Abstract
Lamins are the major components of the nuclear lamina and serve not only as a mechanical support, but are also involved in chromatin organization, epigenetic regulation, transcription and mitotic events. Despite these universal tasks, lamins have so far been found only in metazoans. Yet, recently we have identified Dictyostelium NE81 as the first lamin-like protein in a lower eukaryote. Based on the current knowledge, we draw a model for nuclear envelope organization in Dictyostelium in this Extra View and we review the experimental data that justified this classification. Furthermore we provide unpublished data underscoring the requirement of posttranslational CaaX-box processing for proper protein localization at the nuclear envelope. Sequence comparison of NE81 sequences from four Dictyostelia with bona fide lamins illustrates the evolutional relationship between these proteins. Under certain conditions these usually unicellular social amoebae congregate to form a multicellular body. We propose that the evolution of the lamin-like NE81 went along with the invention of multicellularity.
Introduction
It is a common and well-accepted hypothesis in cell biology that intermediate filament (IF) proteins are present only in metazoan cells, whereas they are absent in unicellular eukaryotic organisms and plants.Citation1,Citation2 This consensus may need to be reconsidered, since our group very recently succeeded in the characterization of a novel nuclear protein, Dictyostelium NE81, as the first lamin-like protein in a unicellular organism.Citation3
Lamins are type V intermediate filament (IF) proteins forming the nuclear lamina in metazoan cells.Citation4 They are present in even the most primitive metazoans such as sponges, Placozoans and Coelenterates, which express no other IF protein. Thus, they are considered the founding members of the IF multigene family. Lamin functions are usually fulfilled by a single B-type lamin in most invertebrates. Yet, vertebrates express up to five different lamin proteins encoded by up to four different genes LMNB1, LMNB2, LIII and LMNA.Citation5 While lamin B1 is expressed in almost all somatic cells, the other lamins are more tissue specific and/or developmentally regulated. Like all IF proteins lamins are characterized by a tripartite structure with a long, central α-helical rod domain flanked by a short, globular head domain and a longer tail domain. As well as other IFs, lamins are capable of self-organization to higher order filaments. The rod domains of two lamin monomers form an α-helical coiled coil structure and the resulting dimers assemble in a head-to-tail fashion. Two of these head-to-tail chains align in an antiparallel manner to form protofilaments. In C. elegans, the association of three to four protofilaments makes up 10 nm filamentsCitation6 forming a network beneath the inner nuclear membrane.Citation7,Citation8 However, these 10 nm-filaments have not yet been visualized in many species and their supramolecular organization within the nuclear lamina is still a matter of debate. Among the IFs lamins are characterized by a nuclear localization sequence (NLS) located right behind the rod domain, a CDK1 phosphorylation consensus sequence directly preceding the rod domain and a CaaX-box at their C-terminus. The NLS is required for transport into the nucleus. As a prerequisite for nuclear envelope breakdown (NEBD), phosphorylation by CDK1 mediates disassembly of the lamin network at the onset of mitosis.Citation9 Finally, the CaaX-box serves as a signal for posttranslational lipidation and processing.Citation10 Lamins are prenylated by farnesyl transferase at the cysteine residue of the CaaX-box (“C” stands for cysteine, “a” for aliphatic amino acid residues, “X” for the amino acid residue determining the type of prenyl moiety). Processing proceeds with cleavage of the three C-terminal amino acid residues (= aaX) either by Ras converting enzyme 1 (RCE1) or by ZMPSTE24 and carboxymethylation of the farnesylated cysteine residue by isoprenylcysteine methyl transferase (ICMT). While this is the last processing step in case of B-type lamins, prelamin A is cleaved once again by ZMPSTE24, removing the last 15 (in case of human lamin A) amino acid residues including the farnesyl anchor. Thus, B-type lamins stay associated with the nuclear envelope (NE) due to their lipid anchor, while A-type lamins require an interaction with inner NE proteins or with B-type lamins for their recruitment to the NE. The lamin-based nuclear lamina is intimately linked to the