation49 The recruitment of Nup62 (and its complex partners) to the plasma membrane and to membrane ruffles is mediated by Exo70, a component of the exocyst complex. Depleting Nup62 significantly impairs cell motility due to disruption of the Nup62-Exo70 complex at the leading edge of the cell.Citation49
Table 1. Table summarizing nucleoporin functions in cell migration, cell signaling as well as tissue specific functions
As outlined above, the C-terminal FG domain of Nup153 was found to be highly dynamic within the NPC in NRK cells and Xenopus oocyte nuclei,Citation26,Citation28 while in a heterokaryon assay Nup153 was found to shuttle very slowly, suggesting that it rarely moves sufficiently far out of the NPC toward the cytoplasm.Citation26 This notion has recently been challenged as Nup153 was detected in the cytoplasm of oligodendrocyte precursor cells derived from patients with multiple sclerosis (MS).Citation50 The functional relevance of this observation has remained elusive, but might have an impact on the inefficient differentiation of the oligodendrocytes seen in MS.Citation50 Furthermore, Nup153 accumulates at the vegetal tip of actin bundles emanating from germinal vesicles of the sea pineapple Halocynthia roretzi during their breakdown in meiosis.Citation51 A link between Nup153 and the actin cytoskeleton is further supported by the finding that depletion of Nup153 from HeLa and human breast cancer MDA231 cells led to reorganization of the actin cytoskeleton and impaired lamellipodia formation, which coincided with defects in cell migration and reduced wound-healing rates.Citation52
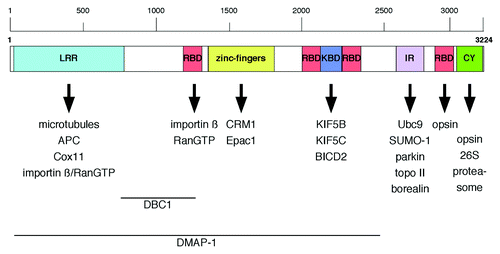
A third nucleoporin linked to cell migration is Nup358, the largest vertebrate nucleoporin, also known as RanBP2.Citation53 Nup358 is a multi-domain nucleoporin () that localizes to the cytoplasmic filaments of the NPCCitation54,Citation55 and it is involved in a multitude of cellular processes ( and ), that range from more general functions in nucleocytoplasmic transport,Citation56-Citation59 mitosis,Citation60-Citation62 and cellular signaling due to its E3 SUMO ligase activityCitation63-Citation66 to tissue-specific functions particularly in neurons and muscle cells.Citation67-Citation72 Moreover, Nup358 interacts with interphase microtubules (MTs) through its N-terminal, leucine-rich domain and overexpression of this MT-targeting domain in CHO cells increased MT bundling and stability, whereas depletion of Nup358 led to a decrease in polarized cell migration and stable MTs.Citation73 Endogenous Nup358 and a GFP-fusion of Nup358’s N-terminal domain were found in the cytoplasm and to co-localize with MTs at cell extensions.Citation73 The recruitment of Nup358 to MTs appears to depend on the adenomatous polyposis coli (APC) protein, a MT plus-end binding tumor-suppressor protein.Citation74 APC exhibits several MT-binding domains and, through its middle region, APC interacts with both MTs and Nup358.Citation74 Ectopic expression of this middle region is sufficient to recruit Nup358 to MT plus ends, and binding of Nup358 to APC as well as the MT-motor kinesin-2 are important for APC’s localization at the cell cortex.Citation74 Interestingly, in migrating neurons and rat embryonic fibroblasts, APC was found to associate with Nup153 at the nuclear membrane.Citation75 The APC-Nup153 complex appears to serve as anchoring site for MTs emanating from the centrosome.Citation75 Whether or not the APC-Nup153 interaction is important for cell migration has not been analyzed specifically, but the APC-Nup358 as well as the APC-Nup153 interaction are important for centrosome reorientation during migration after wound-scratching,Citation74,Citation75 suggesting that Nup153 acts in cell migration via both the actin cytoskeletonCitation52 and MTs.Citation75
Figure 2. Schematic representation of Nup358 domain organization and binding partners. Regions within Nup358 important for nuclear import of DBC1 and DMAP-1 are indicated. LRR, leucine-rich region; RBD, Ran-BP1-like RanGTP binding domain; KBD, kinesin binding domain; IR, internal repeats; CY, cyclophilin A homology domain.
