Abstract
In the past 15 years our perception of nuclear envelope function has evolved perhaps nearly as much as the nuclear envelope itself evolved in the last 3 billion years. Historically viewed as little more than a diffusion barrier between the cytoplasm and the nucleoplasm, the nuclear envelope is now known to have roles in the cell cycle, cytoskeletal stability and cell migration, genome architecture, epigenetics, regulation of transcription, splicing, and DNA replication. Here we will review both what is known and what is speculated about the role of the nuclear envelope in genome organization, particularly with respect to the positioning and repositioning of genes and chromosomes within the nucleus during differentiation.
Introduction
The nuclear envelope (NE) is a double-membrane system with the outer nuclear membrane (ONM) continuous with the endoplasmic reticulum (ER) and facing the cytoplasm while the inner nuclear membrane (INM) faces the nucleoplasm ().Citation1 Both membranes are joined in points where the nuclear pore complexes (NPCs) are inserted. These are >40 MDa gateways built from multiple copies of roughly 30 proteins called nucleoporins that mediate transport of proteins and RNA between the nucleus and cytoplasm.Citation2 Both the INM and ONM harbor specific sets of proteins including several integral membrane proteins generally referred to as NETs for Nuclear Envelope Transmembrane protein.Citation3 Cytoplasmic filament systems are tethered to the nucleus through several NETs of the ONM. These in turn form connections across the lumen of the NE to INM proteins that are grounded in the lamin type V intermediate filament polymer that lines the INM.Citation4 Lamin mutations reduce nuclear resistance to mechanical stress, indicating the polymer confers structural support; however, the baseline levels of this structural support vary among different cell types.Citation5–Citation9 This is because ratios of the different lamin isoforms also vary among cell typesCitation10 and each isoform has a different binding strength.Citation7 NPC proteins, lamins, and several NETs of the INM interact directly with DNA, chromatin and/or chromatin-associated proteins.Citation11
Within the interphase nucleus the 3-dimensional architecture of the genome is not random. For example, denser chromatin observed by electron microscopy tends to be concentrated at the nuclear periphery and around nucleoli and centromeres.Citation12 Individual chromosomes also have a non-random distribution.Citation13 Each chromosome occupies a discrete non-overlapping territory in the interphase nucleus, yet certain specific chromosomes tend to be at the nuclear periphery while others tend be in the interior.Citation14 Such preferred positioning is observed in most cell types for some chromosomes, while other chromosomes favor peripheral or internal positioning only in certain cell types. Most of the non-random genome positioning that has been observed is in relation to the nuclear periphery, suggesting that the NE is a tethering point for chromatin. The NE is perhaps the most logical choice for a structure from which to establish 3-dimensional genome architecture because the lamin polymer gives it structural stability to serve as an anchor point and also the dynamic stability to serve as a reference point. The interaction of NPC proteins, lamins and NETs with chromatinCitation11 is moreover consistent with this idea; however at this point the interactions observed tend to be very general and cannot adequately explain the tissue specific aspects of spatial genome architecture.
Likewise our understanding of the functional relevance of such 3-dimensional genome organization remains obscured. The central idea embraced is that it adds an additional layer to genome regulation.Citation15,Citation16 Many theories have been proposed for the detailed mechanism of gene regulation from sterically blocking access to genes to propagating silencing epigenetic marks to bringing trans regulatory elements together. Yet a greater number of contradictory results have been reported than there are theories and results have contradicted even on the basic question of whether the spatial organization of the genome contributes at all to regulation of gene expression. Thus the questions of how particular genome organizational patterns are established and what the consequences are for genome regulation remain the central questions in this area. We will focus on the former.
Patterns of Genome Organization
In the late 1800s Carl Rabl made the earliest scientific observation indicating that genome organization is not random, noting that the centromeres in nuclei from salamander larvae were located at the nuclear periphery.Citation17 In mammals centromeres are not typically located at the periphery, but they do accumulate at the NE in certain cell types such as human neutrophils.Citation18 It is also more common to find telomeres at the periphery than centromeres in mammalian cells; however, this non-random positioning of telomeres tends to be transient, occurring just prior to or during meiotic recombination. The structure known as the meiotic bouquet, where telomeres are tethered to one side of the NE, could orient chromosomes so as to facilitate synaptonemal complex formation and homologous recombination.Citation19 Though NE tethering of telomeres is usually transient, this connection appears to be permanent in yeastCitation20,Citation21 and is maintained throughout sperm development in mammals.Citation22
While the non-random positioning of general repetitive sequences such as centromeres and telomeres that occur on all chromosomes tends to be transient in mammals, the non-random distribution of individual chromosomes is maintained and heritable. Studies using whole chromosome painting techniques revealed that specific chromosomes tend to have characteristic positions within the 3-dimensional framework of the interphase nucleus (). For example, in human fibroblasts chromosome 18 tends to be located at the nuclear periphery while chromosome 19 tends to be internal.Citation14 As chromosome 18 has a lower density of genes compared to chromosome 19 it was proposed that this non-random distribution might reflect the tendency to observe more electron dense/silent chromatin at the periphery and indeed this is partially supported by other chromosomes that tend to be peripheral. However, differences were observed for the favored positioning of the same individual chromosome between cell types. For example mouse chromosome 5 tends to be at the periphery in lung cells while being internal in blood and liver cells.Citation23 Even among different blood cell lineages certain chromosomes favor peripheral or internal positioning: for example, chromosome 6 was peripheral in CD8+ T-cells but internal in CD4+ T-cells.Citation24 Where many chromosomes have been assayed, typically just a subset are observed to change in position. In addition to this distribution with respect to the periphery some internal chromosomes also tended to be located next to one another in particular tissues. In fact the tendency for two chromosomes to be adjacent in interphase in a particular tissue was found to correlate with types of translocations common to tumors of those tissues.Citation25
