Abstract
Nuclear structures ND10/PML NBs are linked to multiple processes, including the maintenance of intranuclear homeostasis by sequestering proteins into “nuclear depot.” This function presumes release of proteins from PML NBs and their redistribution to the alternative, supposedly “active” locations, in response to the external stress application. To further investigate this nuclear depot function, we focused on the intranuclear distribution of protein Daxx that in normal conditions is mainly accumulated at PML NBs, and has a minor association with centromeres and pericentromeres (CEN/periCEN). Here we report that application of physiological Heat Shock (HS) changes this balance forcing very robust and reversible accumulation of Daxx on CEN/periCEN heterochromatin.
Heterochromatin architecture is essential for the proper orchestration of nuclear processes, while transcription from this part of genome is required for its maintenance. To understand functional consequences of Daxx deposition at CEN/periCEN, we tested for Daxx-dependency of heterochromatin transcription. Depletion of Daxx reduces accumulation of CEN RNA in normal conditions and periCEN RNA after HS application. Searching for the mechanism of Daxx-dependent regulation of heterochromatin transcription, we found that depletion of Daxx decreases incorporation of transcription-associated histone H3 variant, H3.3, into both CEN and periCEN. Surprisingly, HS-induced deposition of Daxx does not further elevate incorporation of H3.3 into CEN/periCEN that remained steady during stress and recovery. Instead, depletion of Daxx leads to HS-induced changes in the balance of epigenetic modifications at heterochromatin, most dramatically elevating levels of active H3K4Me2 modification at periCEN. We propose dualistic function of Daxx-containing complexes at CEN/periCEN: (1) regulation of H3.3 loading in normal conditions and (2) protection of epigenetic status upon stress-induced accumulation, thus collectively guarding epigenetic identity of CEN/periCEN heterochromatin.
Introduction
Architecture of chromatin regulates numerous cellular processes, including gene transcription, DNA replication/repair and export of RNA. Heterochromatin is the key element in this architecture and is involved in regulation of genome transcription, chromosome segregation and genome stability. Heterochromatic regions are predominantly present in the centromeric (CEN), pericentromeric (periCEN) and telomeric regions of chromosomes and consists of tandem repeats (TRs) comprising up to 10% of genomes in majority of higher organisms.Citation1 The CEN regions of human chromosomes contain the largest TR family in the human genome called α-satellite DNA.Citation2 It was proposed that transcription from this region is necessary for proper formation and function of centromeres.Citation3,Citation4 The periCEN region in humans consists of simple sequence satellites arrays; one of examples of these arrays is human satellite DNA 3 (HS3). HS3 arrays of different size are located at periCEN regions of most human chromosomes.Citation5 Several chromosome-specific HS3 subfamilies have been reported in reference Citation6; among them, HS3 of chromosome 9 (HS3-9) was found to transcribe upon external stress application, including heat shock (HS);Citation7 yet, functional significance of transcription and/or transcripts and chromatin remodeling processes associated with the HS-induced periCEN transcription remains incompletely characterized. The key factor that mediates stress-induced transcription is the heat shock factor 1 (HSF1). Upon stress, HSF1 forms nuclear stress bodies (nSBs) at actively transcribed periCEN regions.Citation8
Daxx is a multi-functional nuclear proteinCitation9,Citation10 that is mainly associated with nuclear structure ND10/PML NBs in normal conditions.Citation11 PML NBs are linked to several processes, including the maintenance of intranuclear homeostasis by sequestering proteins into “nuclear depot” and releasing associated proteins into nucleoplasm upon stress application.Citation12 Thus, according to proposed “nuclear depot” model,Citation12,Citation13 PML NBs may not represent site of Daxx activity but predominantly are sites of sequestration and segregation whereas Daxx is active outside of this structure. In agreement with this model, besides the major accumulation at PML NBs, Daxx is also found associated with a subset of interphase centromeres in human cells;Citation14 in mouse cells, Daxx is enriched at condensed heterochromatin (MaSat) at the end of S/beginning of G2 phase,Citation15 suggesting CEN/periCEN heterochromatin as potentially “active” location of Daxx.