Nucleoporins and Cellular Signaling
Due to their central location at the transit routes between the cytoplasm and the cell nucleus, nucleoporins often act as scaffold for proteins being involved in cellular signaling, as seen in transforming growth factor β (TGF-β) and cyclic AMP signaling, as well as DNA damage response. TGF-β cytokines critically regulate a variety of developmental processes and cell homeostasis by an evolutionary conserved mechanism involving Smad transcription factors, which upon phosphorylation translocate into the nucleus to activate TGF-β-responsive genes.Citation76 Drosophila Nup154 is ubiquitously expressed, similar to its human homolog Nup155,Citation77 but exhibits germ-line specific functions.Citation78,Citation79 Consequently, hypomorphic mutations in Nup154 cause male and female sterility and reduction of cell proliferation in the adult fly.Citation78 Whereas Nup154 function during gametogenesis involves its association with Cup, a key player in female germ-line development in Drosophila,Citation78 Nup154 function during spermatogenesis is linked to TGF-β signaling. In testes and in cultured Drosophila cell lines, depletion of Nup154 prevented nuclear accumulation of Mad (mothers against decapentaplegic; mammalian Smad1) and Medea (mammalian Smad4),Citation80 two Smad-like signal transducers in Drosophila. As Nup154 shows no direct interaction with phosphorylated/activated Mad, it indirectly controls Mad’s subcellular localization.Citation80 In contrast to Nup154, the scaffold nucleoporins Sec13 and Nup93 preferentially interact with the phosphorylated/activated form of Mad in Drosophila S2 and S2R+ cells, while they play no role in the nuclear import of classical NLS cargoes.Citation81 Importin 7 and 8 are the two import receptors for Drosophila and mammalian Smad proteins,Citation82,Citation83 and Nup93 is important for their recruitment to the nuclear periphery, whereas Sec13 operates independently of the receptors.Citation81 In human cells, TGF-β signal transduction is dependent on a direct interaction of Smad2 with Nup153 and Nup214Citation84 as well as Smad3 and Smad4 binding to Nup214.Citation85 Similarly, Nup153 and Nup214 appear specifically involved in Mad nuclear import in Drosophila.Citation81 All together this suggests that nucleoporins control Smad/Mad nuclear import, directly and/or indirectly, and consequently TGF-β signal transduction and developmental processes.
Nup358 acts as negative regulator of the second messenger cyclic adenosine monophosphate (cAMP) signaling through Epac1,Citation86 a cAMP-regulated guanine nucleotide exchange factor (GEF) for Rap GTPases.Citation87 Epac1 functions in cellular processes ranging from exocytosis to cell-cell-junction formation and cell-extracellular matrix adhesion, and its activity is directly regulated by cAMP.Citation87 In a yeast two-hybrid screen using Epac1 as bait, Nup358 was identified as novel interacting partner. Epac1 binds directly to the zinc-finger (ZNF) domain of Nup358 (), in a phosphorylation-dependent but Ran-independent manner. Binding to the ZNF of Nup358 mediates the anchoring of Epac1 to the NE during interphase, thereby inhibiting the activity of Epac1 toward Rap GTPases. Depletion of Nup358 by RNAi coincides with a complete loss of YFP-Epac1 from the NE and its displacement to the cytosol and nucleus, resulting in enhanced Epac1 activity. Nup358 hence functions as negative regulator of Epac1 by establishing an inactive pool of this Rap GEF at NPCs.Citation86
A strict dependency on a particular nucleoporin may evolve as a theme by which cellular signaling is further regulated. This became evident from recent studies on Nup358 and Nup153. While neither the depletion of Nup358Citation54 or of Nup153Citation88 impairs general nucleocytoplasmic transport, both nucleoporins have been found to be critical for the nuclear import of specific cargoes. In this context, the DNA methyltransferase 1 associated protein 1 (DMAP-1) and the putative tumor suppressor DBC-1 (deleted in breast cancer 1) have recently been identified as Nup358-dependent nuclear import cargoes.Citation89 DMAP-1 and DBC-1 are directly interacting with the N-terminal region of Nup358 in an import receptor-independent manner and strongly accumulate in the cytoplasm upon depletion of Nup358 by siRNAs.Citation89 Nup358 has a critical function in capturing RanGTP-importin β-complexes at the cytoplasmic filaments of NPCs upon their exit from the nucleus to allow efficient recycling of importin β and in turn importin β-dependent nuclear import.Citation59 Therefore, Nup358 appears to promote the formation of import complexes between DMAP-1 and DBC-1 and (most likely) importin β at the NPC, thereby stimulating their nuclear import.Citation89
Nup153 has been shown to be specifically required for the nuclear import of 53BP1, a mediator of DNA damage response (DDR) that promotes DNA repair and enhances DDR signaling.Citation90,Citation91 Depletion of Nup153 from either HeLa or U2OS cells prevents nuclear entry of 53BP1, but not several other DDR factors, after NE reformation at the end of mitosis. This results in a decreased number of 53BP1 foci after irradiation-induced DNA damage, coinciding with impaired cell survival and delayed DNA repair,Citation90,Citation91 especially non-homologous end joining.Citation91 Nup153 depletion furthermore intensifies DNA damage caused by replication stress. 53BP1 is imported to the nucleus through an interplay between Nup153 and importin β, implicating this specific 53BP1-Nup153-importin β complex in maintenance of genome surveillance.Citation90
Tissue-Specific Nucleoporin Functions