In addition to this potential link to cancer the positioning of chromosomes in the nucleus may play a role in the pathology of several diseases linked to the NE. There are now over twenty diseases linked to mutations in NE proteins, both lamins and NETs.Citation26–Citation28 Just as patterns of chromosome distribution are tissue-specific, so typically is the focus of pathology in NE diseases that range from muscular dystrophy to lipodystrophy, restrictive dermopathy, neuropathy and premature aging progeroid syndromes. A mutation in the LMNA gene (that encodes Lamin A) causing Hutchison-Gilford progeria syndrome, E145K, yielded an abnormal distribution of telomeres and clustering of centromeres,Citation29 while several other mutations in LMNA that cause variously a neuropathy, lipodystrophy and multiple muscular dystrophies reposition chromosomes 13 and 18 away from the nuclear periphery.Citation30 Mutations in the NET emerin that also cause muscular dystrophy had a similar effect on these chromosomes.Citation30 Nonetheless, the link between chromosome repositioning and disease pathology is not clear because different mutations that cause the same disease can yield different effects on chromosome positioning: a LMNA mutation causing cardiomyopathy, E161K, results in chromosome 13 losing its normal peripheral localization, while another mutation, D596N, causing the same disease maintains chromosome 13 at the periphery.Citation31 It is important to note that the positioning of only a small subset of chromosomes are affected in these disorders and that just as the diseases focus pathology in particular tissues the effects may be cell-type specific as changes in chromosome positioning were not observed in all studies.Citation32,Citation33
Individual genes on chromosomes also reposition during differentiation. For example, the immunoglobulin H (IgH) locus moves from the nuclear periphery to the nuclear interior during B lymphocyte development.Citation34 In such studies the repositioning of the chromosome containing the gene was often not tested to determine if gene repositioning correlated with chromosome repositioning. However this was done in a few recent studies where the gene locus of interest moved within and sometimes beyond the general chromosome territory (as defined by whole chromosome painting) while the chromosome itself typically did not correspondingly change position with respect to the NE.Citation35,Citation36 Nonetheless, in one of these studies that focused on repositioning in adipocyte differentiation, the FABP4 gene and its host chromosome both strongly shifted from the nuclear periphery to the nuclear interior.Citation36 There is too little data available correlating gene and chromosome repositioning to determine whether gene repositioning can drive chromosome repositioning or vice versa, but the several instances where gene positioning changed without corresponding changes in chromosome positioning suggest that chromosome repositioning might be a fortuitous consequence of global developmental restructuring of gene positions. During neurogenesis the Mash1 (Ascl1) locus also moves away from the periphery.Citation37 In this case the state of epigenetic marks on the locus was also followed during the differentiation process. Interestingly, when at the periphery the locus had accumulated silencing marks and these had largely disappeared when the locus was in the nuclear interior.Citation37
This observation introduces yet another aspect of non-random positioning in the nucleus: the distribution of heterochromatin. The original definition of heterochromatin was the presumed denser material observed in negative stain electron microscopy. Long before capabilities existed for identifying individual chromosome territories or gene positions it was noted that a majority of this electron-dense chromatin tended to be at the nuclear periphery in most interphase cells. Focused studies moreover suggested that this chromatin is in direct contact with the mammalian NE.Citation38 These direct interactions, presumed to be with the lamin polymer, were moreover supported by NE retention of chromatin after extraction at high ionic strengths.Citation39
While the tendency for this dense chromatin to accumulate at the periphery is common to nearly all cell types, there are particular patterns and degrees characteristic of distinct cell types. For example, fibroblasts tend to have a more uniform distribution of dense chromatin throughout the periphery while epithelial cells tend to have a more patchy distribution. Neurons tend to have little observable dense chromatin while lymphoblasts tend to have an enormous amount that extends several microns into the nucleus from the periphery.Citation12 Thus the distribution of chromatin within the nucleus, particularly with respect to the nuclear periphery, is both non-random and tissue-specific.
The importance of these patterns of electron-dense peripheral chromatin distribution is underscored by the fact that, just like chromosome positioning, they are perturbed in cells from patients with NE diseases. Normal fibroblasts tend to have a reasonable amount of this dense peripheral chromatin generally distributed throughout the periphery. In contrast, in patients with NE mutations linked to muscular dystrophy much of this dense chromatin appears to have broken away from the nuclear periphery,Citation40–Citation44 while in patients with progeroid syndromes and mandibuloacral dysplasia it appears to have completely dissipated.Citation45,Citation46 Patients with familial partial lipodystrophy, Dunnigan type still have dense chromatin at the periphery, but it is no longer uniformly distributed.Citation46 That the patterns are not just disrupted, but disrupted in reproducible ways for each disorder suggests that these spatial genome organizational patterns are functionally relevant.
One suggested function can be found in observations that the lymphoblast dense peripheral chromatin diminished upon activation, consistent with the idea that the strongly negatively stained material was silent chromatin.Citation47,Citation48 However, immunostaining with antibodies to epigenetic markers of silent chromatin have not revealed a similarly strong enrichment at the periphery with the exception that heterochromatin protein 1 alpha (HP1α) seems to have a distinct subpopulation at the NE.Citation49 Nonetheless, several specific interactions have been reported between NE proteins and epigenetically silent marks on chromatin (see below) and the majority of chromatin found in contact with the NE by high-throughput studies is in a silent configuration.Citation50,Citation51
How these patterns are established is an important question in cell biology. As many specific chromosome regions appear to be in direct contact with the periphery by electron microscopy it logically follows that they are driven at least in part by physical tethering to NE proteins. Consistent with this idea several NE proteins have been found to interact with DNA and specific chromatin proteins.