cytosolic cytoskeleton through LINC complexes (linker of nucleoskeleton and cytoskeleton), which consist of Sun-family proteins in the inner nuclear membrane interacting with so-called KASH domain proteins in the outer nuclear membrane.Citation11 The latter interact directly or indirectly with actin filaments, cytoplasmic intermediate filaments and microtubules. In vegetative cells, LINC complexes are also responsible for the tight linkage of the centrosome, i.e., the main microtubule-organizing center, to the nucleus. Through these interactions the nuclear lamina not only influences the mechanical properties of the nucleus, but also that of the whole cell and, thus, lamins are thought to protect cells against mechanical stress.Citation11-Citation13 Yet, with regard to the diverse interactions of the nuclear lamina with chromatin-associated proteins such as histones, transcriptional regulators and chromatin modifiers, it is obvious that they do not only serve as a mechanical support, but that lamins are also involved in chromatin organization, epigenetic regulation, transcription and mitotic events.Citation14 These various important functions are also reflected by the symptoms of laminopathies. These devastating, inheritable diseases including muscular dystrophy, cardiomyopathy, partial lipodystrophy and progeria are caused by mutations of the lamin A gene, or of genes encoding lamin-binding or lamin-processing proteins.Citation15 Bearing in mind these universal tasks for all eukaryotic cells, it should be expected that non-metazoan cells need a functional equivalent of a nuclear lamina or even lamin-like proteins as well. Yet, none of the sequenced genomes of plants (e.g., Arabidopsis and Physcomitella), fungi (e.g., Saccharomyces and Aspergillus) or protozoans (Plasmodium and Trypanosoma) disclosed a lamin or lamin-like protein, even though the first observations of a filamentous lamina were made half a century ago in two unicellular eukaryotes, Amoeba proteus and Gregarina melanopli.Citation16-Citation18 Indeed, the existence of lamins in Saccharomyces, Tetrahymena, Physarum and dinoflagelates has been postulated due to cross-reactivity of lamin-specific antibodies.Citation2 However, in all of these cases the nuclear envelope-associated coiled-coil proteins characterized on the molecular level were evolutionary unrelated to lamins. This is why lamins or lamin-like proteins are generally considered to be absent in lower eukaryotes.
NE81 Shares Many Structural and Functional Features of Lamins
Already in the late seventies the existence of a nuclear lamina in Dictyostelium amoebae was proposed, since electron micrographs of Dictyostelium cells had revealed a thin, continuous, low electron-density layer between the inner nuclear membrane and the chromatin. Its appearance in ultrathin sections was very reminiscent of the previously characterized fibrous (= nuclear) lamina of animal cells.Citation3,Citation19 However, as in other lower eukaryotes, the proteins involved in formation of a putative nuclear lamina in Dictyostelium were completely unknown. Recently, we succeeded in identification and characterization of a lamin-like protein in Dictyostelium discoideum.Citation3 It was found serendipitously in a proteomic screen for centrosomal and centrosome-associated proteins.Citation20 We called it NE81 since is has a calculated molecular mass of 81 kDa and a corresponding GFP fusion protein is targeted to the nuclear envelope.Citation21 The fact that it could be copurified with isolated centrosomes already indicated a putative role in the centrosome/nucleus linkage, which will be discussed below. Sequence analysis of the 716 aa protein instantly revealed striking similarity to lamins with a central, α-helical coiled-coil rod domain flanked by a head and long tail domain. With a length of 370 aa the rod domain has a comparable length to that of lamins and it is also predicted to be interrupted by non-helical linkers. Moreover, the rod domain is directly preceded by a CDK1 phosphorylation consensus sequence and is followed by a basic nuclear localization sequence. The position of these signals with respect to the rod domain matches exactly to those in lamins. Finally, the NE81 protein sequence ends with a CaaX-box (CLIM). The methionine at the X-position is characteristic for lamins, and indicates farnesylation instead of geranylgeranylation.Citation22 Taken together, the sequence features already strongly suggested that NE81 could be a lamin-like protein, since the only known inner NE proteins with a CaaX-box are lamins and Drosophila kugelkern, another lamina protein, which is clearly distinguished from lamins by its much shorter α-helical coiled-coil region.Citation23