Thus far little is known about tissue-specific expression pattern and activity of nucleoporins.Citation72,Citation92,Citation93 In particular, mice models and studies in Drosophila, however, have recently provided some interesting novel insights in this respect. In Drosophila retinal neurons, Nup358 acts as a chaperone for red/green opsin, to which it binds via its Ran-binding domain (RBD) 4 and cyclophilin-like domain ().Citation94 Similarly, Nup358 interacts with red/green opsin in human and bovine cells but not with closely related blue-cone or rod opsin.Citation95 In mice, Nup358 acts as a chaperone for the mitochondrial metallo-chaperone Cox11.Citation96 The association of Nup358 with Cox11 is mediated by Nup358’s leucine-rich domain (). Both proteins co-purify from retinal extracts and co-localize to mitochondria in several classes of neurons, including photosensory neurons and neurons of the central nervous system (CNS). Furthermore, Nup358 is able to suppress the inhibitory activity of Cox11 over hexokinase I (HKI), the major regulator of glycolysis.Citation96 Haploinsufficiency in Nup358 causes a pronounced decrease of HKI and ATP levels in the CNS. This decrease in HKI and ATP levels coincides with defects in glucose clearance and the electrophysiological response of photosensory and postreceptoral neurons, as well as the delocalization of mitochondria in the photosensory neurons.Citation96,Citation97 Nup358 is highly expressed in these retinal neurons and found in the cytoplasm along with RanGTP.Citation69 Furthermore, Nup358 associates with kinesin KIF5B and KIF5C in the cytoplasm via its kinesin-binding domain (; KBD), which binds leucine-rich heptad repeats in the C-terminal coiled-coil domain of the kinesin heavy chain in a RanGTP-dependent manner.Citation68,Citation69,Citation97,Citation98 In the presence of MTs and ATP, binding of Nup358 enhances the low intrinsic ATPase activity of KIF5B, which in addition to the KBD also requires the RBD2 and RBD3 of Nup358.Citation98 Inhibition of the interaction between Nup358 and the KIF5s leads to perinuclear clustering of mitochondria, deficits in the mitochondria membrane potential as well as cell shrinkage, pinpointing to a kinesin-dependent role of Nup358 in mitochondria transport and function.Citation97
In aged mice, haploinsufficiency of Nup358 protects neurons against light-induced oxidative stress.Citation70,Citation71 Prolonged light exposure is a determinant factor in inducing neurodegeneration of photoreceptors by apoptosis. Upon light-elicited stress, aged Nup358+/− mice have suppressed apoptosis and reduced membrane dysgenesis in central retina regions as compared with wild type mice.Citation71 Mechanistically the neuroprotective function of Nup358 is not understood, but reduced levels of Nup358 coincide with reduced levels of free fatty acids, which might be of benefit for compensating the increase in light-elicited oxidative stress in central retina regions.Citation71 Insufficiency in Nup358 furthermore causes upregulation of the orphan transmembrane tyrosine kinase receptor ErbB2 and suppression of ubiquitylation in response to light stress, which may also contribute to apoptosis suppression in central retina regions of Nup358+/− mice.Citation70 Together these studies have revealed a determinant role for Nup358 in glucose, energy and lipid homeostasis in neurons of the CNS and the retina, and implicate Nup358 (and its binding partners) as key player in neuropathic and neurodegenerative diseases.Citation96
Hypomorphic mutant alleles of Nup154 affect female and male fertility in Drosophila (see above).Citation78,Citation79,Citation99 Mutations in Nup155, the human homolog of Nup154, are associated with atrial fibrillation (AF), the most common form of sustained clinical arrhythmia.Citation100 While homozygous Nup155−/− mice die during embryonic development, heterozygous Nup155+/− mice show an AF phenotype. The AF-related mutation in Nup155 in human and the reduction of Nup155 in mice are associated both with inhibition of Hsp70 mRNA export and nuclear import of Hsp70 protein. The heterozygous mice have no overt structural abnormalities in their heart and skeletal muscle, but atrial myocytes showed significantly shortened action potential duration as seen in AF.Citation100 The importance of Nup155 for normal cardiac function is further supported by studies in rat. Here Nup155 was identified as HDAC4-interacting protein in ventricular cardiomyocytes from neonatal rats, which appears important for sarcomere formation and myocyte growth.Citation101 HDAC4-target genes control cardiac growth and truncated mutants of Nup155 that fail to bind HDAC4 suppressed HDAC4-induced gene expression as well as chromatin association of Nup155, suggesting that Nup155-mediated localization of HDAC-4 is required for HDAC4’s effect on gene expression in cardiomyocytes.Citation101
Last, but not least, Nup96 plays a role in innate and adaptive immune response.Citation102 Nup96 and Nup98 are encoded by a single gene, their expression is induced by interferons and Nup98 plays important roles in mRNA export from the nucleus, which is targeted by viruses and regulated by interferon.Citation103,Citation104 Heterozygous Nup96+/− mice show selective alterations of the immune system with decreased expression of IFN-regulated gene products, impaired antigen presentation and impaired T cell proliferation upon immunization. Moreover, Nup96+/− mice and cells derived from these mice are highly susceptible to viral infection, indicating that Nup96 is not only regulated by interferons, but actively participates in interferon-mediated immune response.Citation102 Together these studies show that animal models are prerequisite to unravel tissue-specific functions of nucleoporins and that nucleoporins can influence pathways and processes beyond primary anticipations.