Specific NE Interactions with Chromatin and DNA
NPCs, lamins and NETs all can interact with a variety of nuclear components (). These include DNA, chromatin proteins such as histones, epigenetic marks on chromatin and transcriptional regulators. The first NPC interaction with chromatin described was the tethering of yeast telomeres that required the function of Mlp, which is a homolog of the mammalian nucleoporin Tpr.Citation20,Citation21 Yeast NPCs also bind transcription factorsCitation52,Citation53 and it is thought that such interactions promote activation of genes where an epigenetic regulation referred to as boundary activity segregates active from inactive chromatin.Citation54 NPCs are likely to be the main tethering point for chromatin in yeast and other fungi because these organisms lack a nuclear intermediate filament lamin polymer (see below). Mammalian NPCs also interact with chromatin, but, whereas yeast NPCs were strongly activating, a preponderance of silencing epigenetic marks associated with mammalian nucleoporins.Citation55 Interpretation of NPC results was confounded by there being separate nucleoporin pools assembled into the megadalton NPC transport channels or distributed throughout the nucleoplasm. Fusing nucleoporins to a membrane span to keep them at the NPC/NE clarified that the nucleoplasmic pools interact with active chromatin while the NPC/NE pools tend to interact with silenced chromatin.Citation56,Citation57
Biochemical efforts to identify chromatin proteins interacting with lamins similarly suffer from the inability to properly distinguish nucleoplasmic pools from those assembled into the polymer at the NE. Because the vast majority is thought to be in the polymeric form based on the intensity of signals for the different pools by immunofluoresence microscopy, the interactions observed are considered to reflect the NE pool; however, the abundance may be countered by the greater facility in isolating the unpolymerized population. It is also noteworthy that studies have generally NOT tested whether the different lamin subtypes bind to or have different affinities for the same chromatin proteins. There are three different lamin genes, LMNA, LMNB1 and LMNB2, each of which has multiple splice variants and the ratios of these proteins vary in different cell types.Citation10 Accessibility is also an issue as studies expressing different lamin subtypes in Xenopus oocytes revealed that the subtypes assembled different layered polymers such that Lamin A would be most proximal to chromatin;Citation58 however, lamin mutants of one subtype can disrupt the distribution of other subtypes in vivo, different subtypes are capable of heterotypic interactions in vitro, and polymer structure is less clear in somatic cells so that it remains possible that different subtype interactions might occur in some cell types.Citation7,Citation8,Citation59
Bearing in mind the above-mentioned caveats, lamins have been shown to interact with DNA at matrix or scaffold attachment regions (MARs and SARsCitation60,Citation61), specific chromatin structures such as centromeres and telomeres,Citation62,Citation63 and core histones, specifically H2A and H2B.Citation64–Citation66 On the one hand the interaction of lamins with multiple types of general chromatin suggests that they would not be involved in tissue-specific aspects of spatial genome organization, but if each lamin isoform has a different affinity for different types of chromatin, it could potentially contribute to cell-type differences in genome architecture.
Unlike nucleoporins and lamins, NETs are restricted to the NE because they are integral to the membrane. NETs, like lamins, can interact directly with DNA. LAP2β binds DNA using a domain on its amino-terminusCitation67 while MAN1 binds via a winged Helix fold on its carboxyl-terminus.Citation68 However, the majority of NET interactions identified have been with chromatin proteins or chromatin-associated proteins.
In mammals the Lamin B Receptor (LBR) binds core histones H3/H4 directly.Citation69 LBR can also bind heterochromatin through direct interaction with HP1α and HP1γ.Citation70 Chromatin that co-immunoprecipitated with LBR was highly enriched in silent epigenetic marks.Citation71 LEM-domain NETs that include LAP2, emerin and MAN1 all interact with chromatin through the DNA crosslinking protein BAF (barrier-to-autointegration factor).Citation72–Citation74 In addition to bridging chromatin-NE interactions, BAF also compacts chromatin through promoting DNA looping.Citation75,Citation76 LAP2 has multiple splice variants:Citation77,Citation78 LAP2β is the predominant membrane bound form, but there are other both soluble and transmembrane variants that also bind to BAF.Citation79–Citation81 One of these, LAP2ζ, occurs in the cytoplasm and its overexpression causes BAF to be captured in the cytoplasm thus reducing intranuclear pools.Citation81 As many NETs have multiple splice variants, it is likely that this type of competitive inhibition is common.
Finally, similar to NPC proteins, some NETs interact with transcription factors such as Lmo7 and Smads.Citation82–Citation84 However, whereas the NPC interactions are thought to positively promote transcription, the NET interactions are generally thought to sequester transcription factors away from their targets.
After several proteomic analyses of the NECitation85–Citation88 the number of NETs has grown from roughly a dozen to many hundreds with many being tissue-specific and only a small number tested for interactions with chromatin. Running just ten novel NETs in two screens for effects on genome architecture yielded one that promoted chromatin condensation and two others that repositioned a gene locus.Citation86 Thus there is a strong likelihood that the complexity of NE interactions with the genome is exponentially greater than our current understanding permits. Nonetheless, the fact of these interactions argues that specific tethering of particular genes or chromatin underlies the many distinctive patterns of spatial genome organization observed in cells.
Mechanism for Establishment of Different Organizational Patterns
If the hypothesis that specific tethering interactions at the NE direct spatial genome organization is correct, then it follows that changing NE interactions with the genome should correspondingly change genome spatial organization. Replacing the players or adding post-translational modifications that alter affinities could change NE-genome interactions. It also follows that the interactions driving specificity in the system must themselves exhibit both specificity and dominance. To test this, one could introduce a new extremely high affinity interaction into the system.
Three recent studies did just this, introducing into mammalian cells an artificial high-affinity interaction between a specific genome region and a specific NE protein and finding that it could indeed dominantly alter genome spatial organization.Citation89–Citation91 In all three studies an array of bacterial lac operator repeats (LacO) was first inserted into a mammalian genome in a region that tended to be in the nuclear interior. Separately the bacterial lac repressor (LacI) that specifically binds these repeats was fused to a NE protein and expressed in the cells carrying the array. Expression of the LacI-NE protein fusions in all cases resulted in the repositioning of the LacO array from the nuclear interior to the nuclear periphery. The binding of LacI to LacO sequences can be disrupted by addition of IPTG (isopropyl β-D-1-thiogalactopyranoside) and this correspondingly released the locus from the periphery.