This idea was confirmed by all experimental data. NE81 was localized at the nuclear envelope during the entire cell cycle. Immuno-electron microscopy using anti-NE81 antibodies revealed an association of the protein with the inner nuclear envelope. As it holds true for lamins, proper localization of NE81 along the nuclear envelope requires the presence of a functional CaaX-box. A first indication for this requirement came from the analysis of a Dictyostelium icmA knockout strain lacking the isoprenylcysteine methylase IcmA.Citation24 Although NE81 was still evenly distributed at the nuclear envelope in these cells, we also observed clusters of NE81 within the nucleus in about 50% of these cells (). These clusters were never observed in control cells. This suggests that processing of NE81 including isoprenylcysteine methylation is required for proper localization of the protein at the nuclear envelope. If the CaaX-box is deleted completely, the respective GFP fusion proteins (GFP-NE81ΔCLIM) are exclusively present in clusters within the nucleus that were reminiscent of those of endogenous NE81 in IcmA deficient cells. This behavior was also observed in a new strain where the CaaX-box cysteine was point mutated to serine (). In EM images, these protein clusters appeared as sponge-like structures of low electron density. Both CaaX-box mutated GFP fusion proteins (GFP-NE81ΔCLIM and GFP-NE81SLIM) showed the same cell cycle-dependent behavior, i.e., the GFP-fluorescent clusters in the nucleus disappeared at the G2/M transition () and reappeared in telophase. This behavior and the low density of GFP-NE81ΔCLIM clusters showed that GFP-NE81ΔCLIM clusters were no aggregates of misfolded protein. Rather they were likely to represent three-dimensional assemblies that were formed and dissociated in a cell cycle-dependent fashion. Fluorescence recovery after photobleaching (FRAP) experiments disclosed a similar behavior of the full-length GFP-NE81 protein. It was almost immobile during interphase, which was in agreement with the idea that it forms a stable nuclear lamina composed of assembled NE81 chains. However, between early mitosis and karyokinesis in early telophase GFP-NE81 was highly mobile along the nuclear envelope. Lamin B1 behaves very similar to NE81 in FRAP experiments. Integrated into the nuclear lamina it shows little or no mobility.Citation26,Citation27 Yet at the G2/M transition, as an early event in nuclear envelope breakdown (NEBD), the nuclear lamina disassembles, a process initiated by CDK1 phosphorylation of lamins.Citation9 Like in many fungi and lower eukaryotes, there is no NEBD in Dictyostelium amoebae, however, the postulated nuclear lamina needs to become softer in order to allow constriction during karyokinesis. This could be achieved by partial disassembly of NE81 resulting in an increased mobility. The timing of disappearance and re-formation of GFP-NE81ΔCLIM and GFP-NE81SLIM clusters as well as the increased mobility of full length GFP-NE81 fits exactly to the activity window of the master regulator of mitosis, CDK1. Indeed we could show that the serine residue within the putative CDK1 phosphorylation site at position 122 is required for this dynamic behavior. In the non-phosphorylatable S122A point mutant of GFP-NE81ΔCLIM, the GFP fluorescent clusters in the nucleus persisted throughout mitosis. We concluded that, similarly to lamins, phosphorylation negatively regulates an inherent capacity of NE81 to form high order polymers. Endogenous NE81 becomes isoprenylated and anchored to the inner nuclear envelope favoring polymerization in two dimensions. However, CaaX-box mutations preclude prenylation and processing, and therefore prevent proper attachment to the inner nuclear membrane. This, in turn, could cause cluster formation by polymerization in three dimensions within the nuclear interior.
Figure 1. Phenotypes of NE81 mutant cells. Immunofluorescence microscopy of icmA null cells (A), GFP-NE81SLIM cells (B), NE81 null cells (C), GFP-NE81 overexpressing cells (D) and control cells (E) fixed with glutaraldehyde and stained with anti-NE81/anti-rabbit-AlexaFluor 488 and anti-tubulin YL1/2,Citation25/anti-rat-AlexaFluor 568. Mitotic stages in (B) are indicated. The montage shows GFP fluorescence in green, tubulin in red and nuclei in blue. Maximum intensity projections of widefield deconvolution image stacks are shown. Bar = 2.5 μm.