Nucleoporins and Differentiation
In an in situ muscle differentiation system using mouse C2C12 cells, Nup358 was implicated in myogenesis.Citation72 Depletion of Nup358 in myoblasts suppressed myotube formation without affecting cell viability. This study further revealed that a general change in expression levels of nucleoporins during skeletal muscle differentiation, indicating that NPCs undergo a remodeling process during differentiation.Citation72 In mice, a null allele of Nup133, a component of the Nup107-160 complex (), is disrupting terminal differentiation of neurons.Citation92 In the mouse embryo, Nup133 expression levels vary in between tissues, developmental stages and axial positions within the same tissue. Nup133-deficient epiblasts and embryonic stem cells maintain features of pluripotency and differentiate inefficiently along the neural lineage, whereas NPC assembly in the mouse embryonic tissue is not affected, suggesting that in mice Nup133 may modulate NPC activity rather than acting as a core structural component in NPC assembly.Citation92 The underlying molecular mechanisms that lead to defects in muscle and neuronal differentiation in the absence of Nup358 and Nup133, respectively, remain to be elucidated.
Conclusions
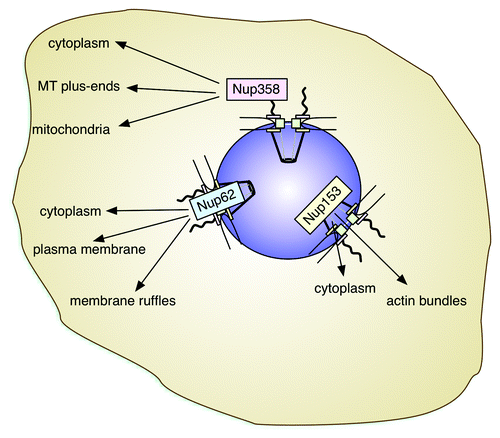
Recent progress has shed light on the role of nucleoporins beyond general nucleocytoplasmic transport. The versatility of nucleoporins makes them integral players not only in mitotic events and gene expression control, but also genome maintenance, cell migration and cellular signaling. Thereby they are acting from their location within the NPC or, as for example Nup358, Nup153 and Nup62, in the cytoplasm (). Their multifunctional properties further implicate nucleoporins in tissue-specific cascades, which, for example, control glucose, energy and lipid homeostasis and cellular differentiation. Future studies will doubtlessly address in more detail the molecular mechanisms that allow nucleoporins to act this many-sided and it will be exciting to see what other surprises go along with that.
Figure 3. Nucleoporins and their localizations outside the NPC. Nup358, Nup153 and Nup62 have been detected in the cytoplasm, frequently associated with cytoskeletal elements. Their roles in the cytoplasm appear tissue-specific. The color-code used for the nucleoporins corresponds to the one used in and . Within the NPC, Nup358 localizes to the cytoplasmic filaments (rose), Nup62 is one of the central channel nucleoporins (blue), and Nup153 a component of the nuclear basket (yellow).
Acknowledgments
This work was supported by a “Mobilité Ulysse” grant (No. F.6006.10) from the FNRS Belgium to B.F., and by the Université Libre de Bruxelles. G.C. is a “collaborateur scientifique” of the FNRS Belgium.