Both recruiting the locus to the periphery and releasing it from the periphery required passage of the cells through mitosis. The density of proteins and established chromosome territories might be expected to block such major repositioning in interphase cells; however, in mitosis the mammalian NE breaks down early in prophase as the chromosomes condense, allowing the chromosomes to move freely. Moreover, the condensed chromosome state together with unrestricted space makes most chromosome surfaces available for interactions that would be established when the NE begins to reform in telophase. The form the broken down NE takes during mitosis is still not entirely clear: some studies are consistent with its streaming into the ERCitation92–Citation95 while others suggest it breaks down into vesicles.Citation88,Citation96–Citation102 It is also possible that both occur in the same or different cell types. For the purposes of drawing the following model () we will assume that it forms vesicles, though the critical interaction with chromatin would be essentially the same whether it has flowed into the ER or vesiculated. Thus we postulate that in telophase vesicles containing NETs from the disassembled NE would bind to partner proteins on the accessible surfaces of mitotic chromosomes (). Then as vesicles fuse to reform the NE the chromosomes with high affinity interactions would remain at the nuclear periphery while those that lack such high affinity interactions might slip into the interior (). As the chromosomes with high affinity interactions decondense taking up more of the reforming NE surface area, those lacking strong tethering interactions would likely be sterically pushed into the interior ( and D). Indeed, in the affinity studies using the LacO-LacI system, whole chromosome painting revealed that the entire chromosome moved to or from the periphery along with the LacO array.Citation89
The LacO-LacI system has an extremely high affinity as the LacO sequence was amplified 128–256 times in the array. Though this is certain to be higher than any individual interactions in mammalian cells, interaction sites on human chromosomes would likely be both abundant and widely distributed thus providing many tethering points that would in the end have the same effect as the concentrated array. Alternatively, large gene clusters such as at the IgH locus might provide unique binding sites that would create distinct microenvironments at the NE. Viewing the current set of NETs known to bind chromatin by microscopy gives no indication of microenvironments in the interphase nucleus, but, interestingly, LBR and emerin bind to distinct positions on chromosomes in telophase.Citation103 This would be consistent with some kind of microenvironment at least on mitotic chromosomes.
Once the high-affinity interactions have tethered genes/chromosomes to the NE the many general interactions such as those between lamins and histones might further cement a particular pattern of spatial genome organization. With roughly 3 million copies of lamins per mammalian nucleus and many more histones, this would provide thousands of binding sites at the NE for each chromosome at the periphery. This additional tethering could help to maintain the distinctive chromosome territories observed in interphase cells. Furthermore, some NETs can also bind to chromatin-modifying enzymes and so could function to propagate silencing through chromatin already at the NE or promote release through adding active marks. LBR and LAP2β also associate with HA95, a kinase involved in the regulation of NE and chromatin interactions.Citation104 For LAP2 this interaction is splice-variant specific and thus does not apply to the soluble splice forms. NETs can also interact with enzymes that add epigenetic marks to chromatin. The histone deacetylase HDAC3 binds to LAP2βCitation105 while the histone acetyltransferase hALP1 binds the NET SUN1.Citation106 An interaction has also been reported for LBR with the DNA methylating enzyme MeCP2.Citation107
As the overall spatial patterns differ among cell types and tissues, the establishment of particular patterns of spatial genome organization requires some tissue-specific components. General interactions such as those described between lamins and histones could not drive the tissue-specific organizational patterns as both protein sets are ubiquitously expressed in all tissues. This specificity could come in the form of epigenetic modifications of chromatin that alter affinities for NE proteins, tissue-specific chromatin binding proteins or transcriptional regulators sitting on DNA that interact with NE proteins, or tissue-specificity in the complement of NE proteins that interact with particular types of chromatin. The first possibility is supported by the changes in the distribution of epigenetic marks that occur in different cell types and during differentiationCitation108 such as changes in epigenetic marks on Mash1 between when it was at the periphery or interior.Citation37 The second possibility is consistent with the finding that several NETs can bind transcriptional regulators, which would enable genes with bound tissue-specific transcriptional regulators to be recruited to the periphery.Citation82–Citation84,Citation109 Finally, the possibility that tissue-specific NETs drive the specific chromatin interactions is supported by the identification of many tissue-specific NETs in two recent proteomic studies.Citation86,Citation88
The Dynamic Scaffold
The ability of the NE to anchor chromosomes depends on the structural stability of the lamin polymer. However, the dynamic of this interaction is very different from that of an anchor sitting on the ocean floor. Though historically both the genome and the lamin polymer were viewed as being quite rigid, live cell microscopy has revealed that nuclei move and exhibit frequent morphological aberrations while chromatin also moves dynamically. Rather than being rigid, the polymer that lines the NE is made entirely of intermediate filaments that are highly elastic and have properties like a spider's web, tough yet elastic. In contrast intermediate filaments in the cytoplasm combine with the more brick-like qualities of microtubules and actin microfilaments to form the cytoskeleton. Moreover, the lamin polymer associates with many NETs to connect it strongly to the lipid bilayer. This enables the lamina to keep chromatin tethered while still being able to stretch in response to other forces placed on the polymer by genome movements (). If the peripheral lamina nucleoskeleton were stiff and rigid it would likely break in response to such strong forces and the chromosomes would lose their tethering ( and E). Similarly, if tethered merely by transmembrane proteins strong forces from chromosomes might rip interacting NETs out of the lipid bilayer. Thus the use of both lamins and NETs is a sensible strategy to support the many dynamic movements of chromatin within the interphase nucleus.
Though whole chromosome territories are generally maintained during interphase,Citation110 within these territories individual loci can move rapidly over large distances.Citation111 Often upon transcriptional activation a locus will decondense and move until it associates with PML bodies to maximize transcriptional output.Citation112 Moreover, recent chromatin conformation capture studiesCitation113,Citation114 indicate that some loci on chromosomes move large distances to engage with other regions as far as 10 MB away and there are thousands of such interactions, some even occurring between two different chromosomes.Citation115–Citation117 Such interactions might bring enhancers together to function in trans and so additionally contribute to gene regulation as has been proposed to occur in what are termed transcription factories (). Additionally such higher order collections of active genes would increase the likelihood that processive transcriptional proteins would rapidly find new substrates after concluding one transcriptional round, effectively increasing the localized concentration of the transcriptional proteins. While large-scale movements have been observed for some loci, those at the nuclear periphery are much less mobile than those residing in the nuclear interior.Citation118 Chromatin is often considered as a rope. To carry this analogy further, one might consider the NE connections as the cowboy throwing a lasso who can then partially restrict the movements of an animal far away that is caught in the lasso. Thus NE tethering at various points along the chromosome rope could both restrict and facilitate these long-range movements with the result of enabling or preventing chromosome regions from participating in higher order structures such as the transcription factories ().