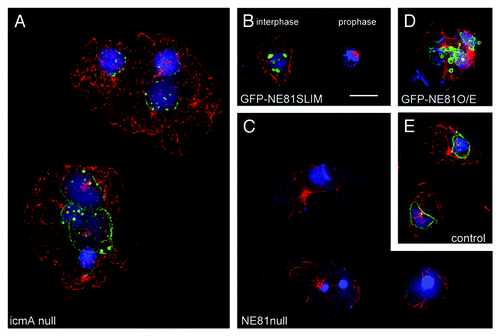
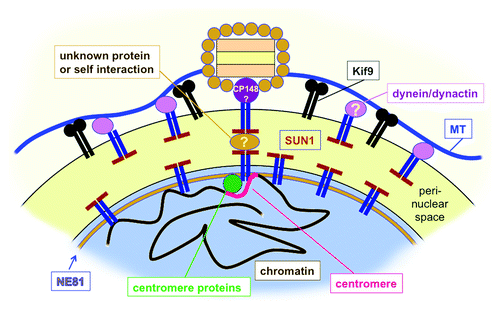
Overexpression of GFP-NE81ΔCLIM revealed a further function shared by NE81 and lamins. GFP-NE81ΔCLIM cells were unable to grow in shaking culture indicating an increased sensitivity to mechanical stress and were hypersensitive against mechanical shear forces. Increased mechanosensitivity of GFP-NE81ΔCLIM cells supports the idea that an NE81-based nuclear lamina could serve as a mechanical abutment for cytosolic cytoskeletal elements just as in cells of higher eukaryotes, where the nuclear lamina is connected to the cytoskeleton through LINC complexes. Although no LINC complexes have been characterized in Dictyostelium yet, several components of the interaction of the nucleus with the cytoskeleton are known. These are Sun1, Kif9, LIS1, CP148, interaptin and centrin B (cenB). While interaptin is an actin-binding outer nuclear envelope protein linking the nucleus to the actin system,Citation28 all others are related to the microtubule system. Although not binding to Sun1, interaptin functionally interacts with Sun1, the classical LINC complex component.Citation29 Bona fide KASH domain proteins of the outer nuclear membrane have not been identified so far in Dictyostelium. Instead, Sun1 resides in both nuclear membranes and is thought to link the centrosome and its microtubule arrays on the cytosolic side of the nucleus with the clustered centromeres inside the nucleusCitation30 (). At the centrosomal side, the centrosomal component CP148 appears to be involved in this linkage,Citation31 while the binding partners at the centromere cluster are unknown. Interference with dynein function by point mutation of its regulator LIS1,Citation32 or knockout of the membrane-associated orphan kinesin Kif9,Citation33 also disrupted the linkage of the centrosome to the nucleus underscoring an important role of the microtubule system for this process (). The unusual, nuclear envelope-associated centrin B may be involved in anchoring of the nucleus/centrosome link to chromatinCitation34 and/or an NE81-based nuclear lamina. The requirement of NE81 for the centrosome/nucleus linkage became clear in NE81 knockout cells, in which the centrosome was detached from the nucleus in more than 50% of all cases (). Moreover, we observed misshapen nuclei with irregularly condensed chromatin (32%), supernumerary centrosomes (7%) and missing centrosomes (5%). All of these are phenotypes virtually absent in control cells (). Overexpression of GFP-NE81 also caused irregularly shaped nuclei, blebbing of the nuclear envelope and disorganized chromatin (). Thus, all phenotypes observed upon NE81 overexpression or knockout indicate a role of NE81 in functions addressed to lamins in metazoan cells such as maintenance of nuclear architecture, chromatin integrity and crosstalk of nuclear structures with the cytoskeleton.
Figure 2. Hypothetical model of nuclear envelope organization in the pericentrosomal region in Dictyostelium. In both nuclear membranes (black line) Sun1 is involved in linkage of the centrosome to an NE81-based nuclear lamina. On the centrosomal side, the centrosomal corona protein CP148 is directly or indirectly associated with Sun1, on the nuclear side Sun1 is directly or indirectly bound to NE81. Centromere proteins are also associated with Sun1 and mediate clustering of centromeres in the pericentrosomal region. The tight linkage between the cytosolic centrosome and the centromere cluster and nuclear lamina could be mediated by a self-interaction between SUN-domains or by an unknown protein within the perinuclear space. At the outer nuclear membrane both the kinesin Kif9 and Sun1-associated dynein/dynactin bind to microtubules and help to hold the centrosome close to the nucleus.
Is NE81 a Lamin or a Lamin-Like Protein?