References
- Wälde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol 2010; 20:461 - 9; http://dx.doi.org/10.1016/j.tcb.2010.05.001; PMID: 20627572
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2010; 2:a000562; http://dx.doi.org/10.1101/cshperspect.a000562; PMID: 20630994
- Lim RY, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol 2008; 129:105 - 16; http://dx.doi.org/10.1007/s00418-007-0371-x; PMID: 18228033
- Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr Opin Cell Biol 2006; 18:342 - 7; http://dx.doi.org/10.1016/j.ceb.2006.03.006; PMID: 16631361
- D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 2008; 18:456 - 66; http://dx.doi.org/10.1016/j.tcb.2008.07.009; PMID: 18786826
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature 2007; 450:695 - 701; http://dx.doi.org/10.1038/nature06405; PMID: 18046406
- Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J 2009; 59:243 - 55; http://dx.doi.org/10.1111/j.1365-313X.2009.03865.x; PMID: 19392704
- Fahrenkrog B, Hurt EC, Aebi U, Panté N. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J Cell Biol 1998; 143:577 - 88; http://dx.doi.org/10.1083/jcb.143.3.577; PMID: 9813081
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 2004; 306:1387 - 90; http://dx.doi.org/10.1126/science.1104808; PMID: 15514115
- Elad N, Maimon T, Frenkiel-Krispin D, Lim RY, Medalia O. Structural analysis of the nuclear pore complex by integrated approaches. Curr Opin Struct Biol 2009; 19:226 - 32; http://dx.doi.org/10.1016/j.sbi.2009.02.009; PMID: 19327984
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell 1998; 1:223 - 34; http://dx.doi.org/10.1016/S1097-2765(00)80023-4; PMID: 9659919
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol 2010; 395:578 - 86; http://dx.doi.org/10.1016/j.jmb.2009.11.010; PMID: 19913035
- Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol 2003; 328:119 - 30; http://dx.doi.org/10.1016/S0022-2836(03)00266-3; PMID: 12684002
- Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol 2005; 15:221 - 6; http://dx.doi.org/10.1016/j.sbi.2005.03.003; PMID: 15837182
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, et al. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A 2006; 103:2172 - 7; http://dx.doi.org/10.1073/pnas.0506345103; PMID: 16461911
- Chatel G, Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal 2011; 23:1555 - 62; http://dx.doi.org/10.1016/j.cellsig.2011.05.023; PMID: 21683138
- Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci 2010; 67:2215 - 30; http://dx.doi.org/10.1007/s00018-010-0325-7; PMID: 20372967
- Nakano H, Wang W, Hashizume C, Funasaka T, Sato H, Wong RW. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle 2011; 10:425 - 33; http://dx.doi.org/10.4161/cc.10.3.14721; PMID: 21270521
- Arib G, Akhtar A. Multiple facets of nuclear periphery in gene expression control. Curr Opin Cell Biol 2011; 23:346 - 53; http://dx.doi.org/10.1016/j.ceb.2010.12.005; PMID: 21242077
- Capelson M, Doucet C, Hetzer MW. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb Symp Quant Biol 2010; 75:585 - 97; http://dx.doi.org/10.1101/sqb.2010.75.059; PMID: 21502404
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009; 136:284 - 95; http://dx.doi.org/10.1016/j.cell.2008.11.037; PMID: 19167330
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol 2001; 154:71 - 84; http://dx.doi.org/10.1083/jcb.200101089; PMID: 11448991
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004; 6:1114 - 21; http://dx.doi.org/10.1038/ncb1184; PMID: 15502822
- Ball JR, Ullman KS. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 2005; 114:319 - 30; http://dx.doi.org/10.1007/s00412-005-0019-3; PMID: 16133350
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell 2004; 15:1991 - 2002; http://dx.doi.org/10.1091/mbc.E03-10-0743; PMID: 14718558
- Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 1999; 18:1982 - 95; http://dx.doi.org/10.1093/emboj/18.7.1982; PMID: 10202161
- Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol 2003; 4:757 - 66; PMID: 14570049
- Fahrenkrog B, Maco B, Fager AM, Köser J, Sauder U, Ullman KS, et al. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol 2002; 140:254 - 67; http://dx.doi.org/10.1016/S1047-8477(02)00524-5; PMID: 12490173
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J 2001; 20:5703 - 14; http://dx.doi.org/10.1093/emboj/20.20.5703; PMID: 11598013
- Panté N, Thomas F, Aebi U, Burke B, Bastos R. Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes. J Struct Biol 2000; 129:306 - 12; http://dx.doi.org/10.1006/jsbi.2000.4232; PMID: 10806081
- Paulillo SM, Phillips EM, Köser J, Sauder U, Ullman KS, Powers MA, et al. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol 2005; 351:784 - 98; http://dx.doi.org/10.1016/j.jmb.2005.06.034; PMID: 16045929