In addition to the dynamics of motion, much of the chromatin in association with the NE may dynamically exchange their tethers. A method that uses bacterial Dam methylase to label DNA in contact with particular proteinsCitation114 revealed that roughly 40% of the genome interacts with lamins.Citation119 This is obviously impossible if the same tethering points were always used as the NE has only ∼1/40th the volume of the nucleoplasm. This discrepancy likely reflects to some degree differences in the genes tethered at the NE within a population of cells and the interactions of the more minor intranuclear lamin pools, but also dynamic exchanges in tethering points within individual cells. It makes sense that some NE-chromatin interactions would exchange during the cell cycle because interactions likely must be broken when DNA is replicated. In this regard it is interesting that nearly all DNA at the nuclear periphery replicates late.Citation120
Would the NET, lamin and NPC proteins and corresponding chromatin proteins involved in tethering also exhibit dynamic properties? This cannot be addressed until specific proteins are identified, but FRAP on NPC proteins suggested that they are highly dynamic despite their assembly into >60 MDa structures.Citation121 Additionally, a recent systems modeling analysis of several NETs found that their extremely varied dynamics could be attributed to differences in the half-time of tethering in the INM.Citation122 Interestingly, the longest half-time estimated from this study for a known chromatin-binding NET was on the order of 12 minutes, much shorter than a mammalian cell cycle. Such dynamic exchange may be driven by post-translational modifications on NETs or chromatin as the CFTR gene locus is peripheral in some cells and can be repositioned into the nuclear interior upon treatment with trichostatin A, which promotes histone hyperacetylation.Citation123 Thus, the many chromatin-modifying proteins discussed above could rapidly direct changes in the specific chromosome loci tethered during interphase without notably changing the spatial positioning of the chromosome territory.
Concluding Observations
There is much left to be worked out about the molecular mechanisms supporting spatial genome organization. Not the least of these is identifying the endogenous NE and chromatin proteins that direct specific spatial genome organization patterns. The only published protein thus far linked to chromosome positioning is Lamin B1,Citation124 but it is hard to conceptualize how any of the interactions known for this protein could support a tissuespecific genome organization pattern because both lamin B1 and its known binding partners are widely expressed. The best candidates on the chromatin side are transcriptional regulators because they have been shown to bind to all NE components (NPC, lamin and NET proteins) and could provide the needed tissue-specificity to the system. This idea is consistent with observations from chromatin conformation capture studies that long-range genome interactions reported have been found to be bridged by several transcriptional regulators and the insulator protein CTCF.Citation125,Citation126
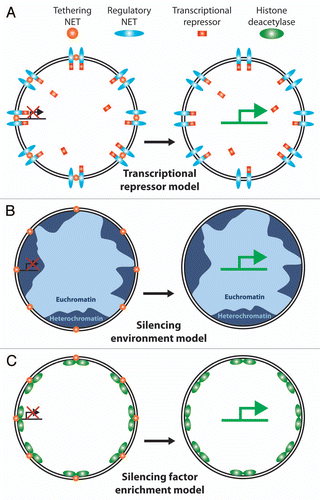
The other outstanding questions relate to the functional consequences of a particular spatial genome organization. When the gypsy insulator DNA sequence was inserted into a locus in the nuclear interior it relocated this locus to the periphery with a concomitant reduction in gene expression.Citation127 Since this finding, many studies have attempted to correlate changes in gene expression with spatial changes in gene or chromosome position. The most common mechanisms proposed for how this could work are: (1) NE tethering of a gene and/or a transcriptional regulator could be controlled to either bring the two together or keep them apart (); (2) NE recruitment of a gene could bring it to the generally silencing environment of the periphery (); or (3) NE recruitment of both a gene and a histone/DNA modifying enzyme could add to the transcriptional regulation of the gene (). However, many studies over many years have tendered many contradictory results. Even among the three studies mentioned earlier that all repositioned a LacO array to the NE, one found general gene repression as a consequence,Citation91 one found a mixture of repressed and unchanged genes,Citation89 and one found no repression at all.Citation90 Many possible explanations can be proposed to explain these discrepancies, but it is likely that such controversies will persist until the endogenous players have been identified as absent this information all these studies must use highly artificial reporter systems. Determining which proteins normally tether a particular gene or chromosome to the periphery should provide the necessary tools to answer this question.
Figures and Tables
Figure 1 NE proteins interact with chromatin proteins. The NE consists of outer (ONM) and inner (INM) nuclear membranes that fuse where the nuclear pore complexes (NPCs) are inserted. Both ONM and INM contain membrane spanning proteins that are generally referred to as NETs for Nuclear Envelope Transmembrane protein. Underlying the INM is the lamin intermediate filament polymer (green). The INM harbors a specific set of NETs (red), which together with the lamins are referred to as the lamina. Lamins, NETs and NPCs can all interact with chromatin components like Barrier-to-Autointegration Factor (BAF), Heterochromatin Protein 1 (HP1) and histones.
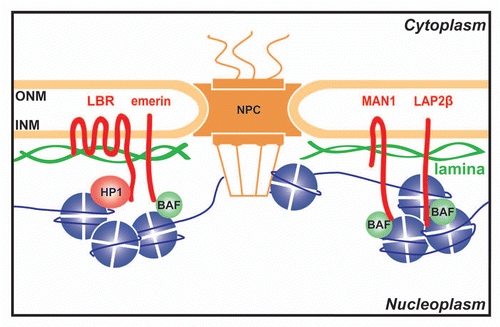
Figure 2 Each chromosome has a distinctive positioning in the nucleus with respect to the nuclear periphery. In human HT1080 fibroblast cells chromosomes 5 and 17 tend to be in the nuclear interior while chromosome 13 tends to be at the periphery. The chromosome is shown in green and the DNA from DAPI staining in blue delineates the nuclear boundary. Cells were fixed with formaldehyde prior to processing for 2D FISH so that much of the 3D structure is maintained. Deconvolved sections from z-series through the nucleus are shown.
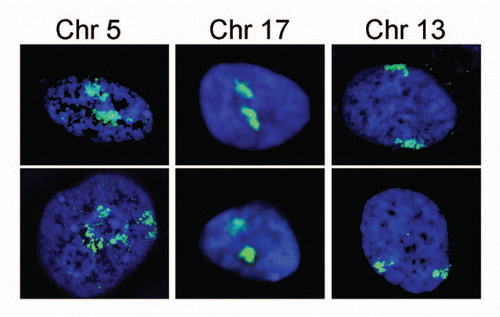
Figure 3 Speculative affinity mechanism for establishment of spatial chromosome organizational patterns. (A) During mitosis the NE either breaks down into ER /NE vesicles or diffuses into the ER. The vesicle model is shown here. Distinct vesicles contain specific components (red and green triangles representing different NETs). Components of some vesicles interact with regions of particular condensed chromosomes. (B) At the end of mitosis the NE starts to reform from vesicles with some specific chromosomes still being attached to particular vesicles. (C) The NE has reformed with some chromosomes being trapped at the NE due to specific NE components that have a high affinity for these chromosomes. The chromosomes are still largely condensed. (D) During interphase the chromosomes decondense and occupy distinct territories within the interphase nucleus. At this point a multitude of less specific lower affinity interactions from the lamina would be expected to help maintain the positioning established in mitosis.