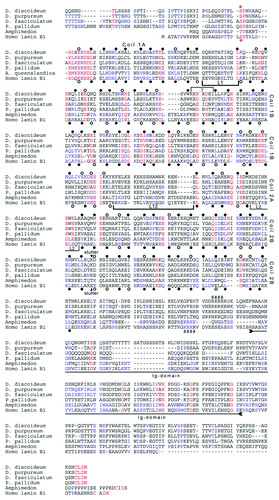
The many similarities to lamins raise the question whether NE81 should be considered a lamin-like protein or a real lamin. Due to the functional similarities regarding its role in nuclear integrity, centrosome/nucleus attachment and mechanical stability of cells as well as its cell cycle-dependent regulation, it is now clear that NE81 functions like a lamin in Dictyostelium. However, regarding structural similarities classification of NE81 as a lamin is a matter of debate. On the one hand, structural hallmarks of lamins such as the tripartite structure with an ~370 aa α-helical rod domain, the positions of the CDK1 phosphorylation site, the NLS and the CaaX-box are conserved. On the other hand the subdivision pattern of the rod domain by non-helical linkers in lamins and NE81 does not match exactly and the IF consensus sequence at the C-terminal end of the rod is not well conserved in NE81 Citation5 (). The presence of the typical Ig-domain of lamins remains unclear as well. Sequence similarities in this region between NE81 sequences from social amoebae and lamins are low (). However, this region is also absent in the bona fide lamins of tunicates and therefore its presence may not be essential for general lamin functions.Citation36 We could show that GFP-NE81 colocalizes with lamin B1 in human cells. It will be very interesting to investigate whether NE1 is capable to copolymerize with lamins or even to functionally compensate knockdown of one of the lamins by RNAi. Whether it may be allowed to call NE81 a lamin will depend on the outcome of further analyses of its structure and supramolecular assembly.
Figure 3. Protein sequence alignment of two bona fide lamins with NE81 orthologs from four Dictyostelidae. Coding sequences from the social amoebae D. discoideum (starting with aa position 99), D. purpureum (starting with aa position 84), D. fasciculatum (starting with aa position 110) and Polysphondylium pallidum (starting with aa position 119) and Amphimedon queenslandica were derived from the respective genome projects (dictybase.org, sacgb.fli-leibniz.de, www.metazome.net/amphimedon). In case of P. pallidum the predicted second intron had to be neglected since otherwise the deduced amino acid sequence were devoid of a CaaX-box. Sequences were aligned using MultAlin software.Citation35 High consensus of amino acid similarity is colored in red, low consensus in blue. Heptad repeats of the human B1 are marked at the 1st and 4th position (a and d positions) below the sequence. Filled circles indicate hydrophobic and uncharged amino acid residues favorable for coiled-coil formation (L, I, V, A, M, N, Q, Y, F and W), open circles indicate less favorable amino acid residues (E, D, K, R, H, S, T, C and G). Deduced from the lamin heptad pattern, tentative heptad positions were assigned to the NE81 sequences (above the sequence). A small linker, marked by a bracket with a (?), had to be introduced into the NE81 sequence to match the heptad pattern of the lamins. Linker regions separating the coils of lamins are marked by brackets. The discontinuity in the heptad pattern in the second half of coil 2B (stutter) is indicated by an arrow above and below the D. discoideum and human lamin B1 sequences, respectively. The region of the Ig-domain within the tail of the lamin sequences is indicated by a double-headed arrow. Three characteristic sequence motifs for lamins are indicated: a CDK1 phosphorylation consensus sequence upstream of the rod domain (*), a putative nuclear localization sequence (####), and the C-terminal CaaX-box (in bold letters). In case of D. fasciculatum the CDK1 target sequence does not match with the consensus (S/T-P-X-R/K).
NE81 in Other Lower Eukaryotes
The many similarities shared between NE81 and lamins raise the question why NE81 was not detected earlier in searches for lamins in the sequenced genomes of social amoebae (Dictyostelia). One reason is that sequence similarities between NE81 and lamins are low and, thus, BLAST searches with lamins as a query yield numerous non-related coiled coil proteins. It appears that formation of a nuclear lamina enforces mainly a structural conservation of its main constituent, while sequence conservation is restricted to a few consensus sequences such as the CaaX-box, the NLS and the CDK1 site. Even among Dictyostelia NE81 sequences vary strongly. The most closely related ortholog in Dictyostelium purpureum exhibits an amino acid identity of only 58% (74% similarity) to D. discoideum NE81. Compared with Dictyostelium fasciculatum and Polysphondylium pallidum NE81 sequence conservation is even lower with amino acid identities of 37% (56% similarity) and 34% (54% similarity), respectively (). These values reflect the phylogenetic distance between the two latter species, which belong to two different groups within the Dictyostelia than D. discoideum and D. purpureum.Citation37 We found no NE81-like protein in the sequenced protozoans including Entamoeba, Plasmodium, Trypanosoma and Paramecium. From an evolutionary point of view Dictyostelia are positioned between protists lacking lamins, and metazoans with a lamin-based nuclear lamina. We believe that the evolution of the lamin-like NE81 went along with a typical attribute of all Dictyostelia, i.e., the capability to form multicellular bodies from single cells under certain conditions.Citation38 Future research may disclose whether the invention of a nuclear lamina based on a lamin-like protein may even have been a prerequisite to reach the multicellular state.