- Schwarz-Herion K, Maco B, Sauder U, Fahrenkrog B. Domain topology of the p62 complex within the 3-D architecture of the nuclear pore complex. J Mol Biol 2007; 370:796 - 806; http://dx.doi.org/10.1016/j.jmb.2007.05.030; PMID: 17544442
- Chatel G, Desai SH, Mattheyses AL, Powers MA, Fahrenkrog B. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J Struct Biol 2012; 177:81 - 9; http://dx.doi.org/10.1016/j.jsb.2011.11.004; PMID: 22100335
- Paulillo SM, Powers MA, Ullman KS, Fahrenkrog B. Changes in nucleoporin domain topology in response to chemical effectors. J Mol Biol 2006; 363:39 - 50; http://dx.doi.org/10.1016/j.jmb.2006.08.021; PMID: 16962132
- Lim RY, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science 2007; 318:640 - 3; http://dx.doi.org/10.1126/science.1145980; PMID: 17916694
- Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell 2002; 110:349 - 60; http://dx.doi.org/10.1016/S0092-8674(02)00836-X; PMID: 12176322
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, et al. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol 2001; 153:1465 - 78; http://dx.doi.org/10.1083/jcb.153.7.1465; PMID: 11425876
- Oka M, Asally M, Yasuda Y, Ogawa Y, Tachibana T, Yoneda Y. The mobile FG nucleoporin Nup98 is a cofactor for Crm1-dependent protein export. Mol Biol Cell 2010; 21:1885 - 96; http://dx.doi.org/10.1091/mbc.E09-12-1041; PMID: 20375145
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 2011; 144:539 - 50; http://dx.doi.org/10.1016/j.cell.2011.01.012; PMID: 21335236
- Kowalczyk SW, Kapinos L, Blosser TR, Magalhães T, van Nies P, Lim RY, et al. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat Nanotechnol 2011; 6:433 - 8; http://dx.doi.org/10.1038/nnano.2011.88; PMID: 21685911
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, et al. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol 2008; 180:857 - 65; http://dx.doi.org/10.1083/jcb.200707026; PMID: 18316408
- Krull S, Thyberg J, Björkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 2004; 15:4261 - 77; http://dx.doi.org/10.1091/mbc.E04-03-0165; PMID: 15229283
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 2002; 13:1282 - 97; http://dx.doi.org/10.1091/mbc.01-11-0538; PMID: 11950939
- Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell 2003; 14:600 - 10; http://dx.doi.org/10.1091/mbc.E02-09-0582; PMID: 12589057
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol 1991; 114:169 - 83; http://dx.doi.org/10.1083/jcb.114.1.169; PMID: 2050741
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J Biochem 1993; 113:377 - 82; PMID: 8486610
- Monette A, Panté N, Mouland AJ. HIV-1 remodels the nuclear pore complex. J Cell Biol 2011; 193:619 - 31; http://dx.doi.org/10.1083/jcb.201008064; PMID: 21576391
- Hubert T, Van Impe K, Vandekerckhove J, Gettemans J. The actin-capping protein CapG localizes to microtubule-dependent organelles during the cell cycle. Biochem Biophys Res Commun 2009; 380:166 - 70; http://dx.doi.org/10.1016/j.bbrc.2009.01.064; PMID: 19166812
- Hubert T, Vandekerckhove J, Gettemans J. Exo70-mediated recruitment of nucleoporin Nup62 at the leading edge of migrating cells is required for cell migration. Traffic 2009; 10:1257 - 71; http://dx.doi.org/10.1111/j.1600-0854.2009.00940.x; PMID: 19552648
- Nakahara J, Kanekura K, Nawa M, Aiso S, Suzuki N. Abnormal expression of TIP30 and arrested nucleocytoplasmic transport within oligodendrocyte precursor cells in multiple sclerosis. J Clin Invest 2009; 119:169 - 81; PMID: 19104151
- Prodon F, Hanawa K, Nishida H. Actin microfilaments guide the polarized transport of nuclear pore complexes and the cytoplasmic dispersal of Vasa mRNA during GVBD in the ascidian Halocynthia roretzi. Dev Biol 2009; 330:377 - 88; http://dx.doi.org/10.1016/j.ydbio.2009.04.006; PMID: 19362546
- Zhou L, Panté N. The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett 2010; 584:3013 - 20; http://dx.doi.org/10.1016/j.febslet.2010.05.038; PMID: 20561986
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem 1995; 270:14209 - 13; http://dx.doi.org/10.1074/jbc.270.23.14209; PMID: 7775481
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, et al. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol 2002; 158:63 - 77; http://dx.doi.org/10.1083/jcb.200202088; PMID: 12105182
- Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, et al. A giant nucleopore protein that binds Ran/TC4. Nature 1995; 376:184 - 8; http://dx.doi.org/10.1038/376184a0; PMID: 7603572
- Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell 2008; 19:2300 - 10; http://dx.doi.org/10.1091/mbc.E07-12-1279; PMID: 18305100