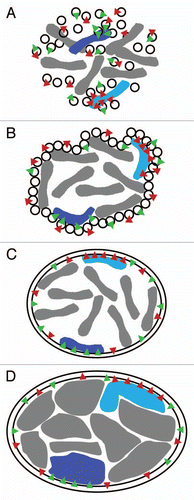
Figure 4 Elastic behavior of the NE. If the intermediate filament lamin polymer supporting the NE were very stiff like other filament systems in the cytoplasm, it would be likely to rupture under the stresses exerted on it by the genome (e.g., growth during replication or rapid movements of regions within chromosome territories (A–C) or the cytoskeleton (D–F). When all components are bound and working together with an elastic nucleoskeleton the whole system can move slightly together while providing a counterforce to that exerted (A and D). In contrast, if the nucleoskeleton functioned like a brick and mortar scaffolding, then components of the system might pull apart from one another or the lamin polymer might physically rupture as do microtubules when subjected to strong bending forces (B, C, E and F). In this case NETs that have strong interactions with chromatin and/or the lamin polymer might even be pulled out of the membrane.
Figure 5 Transcription factories in the nuclear interior can be affected by NE affinity for distinct regions on chromosomes. Because each chromosome is essentially a continuous strand of DNA folded over on itself for compaction into the 10 or 30 nm fibers observed by electron microscopy, it can be unraveled or compacted based on connections to the NE. (A) Recent 3C and 4C chromatin capture studies have revealed interactions within chromosomes and between adjacent chromosomes (e.g., chromosomes A–C in the diagram). Some of these are thought to act as transcription factories where greater local concentrations of transcriptional proteins can optimize transcription (larger green arrows). The availability of chromosome regions to participate in such transcription factories may depend on connections with chromatin and the INM (regions marked i and ii). (B) Changing the pattern of the connections to the INM (e.g., by recruiting also locus iii to the NE) will also affect the transcription factory structure and transcription levels.
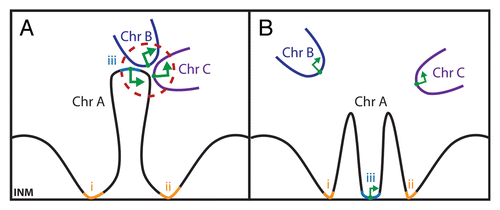
Figure 6 Possible mechanisms for NE interactions to regulate gene expression. Various models have been proposed for how NE tethering of a chromosome or gene could affect gene regulation. (A) Transcriptional regulator sequestration. One NET (orange ball) recruits a gene to the periphery while another NET (blue oval) recruits a transcriptional regulator, in this case a transcriptional repressor (red rectangle). Because the environment of the NE is only ∼1/40 the volume of the nucleus this would effectively increase the local concentration of the transcriptional regulator to keep the gene more tightly repressed. During differentiation expression would shut down for the NET tethering the gene (or the transcriptional repressor) enabling the gene to move away from the high local concentration of the repressor and become more strongly activated. There are obviously many versions of this model. (B) Recruitment to a generally silencing environment. The majority of NE interactions with chromatin identified to date appear to involve heterochromatin by both the original definition of electron dense chromatin observed by electron microscopy and the more modern definition of histones carrying silencing modifications. Thus recruitment of a gene to the periphery could result in its silencing by the general environment. One flaw with this model is that NETs tend to be generally distributed throughout the INM and there are also patches of euchromatin at the periphery so the gene could conceivably move to an active region and not be repressed e.g., tethering NETs (gold balls) in lighter blue regions of euchromatin. (C) Silencing enzyme recruitment model. In addition to binding directly to silenced chromatin, some NETs have been found to recruit factors that modify chromatin to a silent configuration (e.g., the LAP2 interaction with HDAC3 and the LBR interaction with MeCP2). Thus merging aspects of the first two models, co-recruitment of a gene and a chromatin-silencing enzyme to the periphery would effectively shut expression from the gene.
References
- Callan HG, Tomlin SG. Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc R Soc Lond B Biol Sci 1950; 137:367 - 378
- Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly and function. Dev Cell 2003; 4:775 - 789
- Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp Cell Res 2007; 313:2167 - 2179
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008; 22:832 - 853
- Lammerding J, Schulze P, Takahashi T, Kozlov S, Sullivan T, Kamm R, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004; 113:370 - 378
- Liu J, Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, et al. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression and spatial organization of nuclear pore complexes. Mol Biol Cell 2000; 11:3937 - 3947
- Schirmer EC, Gerace L. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J Biol Chem 2004; 279:42811 - 42817
- Schirmer EC, Guan T, Gerace L. Involvement of the lamin rod domain in heterotypic lamin interactions important for nuclear organization. J Cell Biol 2001; 153:479 - 489
- Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes and accumulation of annulate lamellae. J Cell Biol 1997; 137:1001 - 1016
- Broers JL, Machiels BM, Kuijpers HJ, Smedts F, van den Kieboom R, Raymond Y, et al. A- and B-type lamins are differentially expressed in normal human tissues. Histochem Cell Biol 1997; 107:505 - 517
- Mattout-Drubezki A, Gruenbaum Y. Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell Mol Life Sci 2003; 60:2053 - 2063
- Fawcett DW. The Cell 1981; Philadelphia Saunders WB Co.