The functional significance of Daxx association with CEN/periCEN was dim until recent findings that identified Daxx-containing complex as a novel chaperone for histone H3.3.Citation16,Citation17 Incorporation of this H3 variant into chromatin is replication-independent and is associated with elevated gene transcription.Citation18-Citation20 Previously identified histone chaperone HIRA mediates loading of H3.3 into gene-rich areas of genome, specifically into gene regulatory elements.Citation21 Several recent studies have shown enrichment of H3.3 in repetitive regions of genome including telomeric and pericentromeric heterochromatin;Citation16 moreover, Daxx-mediated incorporation of H3.3 into MaSat in mouse cells correlates with transcription elevation from this region of genome.Citation17 It was also suggested that integration of H3.3 is important for the transcription-associated maintenance of heterochromatin structure at these locations.Citation17
Given the major difference between human periCEN and CEN as well as in organization between mouse and human heterochromatin, it is very appealing to investigate function of Daxx in the incorporation of H3.3 into both CEN/periCEN regions of human chromatin and to analyze the consequences of this incorporation on transcription of these regions, in both normal and stress-induced conditions. Pursuing this direction we found that depletion of Daxx in human cells diminishes recruitment of histone H3.3 into both CEN and periCEN and reduces transcription from these regions of genome. Rather unexpectedly we observed that accumulation of Daxx at CEN/periCEN upon HS prevents changes in epigenetic modifications at these locations. Our findings suggest that Daxx-containing complex secures chromatin integrity by H3.3 incorporation in normal conditions and protects CEN/periCEN epigenetic steady-state upon stress application.
Results
HS increases accumulation of Daxx at CEN/periCEN
Daxx is mainly associated with ND10/PML NBs in normal conditions (),Citation11,Citation15 but in small sub-population of cells, Daxx formed additional foci associated with CEN (visualized by CREST antibodies; , inset 2). According to proposed “nuclear depot” model,Citation12 PML NBs response to extracellular stress by releasing associated proteins into nucleoplasm; thus, we tested localization of Daxx upon physiological hyperthermia (heat shock, HS) application. After 1 h of HS at 42°C, number of PML NBs increased 2- to 3-fold in agreement with previously published data (),Citation22,Citation23 most likely due to balance changes in post-translational modification of PML itself and/or other components of PML NBs; Daxx remained nuclear as in others stress conditions,Citation9 still co-localizing with majority of PML NBs (). In addition to PML NBs localization, Daxx also appeared in multiple (20–40) PML-negative domains in the majority of cells (). Number and shape of these domains was reminiscent of CEN. Indeed, we observed that Daxx co-localized with CEN in most cells after 1 h of HS (). Daxx remained associated with CEN at 1 h after HS recovery ( and inset 1) and also accumulated juxtaposed to CEN ( and inset 2). At 2 h after HS recovery, number of PML NBs, Daxx-PML and Daxx-CEN association mostly returned to the pre-stress conditions (). Thus, Daxx, which in normal condition mainly resided at PML NBs and had minor association with CEN, became predominantly associated with CEN in response of HS application, although it was still observed at PML NBs. Statistical analysis is presented in ; stability of Daxx did not change upon stress and recovery (Fig. S1G). Similar HS-induced dynamics of Daxx was observed in HeLa and MCF7 cancer cell lines and MCF10A, untransformed breast epithelial cell line (not shown).