- Hutten S, Wälde S, Spillner C, Hauber J, Kehlenbach RH. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J Cell Sci 2009; 122:1100 - 10; http://dx.doi.org/10.1242/jcs.040154; PMID: 19299463
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 1999; 274:37370 - 8; http://dx.doi.org/10.1074/jbc.274.52.37370; PMID: 10601307
- Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Wälde S, et al. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol 2011; 194:597 - 612; http://dx.doi.org/10.1083/jcb.201102018; PMID: 21859863
- Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 2003; 162:991 - 1001; http://dx.doi.org/10.1083/jcb.200304080; PMID: 12963708
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 2005; 7:626 - 32; http://dx.doi.org/10.1038/ncb1263; PMID: 15908946
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol 2004; 14:611 - 7; http://dx.doi.org/10.1016/j.cub.2004.03.031; PMID: 15062103
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 2002; 108:109 - 20; http://dx.doi.org/10.1016/S0092-8674(01)00633-X; PMID: 11792325
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 2008; 133:103 - 15; http://dx.doi.org/10.1016/j.cell.2008.01.045; PMID: 18394993
- Klein UR, Haindl M, Nigg EA, Muller S. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell 2009; 20:410 - 8; http://dx.doi.org/10.1091/mbc.E08-05-0511; PMID: 18946085
- Kirsh O, Seeler JS, Pichler A, Gast A, Müller S, Miska E, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 2002; 21:2682 - 91; http://dx.doi.org/10.1093/emboj/21.11.2682; PMID: 12032081
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 2010; 8:e1000350; http://dx.doi.org/10.1371/journal.pbio.1000350; PMID: 20386726
- Cai Y, Singh BB, Aslanukov A, Zhao H, Ferreira PA. The docking of kinesins, KIF5B and KIF5C, to Ran-binding protein 2 (RanBP2) is mediated via a novel RanBP2 domain. J Biol Chem 2001; 276:41594 - 602; http://dx.doi.org/10.1074/jbc.M104514200; PMID: 11553612
- Mavlyutov TA, Cai Y, Ferreira PA. Identification of RanBP2- and kinesin-mediated transport pathways with restricted neuronal and subcellular localization. Traffic 2002; 3:630 - 40; http://dx.doi.org/10.1034/j.1600-0854.2002.30905.x; PMID: 12191015
- Cho KI, Yi H, Tserentsoodol N, Searle K, Ferreira PA. Neuroprotection resulting from insufficiency of RANBP2 is associated with the modulation of protein and lipid homeostasis of functionally diverse but linked pathways in response to oxidative stress. Dis Model Mech 2010; 3:595 - 604; http://dx.doi.org/10.1242/dmm.004648; PMID: 20682751
- Cho KI, Yi H, Yeh A, Tserentsoodol N, Cuadrado L, Searle K, et al. Haploinsufficiency of RanBP2 is neuroprotective against light-elicited and age-dependent degeneration of photoreceptor neurons. Cell Death Differ 2009; 16:287 - 97; http://dx.doi.org/10.1038/cdd.2008.153; PMID: 18949001
- Asally M, Yasuda Y, Oka M, Otsuka S, Yoshimura SH, Takeyasu K, et al. Nup358, a nucleoporin, functions as a key determinant of the nuclear pore complex structure remodeling during skeletal myogenesis. FEBS J 2011; 278:610 - 21; http://dx.doi.org/10.1111/j.1742-4658.2010.07982.x; PMID: 21205196
- Joseph J, Dasso M. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett 2008; 582:190 - 6; http://dx.doi.org/10.1016/j.febslet.2007.11.087; PMID: 18070602
- Murawala P, Tripathi MM, Vyas P, Salunke A, Joseph J. Nup358 interacts with APC and plays a role in cell polarization. J Cell Sci 2009; 122:3113 - 22; http://dx.doi.org/10.1242/jcs.037523; PMID: 19654215
- Collin L, Schlessinger K, Hall A. APC nuclear membrane association and microtubule polarity. Biol Cell 2008; 100:243 - 52; http://dx.doi.org/10.1042/BC20070123; PMID: 18042042
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development 2009; 136:3699 - 714; http://dx.doi.org/10.1242/dev.030338; PMID: 19855013
- Zhang X, Yang H, Yu J, Chen C, Zhang G, Bao J, et al. Genomic organization, transcript variants and comparative analysis of the human nucleoporin 155 (NUP155) gene. Gene 2002; 288:9 - 18; http://dx.doi.org/10.1016/S0378-1119(02)00470-5; PMID: 12034489
- Grimaldi MR, Cozzolino L, Malva C, Graziani F, Gigliotti S. nup154 genetically interacts with cup and plays a cell-type-specific function during Drosophila melanogaster egg-chamber development. Genetics 2007; 175:1751 - 9; http://dx.doi.org/10.1534/genetics.106.062844; PMID: 17277377
- Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, et al. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J Cell Biol 1998; 142:1195 - 207; http://dx.doi.org/10.1083/jcb.142.5.1195; PMID: 9732281
- Colozza G, Montembault E, Quénerch’du E, Riparbelli MG, D’Avino PP, Callaini G. Drosophila nucleoporin Nup154 controls cell viability, proliferation and nuclear accumulation of Mad transcription factor. Tissue Cell 2011; 43:254 - 61; http://dx.doi.org/10.1016/j.tice.2011.05.001; PMID: 21696798