- Walter J, Joffe B, Bolzer A, Albiez H, Benedetti PA, Muller S, et al. Towards many colors in FISH on 3D-preserved interphase nuclei. Cytogenet Genome Res 2006; 114:367 - 378
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 1999; 145:1119 - 1131
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 2007; 8:104 - 115
- Mateos-Langerak J, Goetze S, Leonhardt H, Cremer T, van Driel R, Lanctot C. Nuclear architecture: Is it important for genome function and can we prove it?. J Cell Biochem 2007; 102:1067 - 1075
- Rabl C. Über Zelltheilung. Morphol Jahrb 1885; 10:214 - 330
- Aquiles Sanchez J, Karni RJ, Wangh LJ. Fluorescent in situ hybridization (FISH) analysis of the relationship between chromosome location and nuclear morphology in human neutrophils. Chromosoma 1997; 106:168 - 177
- Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 1996; 134:1109 - 1125
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 2000; 403:108 - 112
- Scherthan H, Jerratsch M, Li B, Smith S, Hulten M, Lock T, et al. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell 2000; 11:4189 - 4203
- Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci USA 2007; 104:7426 - 7431
- Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol 2004; 5:44
- Kim SH, McQueen PG, Lichtman MK, Shevach EM, Parada LA, Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet Genome Res 2004; 105:292 - 301
- Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol 2002; 12:1692 - 1697
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet 2006; 7:940 - 952
- Foisner R, Aebi U, Bonne G, Gruenbaum Y, Novelli G. 141st ENMC International Workshop inaugural meeting of the EURO-Laminopathies project “Nuclear Envelope-linked Rare Human Diseases: From Molecular Pathophysiology towards Clinical Applications”, 10–12 March 2006, Naarden, The Netherlands. Neuromuscul Disord 2007; 17:655 - 660
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res 2007; 313:2121 - 2133
- Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture and chromosome organization. Proc Natl Acad Sci USA 2009; 106:20788 - 20793
- Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, et al. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell 2007; 6:139 - 153
- Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, et al. Altered chromosomal positioning, compaction and gene expression with a lamin A/C gene mutation. PLoS One 2010; 5:14342
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerinmutant cells. Hum Mol Genet 2001; 10:211 - 219
- Meaburn KJ, Levy N, Toniolo D, Bridger JM. Chromosome positioning is largely unaffected in lymphoblastoid cell lines containing emerin or A-type lamin mutations. Biochem Soc Trans 2005; 33:1438 - 1440
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 2002; 296:158 - 162
- Morey C, Da Silva NR, Kmita M, Duboule D, Bickmore WA. Ectopic nuclear reorganisation driven by a Hoxb1 transgene transposed into Hoxd. J Cell Sci 2008; 121:571 - 577
- Szczerbal I, Foster HA, Bridger JM. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma 2009; 118:647 - 663
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci 2006; 119:132 - 140
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol 1993; 123:1671 - 1685
- Bouvier D, Hubert J, Seve AP, Bouteille M. Characterization of lamina-bound chromatin in the nuclear shell isolated from HeLa cells. Exp Cell Res 1985; 156:500 - 512
- Sewry CA, Brown SC, Mercuri E, Bonne G, Feng L, Camici G, et al. Skeletal muscle pathology in autosomal dominant Emery-Dreifuss muscular dystrophy with lamin A/C mutations. Neuropathol Appl Neurobiol 2001; 27:281 - 290
- Verga L, Concardi M, Pilotto A, Bellini O, Pasotti M, Repetto A, et al. Loss of lamin A/C expression revealed by immuno-electron microscopy in dilated cardiomyopathy with atrioventricular block caused by LMNA gene defects. Virchows Arch 2003; 443:664 - 671
- Fidzianska A, Toniolo D, Hausmanowa-Petrusewicz I. Ultrastructural abnormality of sarcolemmal nuclei in Emery-Dreifuss muscular dystrophy (EDMD). J Neurol Sci 1998; 159:88 - 93
- Maraldi NM, Squarzoni S, Sabatelli P, Lattanzi G, Ognibene A, Manzoli FA. Emery-Dreifuss muscular dystrophy, nuclear cell signaling and chromatin remodeling. Adv Enzyme Regul 2002; 42:1 - 18
- Ognibene A, Sabatelli P, Petrini S, Squarzoni S, Riccio M, Santi S, et al. Nuclear changes in a case of X-linked Emery-Dreifuss muscular dystrophy. Muscle Nerve 1999; 22:864 - 869
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA 2004; 101:8963 - 8968
- Maraldi NM, Lattanzi G, Capanni C, Columbaro M, Mattioli E, Sabatelli P, et al. Laminopathies: a chromatin affair. Adv Enzyme Regul 2006; 46:33 - 49
- Mirsky AE, Allfrey V. Biochemical activities of the cell nucleus. Dis Nerv Syst 1960; 21:23 - 28
- Hirschhorn R, Decsy MI, Troll W. The effect of PHA stimulation of human peripheral blood lymphocytes upon cellular content of euchromatin and heterochromatin. Cell Immunol 1971; 2:696 - 701
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 1999; 108:220 - 234
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 2006; 38:1005 - 1014
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaeva ES, Christophides GK, et al. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci 2007; 120:2344 - 2351
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell 2006; 21:379 - 391
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 2006; 441:774 - 778
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 2002; 109:551 - 562
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev 2008; 22:627 - 639
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140:372 - 383
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell cycle genes inside the nucleoplasm. Cell 2010; 140:360 - 371
- Goldberg MW, Huttenlauch I, Hutchison CJ, Stick R. Filaments made from A- and B-type lamins differ in structure and organization. J Cell Sci 2008; 121:215 - 225
- Delbarre E, Tramier M, Coppey-Moisan M, Gaillard C, Courvalin JC, Buendia B. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum Mol Genet 2006; 15:1113 - 1122
- Luderus ME, de Graaf A, Mattia E, den Blaauwen JL, Grande MA, de Jong L, et al. Binding of matrix attachment regions to lamin B1. Cell 1992; 70:949 - 959
- Rzepecki R, Bogachev SS, Kokoza E, Stuurman N, Fisher PA. In vivo association of lamins with nucleic acids in Drosophila melanogaster. J Cell Sci 1998; 111:121 - 129
- Baricheva EA, Berrios M, Bogachev SS, Borisevich IV, Lapik ER, Sharakhov IV, et al. DNA from Drosophila melanogaster beta-heterochromatin binds specifically to nuclear lamins in vitro and the nuclear envelope in situ. Gene 1996; 171:171 - 176
- Shoeman RL, Traub P. The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. J Biol Chem 1990; 265:9055 - 9061
- Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, et al. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc Natl Acad Sci USA 1999; 96:2852 - 2857
- Hoger TH, Krohne G, Kleinschmidt JA. Interaction of Xenopus lamins A and LII with chromatin in vitro mediated by a sequence element in the carboxyterminal domain. Exp Cell Res 1991; 197:280 - 289
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol 1995; 131:33 - 44
- Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J 2001; 20:4399 - 4407