Figure 1. Daxx accumulation at CEN/periCEN. HEp2 cells were fixed at 37°C or exposed to 42°C for 1 h and either fixed immediately or recovered at 37°C for 1 h or 2 h. Daxx is mostly associated with PML NBs (visualized by anti-PML ab’s) in control conditions (A); it forms additional domains after HS and after 1 h recovery (B and C) and is mostly returned to pre-stress localization after 2 h recovery (D). Daxx co-localizes with centromeres (visualized by CREST human autoimmune ab’s) in some cells at 37°C (E, inset 2); this co-localization is obvious in most cells after HS and 1 h recovery (F and G, inset 1), and is decreased after 2 h recovery (H); in addition, Daxx accumulates juxtaposed to centromeres upon recovery (G, inset 2). HSF1 is nuclear homogenous in control conditions (I) and forms stress bodies (SBs) upon HS; Daxx is adjacent to SBs after HS/1 h recovery (insets L–K), and co-localizes with majority of SBs at 1 h recovery (K, insets 2 and 3). At 2 h recovery, Daxx is associated with some SB (L; inset 1 high association, inset 2 low association). (M) Colocalization analysis (Pearson’s correlation coefficient upon HS and recovery) has been used to quantify the degree of association for Daxx/PML and Daxx/centromeres (CREST). Data are the means of three experiments, and the standard deviation is shown. (N) ChIP analysis of Daxx association with CEN/periCEN in HEp2 cells stably expressing FLAG-HA-Daxx. Daxx association with CEN (α-satellite) was minor at 37°C, was elevated ~10-fold upon HS (p < 0.001), was still high, but reduced, at 1 h of recovery and returned to pre-stress levels at 2 h and 3 h. Daxx association with periCEN (HS 3-9) was minimal at 37°C, was elevated upon HS (p < 0.001) and 1 h of recovery, and reduced to almost pre-stress levels at 3 h of recovery.
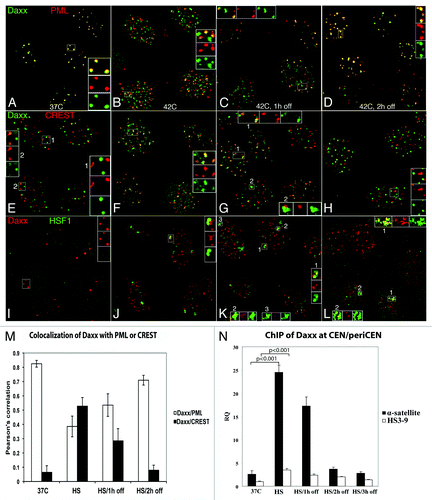
Upon HS recovery, in addition to CEN, Daxx accumulates adjacent to CEN ( and inset 2), potentially at periCEN. Upon HS, transcription factor HSF1 activates transcription from periCEN (satellite HS3–9),Citation7 accumulating at these TRs in so-called stress bodies (SBs).Citation7 Thus, we used HSF-1 as a surrogate marker to monitor Daxx localization relative to the transcriptionally active periCEN. HSF-1, that was nuclear homogenous in normal conditions (), formed SBs upon HS application (); HSF-1-stained SBs were juxtaposed to some of Daxx accumulations (presumably at CEN), reflecting the expected adjacent localization of CEN and periCEN in interphase chromosomal territories (, insets). Daxx co-localized with SBs at 1 h after HS recovery ( and compare insets 2/3 with inset 2 at ). At 2 h after HS recovery, Daxx was associated with subpopulation of SBs (). Thus, in addition to CEN, Daxx was also associated with SBs (that should mark periCEN) upon stress recovery.
To analyze Daxx association with CEN/periCEN TRs more directly, we developed CEN and periCEN probes for fluorescent in situ hybridization (FISH) to combine with immunostaining for Daxx. Specificity of CEN (pan-CEN) and periCEN (HS3–9) probes was tested in mitotic spreads for BJ (human fibroblasts) and HEp2 cells. As expected, pan-CEN produced signal at all chromosomes, while HS3–9 hybridized with two chromosomes in BJ and three chromosomes in HEp2 cells, that are tri-somic for chromosome 9 (Fig. S1); CEN and periCEN probes had adjacent/partly overlapping localization in mitotic spreads and also in interphase cells (Figs. S1 and S2), reflecting neighboring positions of CEN/periCEN at chromosomes. In agreement with immunofluorescence results (), Daxx had minor association with both CEN and periCEN probes in normal condition (Fig. S2A). Daxx co-localized with CEN signal after HS and 1 h of recovery (Fig. S2B and C); it was associated with periCEN mostly at 1 h of stress recovery (Fig. S2C). Thus, DNA FISH/immunofluorescence data proved increase of stress-induced association of Daxx with CEN/periCEN.