- Chen X, Xu L. Specific nucleoporin requirement for Smad nuclear translocation. Mol Cell Biol 2010; 30:4022 - 34; http://dx.doi.org/10.1128/MCB.00124-10; PMID: 20547758
- Xu L, Yao X, Chen X, Lu P, Zhang B, Ip YT. Msk is required for nuclear import of TGF-beta/BMP-activated Smads. J Cell Biol 2007; 178:981 - 94; http://dx.doi.org/10.1083/jcb.200703106; PMID: 17785517
- Yao X, Chen X, Cottonham C, Xu L. Preferential utilization of Imp7/8 in nuclear import of Smads. J Biol Chem 2008; 283:22867 - 74; http://dx.doi.org/10.1074/jbc.M801320200; PMID: 18519565
- Xu L, Kang Y, Cöl S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol Cell 2002; 10:271 - 82; http://dx.doi.org/10.1016/S1097-2765(02)00586-5; PMID: 12191473
- Xu L, Alarcón C, Cöl S, Massagué J. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem 2003; 278:42569 - 77; http://dx.doi.org/10.1074/jbc.M307601200; PMID: 12917407
- Gloerich M, Vliem MJ, Prummel E, Meijer LA, Rensen MG, Rehmann H, et al. The nucleoporin RanBP2 tethers the cAMP effector Epac1 and inhibits its catalytic activity. J Cell Biol 2011; 193:1009 - 20; http://dx.doi.org/10.1083/jcb.201011126; PMID: 21670213
- Gloerich M, Bos JL. Epac: defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol 2010; 50:355 - 75; http://dx.doi.org/10.1146/annurev.pharmtox.010909.105714; PMID: 20055708
- Mackay DR, Elgort SW, Ullman KS. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell 2009; 20:1652 - 60; http://dx.doi.org/10.1091/mbc.E08-08-0883; PMID: 19158386
- Wälde S, Thakar K, Hutten S, Spillner C, Nath A, Rothbauer U, et al. The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic 2012; 13:218 - 33; http://dx.doi.org/10.1111/j.1600-0854.2011.01302.x; PMID: 21995724
- Moudry P, Lukas C, Macurek L, Neumann B, Heriche JK, Pepperkok R, et al. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ 2011; http://dx.doi.org/10.1038/cdd.2011.150; PMID: 22075984
- Lemaìtre C, Fischer B, Kalousi A, Hoffbeck AS, Guirouilh-Barbat J, Shahar OD, et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene 2012; http://dx.doi.org/10.1038/onc.2011.638; PMID: 22249246
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 2008; 14:831 - 42; http://dx.doi.org/10.1016/j.devcel.2008.03.011; PMID: 18539113
- Olsson M, Schéele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res 2004; 292:359 - 70; http://dx.doi.org/10.1016/j.yexcr.2003.09.014; PMID: 14697343
- Ferreira PA, Nakayama TA, Pak WL, Travis GH. Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature 1996; 383:637 - 40; http://dx.doi.org/10.1038/383637a0; PMID: 8857542
- Ferreira PA, Nakayama TA, Travis GH. Interconversion of red opsin isoforms by the cyclophilin-related chaperone protein Ran-binding protein 2. Proc Natl Acad Sci U S A 1997; 94:1556 - 61; http://dx.doi.org/10.1073/pnas.94.4.1556; PMID: 9037092
- Aslanukov A, Bhowmick R, Guruju M, Oswald J, Raz D, Bush RA, et al. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet 2006; 2:e177; http://dx.doi.org/10.1371/journal.pgen.0020177; PMID: 17069463
- Cho KI, Cai Y, Yi H, Yeh A, Aslanukov A, Ferreira PA. Association of the kinesin-binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic 2007; 8:1722 - 35; http://dx.doi.org/10.1111/j.1600-0854.2007.00647.x; PMID: 17887960
- Cho KI, Yi H, Desai R, Hand AR, Haas AL, Ferreira PA. RANBP2 is an allosteric activator of the conventional kinesin-1 motor protein, KIF5B, in a minimal cell-free system. EMBO Rep 2009; 10:480 - 6; http://dx.doi.org/10.1038/embor.2009.29; PMID: 19305391
- Riparbelli MG, Gigliotti S, Callaini G. The Drosophila nucleoporin gene nup154 is required for correct microfilament dynamics and cell death during oogenesis. Cell Motil Cytoskeleton 2007; 64:590 - 604; http://dx.doi.org/10.1002/cm.20206; PMID: 17410542
- Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 2008; 135:1017 - 27; http://dx.doi.org/10.1016/j.cell.2008.10.022; PMID: 19070573
- Kehat I, Accornero F, Aronow BJ, Molkentin JD. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol 2011; 193:21 - 9; http://dx.doi.org/10.1083/jcb.201101046; PMID: 21464227
- Faria AM, Levay A, Wang Y, Kamphorst AO, Rosa ML, Nussenzveig DR, et al. The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 2006; 24:295 - 304; http://dx.doi.org/10.1016/j.immuni.2006.01.014; PMID: 16546098
- Von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell 2000; 6:1243 - 52; http://dx.doi.org/10.1016/S1097-2765(00)00120-9; PMID: 11106761
- Enninga J, Levy DE, Blobel G, Fontoura BM. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 2002; 295:1523 - 5; http://dx.doi.org/10.1126/science.1067861; PMID: 11809937