Acknowledgements
We would like to thank Dr Annette Peter for critical and helpful comments on the manuscript. We are also very grateful to Dr Kyle McQuade (Colorado Mesa University, Grand Junction, CO USA) for providing the Dictyostelium icmA null strain. This work was supported by DFG GR1642/4-1.
References
- Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci 2001; 26:41 - 7; http://dx.doi.org/10.1016/S0968-0004(00)01727-8; PMID: 11165516
- Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res 2007; 313:2157 - 66; http://dx.doi.org/10.1016/j.yexcr.2007.03.004; PMID: 17451683
- Krüger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, et al. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell 2012; 23:360 - 70; http://dx.doi.org/10.1091/mbc.E11-07-0595; PMID: 22090348
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol 2010; 2:a000547; http://dx.doi.org/10.1101/cshperspect.a000547; PMID: 20826548
- Peter A, Stick R. Evolution of the lamin protein family: What introns can tell. Nucleus 2012; 3:44 - 59; http://dx.doi.org/10.4161/nucl.18927; PMID: 22156746
- Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, Aebi U, et al. The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol 2009; 386:1392 - 402; http://dx.doi.org/10.1016/j.jmb.2008.12.024; PMID: 19109977
- Goldberg MW, Fiserova J, Huttenlauch I, Stick R. A new model for nuclear lamina organization. Biochem Soc Trans 2008; 36:1339 - 43; http://dx.doi.org/10.1042/BST0361339; PMID: 19021552
- Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest 2009; 119:1772 - 83; http://dx.doi.org/10.1172/JCI38214; PMID: 19587452
- Foisner R. Cell cycle dynamics of the nuclear envelope. ScientificWorldJournal 2003; 3:1 - 20; http://dx.doi.org/10.1100/tsw.2003.06; PMID: 12806116
- Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest 2009; 119:1825 - 36; http://dx.doi.org/10.1172/JCI37679; PMID: 19587457
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res 2008; 314:1892 - 905; http://dx.doi.org/10.1016/j.yexcr.2008.02.022; PMID: 18396275
- Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 2004; 117:4779 - 86; http://dx.doi.org/10.1242/jcs.01357; PMID: 15331638
- Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol 2011; 23:55 - 64; http://dx.doi.org/10.1016/j.ceb.2010.10.015; PMID: 21109415
- Andrés V, González JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol 2009; 187:945 - 57; http://dx.doi.org/10.1083/jcb.200904124; PMID: 20038676
- Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000760; http://dx.doi.org/10.1101/cshperspect.a000760; PMID: 20182615
- Pappas GD. The fine structure of the nuclear envelope of Amoeba proteus.. J Biophys Biochem Cytol 1956; 2:Suppl 431 - 4; http://dx.doi.org/10.1083/jcb.2.4.431; PMID: 13357581
- Beams HW, Tahmisian TN, Devine R, Anderson E. Ultrastructure of the nuclear membrane of a gregarine parasitic in grasshoppers. Exp Cell Res 1957; 13:200 - 4; http://dx.doi.org/10.1016/0014-4827(57)90071-X; PMID: 13473864
- Mercer EH. An electron microscopic study of Amoeba proteus.. Proc R Soc Lond B Biol Sci 1959; 150:216 - 32; http://dx.doi.org/10.1098/rspb.1959.0016; PMID: 13633977
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat 1966; 119:129 - 45; http://dx.doi.org/10.1002/aja.1001190108; PMID: 6007824
- Reinders Y, Schulz I, Gräf R, Sickmann A. Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J Proteome Res 2006; 5:589 - 98; http://dx.doi.org/10.1021/pr050350q; PMID: 16512674
- Schulz I, Erle A, Gräf R, Krüger A, Lohmeier H, Putzler S, et al. Identification and cell cycle-dependent localization of nine novel, genuine centrosomal components in Dictyostelium discoideum.. Cell Motil Cytoskeleton 2009; 66:915 - 28; http://dx.doi.org/10.1002/cm.20384; PMID: 19466752
- Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem 1996; 271:5289 - 92; http://dx.doi.org/10.1074/jbc.271.10.5289; PMID: 8621375
- Melcer S, Gruenbaum Y. Nuclear morphology: when round kernels do the Charleston. Curr Biol 2006; 16:R195 - 7; http://dx.doi.org/10.1016/j.cub.2006.02.032; PMID: 16431372
- Chen Y, McQuade KJ, Guan XJ, Thomason PA, Wert MS, Stock JB, et al. Isoprenylcysteine carboxy methylation is essential for development in Dictyostelium discoideum.. Mol Biol Cell 2007; 18:4106 - 18; http://dx.doi.org/10.1091/mbc.E06-11-1006; PMID: 17699599
- Wehland J, Willingham MC. A rat monoclonal antibody reacting specifically with the tyrosylated form of alpha-tubulin. II. Effects on cell movement, organization of microtubules, and intermediate filaments, and arrangement of Golgi elements. J Cell Biol 1983; 97:1476 - 90; http://dx.doi.org/10.1083/jcb.97.5.1476; PMID: 6685128
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, et al. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci 1999; 112:3463 - 75; PMID: 10504295
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol 2000; 151:1155 - 68; http://dx.doi.org/10.1083/jcb.151.6.1155; PMID: 11121432
- Rivero F, Kuspa A, Brokamp R, Matzner M, Noegel AA. Interaptin, an actin-binding protein of the alpha-actinin superfamily in Dictyostelium discoideum, is developmentally and cAMP-regulated and associates with intracellular membrane compartments. J Cell Biol 1998; 142:735 - 50; http://dx.doi.org/10.1083/jcb.142.3.735; PMID: 9700162
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, et al. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic 2008; 9:708 - 24; http://dx.doi.org/10.1111/j.1600-0854.2008.00721.x; PMID: 18266910
- Schulz I, Baumann O, Samereier M, Zoglmeier C, Gräf R. Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur J Cell Biol 2009; 88:621 - 38; http://dx.doi.org/10.1016/j.ejcb.2009.06.003; PMID: 19632001
- Kuhnert O, Baumann O, Meyer I, Gräf R. Functional characterization of CP148, a novel key component for centrosome integrity in Dictyostelium. Cell Mol Life Sci 2012; 69:1875 - 88; http://dx.doi.org/10.1007/s00018-011-0904-2; PMID: 22223109
- Rehberg M, Kleylein-Sohn J, Faix J, Ho TH, Schulz I, Gräf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol Biol Cell 2005; 16:2759 - 71; http://dx.doi.org/10.1091/mbc.E05-01-0069; PMID: 15800059
- Tikhonenko I, Nag DK, Robinson DN, Koonce MP. Microtubule-nucleus interactions in Dictyostelium discoideum mediated by central motor kinesins. Eukaryot Cell 2009; 8:723 - 31; http://dx.doi.org/10.1128/EC.00018-09; PMID: 19286984
- Mana-Capelli S, Gräf R, Larochelle DA. Dictyostelium discoideum CenB is a bona fide centrin essential for nuclear architecture and centrosome stability. Eukaryot Cell 2009; 8:1106 - 17; http://dx.doi.org/10.1128/EC.00025-09; PMID: 19465563
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988; 16:10881 - 90; http://dx.doi.org/10.1093/nar/16.22.10881; PMID: 2849754
- Riemer D, Wang J, Zimek A, Swalla BJ, Weber K. Tunicates have unusual nuclear lamins with a large deletion in the carboxyterminal tail domain. Gene 2000; 255:317 - 25; http://dx.doi.org/10.1016/S0378-1119(00)00323-1; PMID: 11024292
- Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum.. Development 2011; 138:387 - 96; http://dx.doi.org/10.1242/dev.048934; PMID: 21205784
- Kessin RH. Dictyostelium: evolution, cell biology and the development of multicellularity. Cambridge: Cambridge University Press 2001.