For quantitative analysis of Daxx association with CEN/periCEN, we next performed ChIP assay using HEp2 cell line modified for stable expression of FLAG-HA-Daxx; microscopy characterization of this cell line confirmed that HS-induced intranuclear dynamics of FLAG-HA-Daxx recapitulates endogenous Daxx protein. Daxx association with CEN (α-satellite, ) was minor at 37°C, was elevated ~10-fold upon HS (p < 0.001), was still high, but reduced, upon 1 h of recovery and returned to pre-stress levels at 2 h and 3 h of recovery. Daxx association with periCEN (HS3-9) was minimal at 37°C, elevated upon HS (p < 0.001) and 1 h of recovery, and reduced to almost pre-stress levels by 3 h of recovery. Thus, alternative method of analysis recapitulated microscopy results for Daxx association with CEN and periCEN; increased accumulation of Daxx at periCEN upon HS measured by ChIP can be explained by higher sensitivity of this method compared with FISH analysis.
Daxx depletion reduces transcription from CEN and periCEN repeats in control and HS conditions
The “nuclear depot” function presumes release of PML NBs-associated proteins from domains in response to the application of external stress,Citation12 and their redistribution to alternative, supposedly “active” locations, that, in case of Daxx, is CEN/periCEN. Therefore, we asked next, what is a functional consequence of Daxx association with CEN/periCEN. The role of Daxx in transcriptional regulation was suggested more than a decade ago;Citation24 Daxx represses transcription through interaction with a number of transcriptional factors and chromatin modifiers.Citation10,Citation25 The ability of Daxx to regulate transcription is affected by its sequestration to PML NBs, confirming the “nuclear depot” model.Citation13 It was previously shown that Daxx is associated with periCEN heterochromatin in mouse cells,Citation15 modulating transcription from major satellite repeats (MaSat);Citation17 Daxx was also found colocalized with a subset of human CENCitation14 suggesting it may regulate transcription from centromeric TRs as well. These facts allow us to hypothesize that Daxx association with CEN/periCEN can affect transcription from CEN/periCEN TRs. To examine whether Daxx association with CEN/periCEN affects transcription from these loci we performed real-time qPCR on RNA collected from control-depleted and Daxx-depleted HEp2 cells (Fig. S3A). RNA was collected at 37°C, immediately after HS (42°C, 1 h) and after different times of HS recovery (37°C, 1–12 h). qPCR analysis was performed using primers for the pan-centromeric α-satellite repeats for CEN and primers specific for the satellite HS3-9 for periCEN (Fig. S3B for specificity; conditions of qPCR were adjusted to produce mostly monomer for product quantization). In control-depleted cells, HS reduced CEN transcripts accumulation. It reached minimum at 2 h of recovery and then gradually increased over time, to almost pre-stress level at 12 h of recovery ( and left part). Daxx depletion reduced CEN transcription already in normal condition (p < 0.01). HS application did not reduce further CEN transcription upon Daxx depletion ( and left part).
Figure 2. Daxx-dependent expression of CEN/periCEN upon HS and recovery. RNA was purified from control-(CTL) and Daxx-depleted HEp2 cells at 37°C, immediately after HS at 42°C for 1 h or after recovery at 37°C for 1–12 h; qPCR analysis of α-satellite repeats transcripts (CEN, left) and HS3-9 transcripts (periCEN, right). Transcript levels were normalized to GAPDH expression levels. Bars represent the mean between replicates (mean ± SD).
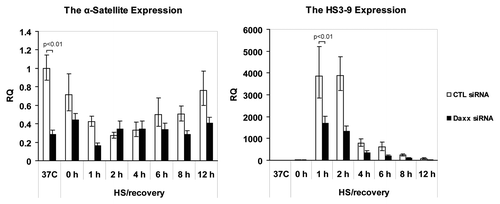
PeriCEN transcripts were elevated during HS, and were strongly accumulated at 1 h and 2 h of stress recovery, and then gradually declined ( and right part). Daxx depletion resulted in reduction of post-stress periCEN transcripts accumulation [, right part, (p < 0.01)]. Thus, depletion of Daxx represses transcription of both TRs: CEN in normal conditions and periCEN upon stress application, suggesting that Daxx activates transcription from both heterochromatin regions.