- Caputo S, Couprie J, Duband-Goulet I, Konde E, Lin F, Braud S, et al. The carboxyl-terminal nucleoplasmic region of MAN1 exhibits a DNA binding winged helix domain. J Biol Chem 2006; 281:18208 - 18215
- Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos PA, Singh PB, et al. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep 2001; 2:920 - 925
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem 1996; 271:14653 - 14656
- Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, Theodoropoulos PA, et al. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem 2004; 279:25567 - 25573
- Furukawa K. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J Cell Sci 1999; 112:2485 - 2492
- Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci 2001; 114:4567 - 4573
- Mansharamani M, Wilson KL. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J Biol Chem 2005; 280:13863 - 13870
- Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci USA 2000; 97:8997 - 9002
- Skoko D, Li M, Huang Y, Mizuuchi M, Cai M, Bradley CM, et al. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc Natl Acad Sci USA 2009; 106:16610 - 16615
- Berger R, Theodor L, Shoham J, Gokkel E, Brok-Simoni F, Avraham KB, et al. The characterization and localization of the mouse thymopoietin/laminaassociated polypeptide 2 gene and its alternatively spliced products. Genome Res 1996; 6:361 - 370
- Harris CA, Andryuk PJ, Cline SW, Mathew S, Siekierka JJ, Goldstein G. Structure and mapping of the human thymopoietin (TMPO) gene and relationship of human TMPO beta to rat lamin-associated polypeptide 2. Genomics 1995; 28:198 - 205
- Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, et al. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J Cell Sci 2004; 117:6117 - 6128
- Suzuki Y, Yang H, Craigie R. LAP2alpha and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J 2004; 23:4670 - 4678
- Shaklai S, Somech R, Gal-Yam EN, Deshet-Unger N, Moshitch-Moshkovitz S, Hirschberg K, et al. LAP2zeta binds BAF and suppresses LAP2beta-mediated transcriptional repression. Eur J Cell Biol 2008; 87:267 - 278
- Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum Mol Genet 2006; 15:3459 - 3472
- Osada S, Ohmori SY, Taira M. XMAN1, an inner nuclear membrane protein, antagonizes BMP signaling by interacting with Smad1 in Xenopus embryos. Development 2003; 130:1783 - 1794
- Pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, Zhou S, et al. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. J Biol Chem 2005; 280:15992 - 16001
- Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci USA 2001; 98:11943 - 11948
- Korfali N, Wilkie GS, Swanson SK, Srsen V, Batrakou DG, Fairley EA, et al. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol Cell Proteomics 2010; 9:2571 - 2585
- Schirmer EC, Florens L, Guan T, Yates JRr, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003; 301:1380 - 1382
- Wilkie GS, Korfali N, Swanson SK, Malik P, Srsen V, Batrakou DG, et al. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol Cell Proteomics 2011; 10:110 - 3129
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 2008; 4:1000039
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 2008; 180:51 - 65
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008; 452:243 - 247
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol 2007; 9:1160 - 1166
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 2008; 182:911 - 924
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 1997; 138:1193 - 1206
- Yang L, Guan T, Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol 1997; 137:1199 - 1210
- Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol 2007; 178:43 - 56
- Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci 2009; 122:2100 - 2107
- Drummond S, Ferrigno P, Lyon C, Murphy J, Goldberg M, Allen T, et al. Temporal differences in the appearance of NEP-B78 and an LBR-like protein during Xenopus nuclear envelope reassembly reflect the ordered recruitment of functionally discrete vesicle types. J Cell Biol 1999; 144:225 - 240
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol 2001; 3:1086 - 1091
- Vigers GP, Lohka MJ. A distinct vesicle population targets membranes and pore complexes to the nuclear envelope in Xenopus eggs. J Cell Biol 1991; 112:545 - 556
- Vigers GP, Lohka MJ. Regulation of nuclear envelope precursor functions during cell division. J Cell Sci 1992; 102:273 - 284
- Salpingidou G, Rzepecki R, Kiseleva E, Lyon C, Lane B, Fusiek K, et al. NEP-A and NEP-B both contribute to nuclear pore formation in Xenopus eggs and oocytes. J Cell Sci 2008; 121:706 - 716
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, et al. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2 and Nup153 to reforming functional nuclear envelopes. J Cell Sci 2000; 113:779 - 794
- Martins S, Eikvar S, Furukawa K, Collas P. HA95 and LAP2 beta mediate a novel chromatin-nuclear envelope interaction implicated in initiation of DNA replication. J Cell Biol 2003; 160:177 - 188
- Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, et al. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci 2005; 118:4017 - 4025
- Chi YH, Haller K, Peloponese JM Jr, Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem 2007; 282:27447 - 27458
- Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp Cell Res 2009; 315:1895 - 1903
- Mattout A, Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr Opin Cell Biol 2010; 22:334 - 341
- Nili E, Cojocaru GS, Kalma Y, Ginsberg D, Copeland NG, Gilbert DJ, et al. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J Cell Sci 2001; 114:3297 - 3307
- Strickfaden H, Zunhammer A, van Koningsbruggen S, Kohler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri's hypotheses revisited. Nucleus 2010; 1:284 - 297
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol 2006; 16:825 - 831
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol 2000; 2:871 - 878
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 2007; 4:895 - 901
- van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol 2010; 28:1089 - 1095
- Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem 2009; 107:30 - 39
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289 - 293
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 2006; 38:1348 - 1354
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 2002; 12:439 - 445
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 2010; 38:603 - 613
- Zink D, Bornfleth H, Visser A, Cremer C, Cremer T. Organization of early and late replicating DNA in human chromosome territories. Exp Cell Res 1999; 247:176 - 188
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004; 6:1114 - 1121
- Zuleger N, Kelly DA, Richardson AC, Kerr AR, Goldberg MW, Goryachev AB, et al. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J Cell Biol 2011; 193:109 - 123
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol 2004; 166:815 - 825
- Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol 2007; 176:593 - 603
- Hakim O, Sung MH, Hager GL. 3D shortcuts to gene regulation. Curr Opin Cell Biol 2010; 22:305 - 313
- Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol 2009; 20:849 - 855
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell 2000; 6:1025 - 1035