Since HS-induced transcription of periCEN TRs is mediated by HSF1 and it was suggested that Daxx participates in HSF1 activation upon stress,Citation26 we tested the ability of Daxx to affect transcription of another HSF1 target, HSP70 (HSPA1). We found that depletion of Daxx in HEp2 cells did not reduce expression of HSP70, but rather slightly increased it (Fig. S3C); thus, Daxx may affect CEN/periCEN transcription not by previously demonstrated HSF1 activation, but via an alternative mechanism.
Daxx is required for H3.3 association with CEN/periCEN
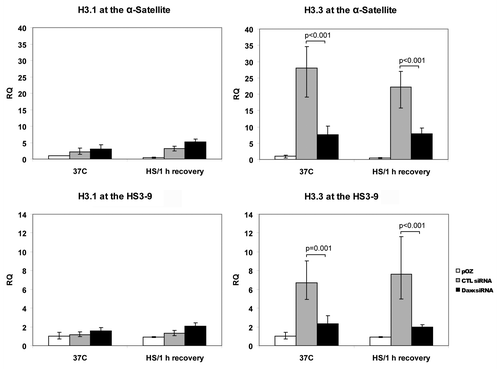
Recently, several groups have identified Daxx as a chaperone of histone H3.3.Citation16,Citation17 Considering that deposition of this variant of histone H3 is usually associated with elevated transcription, we reasoned that Daxx-dependent expression of CEN/periCEN could be explained by differential incorporation of H3.3. To test this idea, control- and Daxx-depleted HEp2 cells were modified for stable expression of FLAG/HA tagged H3.3 or H3.1 by transduction with pOZ vectorCitation27 [(Fig. S4) for cell lines characterization; note that both H3.1 and H3.3 have mostly homogenous nuclear localization]. Next, we used a ChIP assay to analyze association of H3.3 and H3.1 with CEN and periCEN in control- and Daxx-depleted cells (). In normal conditions, incorporation of H3.3 into CEN (α-sattelite) and periCEN (HS3-9) repeats was reduced by Daxx depletion (p < 0.001), while deposition of H3.1 was not affected. Despite robust accumulation of Daxx at CEN/periCEN loci upon HS application, we did not observe major changes in H3.3 incorporation into both TRs in control-depleted (and also in Daxx-depleted) cells upon 1 h of recovery. We concluded that Daxx is required for deposition of H3.3 at CEN and periCEN at 37°C, but not during HS application/recovery.
Figure 3. Association of histone H3.3 and H3.1 with CEN/periCEN. Control- or Daxx-depleted HEp2 cells expressing FLAG/HA-tagged H3.3 or H3.1 were used for FLAG-ChIP in normal condition (37°C) or after 1 h HS/1 h recovery. qPCR data for H3.3 and 3.1 association with α-satellite repeats (CEN) or HS3-9 (periCEN) are presented as fold enrichment IP over input; cells expressing FLAG/HA without an insert were used as negative control (pOZ). Bars represent the mean between replicates (mean ± SD).
Daxx protects epigenetic signature of heterochromatin. We did not observe major changes in H3.3 loading at CEN/periCEN at 1 h of HS recovery (), the time-point of (or immediately after) maximal Daxx association with these TRs ( and S2), indicating a potential alternative role of Daxx at heterochromatin during stress/recovery. In order to examine this novel Daxx function, we analyzed epigenetic modifications at CEN/periCEN by ChIP assay. It was previously shown that periCEN is enriched with repressive chromatin marker H3K9Me3, while active chromatin marker H3K4Me2 is specifically associated with CEN;Citation28 thus, we focused our analysis on these epigenetic markers of chromatin before, during and after HS application.
At 37°C, Daxx depletion resulted in reduction of H3K9Me3 levels at both CEN and periCEN (p < 0.01) that was not further affected by HS ( and right). HS slightly reduced H3K9Me3 at periCEN in control-depleted cells; this data may explain recent observation that loading of HP1 (that is H3K9Me3-dependent) is diminished in response to HS.Citation29
Figure 4. Analysis of epigenetic modifications on CEN/periCEN. ChIP for active (histone H3K4Me2) and repressive (histone H3K9Me3) chromatin marker at α-satellite (CEN) and HS3-9 (periCEN) TRs at normal conditions (37°C) and in post-stress time points (1 h and 12 h recovery). Bars represent the mean between replicates (mean ± SD).
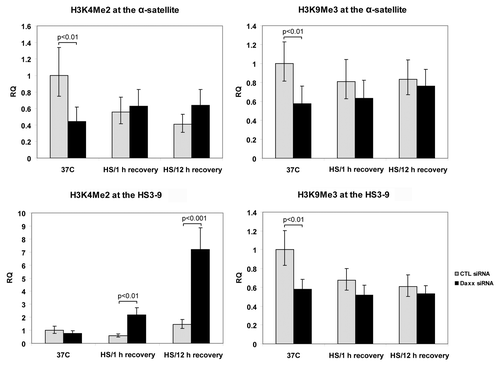
At 37°C, depletion of Daxx resulted in reduction of H3K4Me2 levels at CEN [α-satellite, left top, (p < 0.01)]. The difference disappeared after HS application: H3K4Me2 level in control cells decreased, while it was not affected in Daxx-depleted cells. Level of active chromatin marker H3K4Me2 correlated with the dynamics of CEN transcription (compare to ): at 37°C, relative amount of transcripts was reduced in Daxx-depleted cells; after HS, however, level of H3K4Me2 in control cells—and CEN transcription—declined and become similar in both cell lines. Our data are in agreement with the observation that the level of centromeric transcription is reduced in the absence of H3K4Me2.Citation4 Depletion of Daxx did not affect H3K4Me2 at periCEN (HS3-9) in normal conditions ( and bottom); however, this modification accumulated during stress recovery, at 2- to 3-fold at 1 h (p < 0.01) and almost 10-fold at 12 h (p < 0.001, ). Thus, depletion of Daxx resulted in post-stress enrichment of periCEN with active chromatin marker H3K4Me2. We concluded that depletion of Daxx leads to epigenetic changes of heterochromatin in response to stress, specifically elevating presence of active chromatin marker H3K4Me2 at periCEN. This observation suggests a function for Daxx-containing complex in “epigenetic signature” protection at CEN/periCEN heterochromatin upon stress application.
Discussion
Daxx is a multi-functional ND10/PML NBs associated proteinCitation9,Citation10 that was identified recently as a histone chaperone incorporating histone H3.3 into pericentromeric MaSat in mouse fibroblasts.Citation17 Here we present evidence that Daxx possesses a similar histone chaperone activity incorporating H3.3 in human periCEN, suggesting a conservative chaperone function of Daxx, despite the major differences in pericentromeric heterochromatin structures between mouse and human species.Citation30 In addition, we show that Daxx possesses the same H3.3 loading activity toward CEN chromatin in human cells. Thus, in combination with published data in reference Citation16 and Citation17, we conclude that Daxx incorporates H3.3 into both centromeric and pericentromeric regions in human and mouse cells. This implicates involvement of Daxx in the establishing a “H3 barcode” at heterochromatin regions,Citation18 confirming previously suggested model.Citation31 In agreement with the reported role of H3.3 in transcription activation, Daxx-dependent incorporation of H3.3 may mechanistically explain transcription reduction from both TRs upon Daxx depletion ().
In normal conditions, Daxx is associated with ND10/PML NBs; in agreement with previously published observation,Citation14 we also found minor accumulation at CEN/periCEN in a subpopulation of cells, suggesting potential cell cycle dependence of this transient association. Daxx was previously found at pericentromeric heterochromatin (MaSat) at the end of S/beginning of G2 phases in mouse cells,Citation15 suggesting that Daxx-containing chaperone complex may incorporate H3.3 into human CEN/periCEN upon transient association with these heterochromatic regions, likely at the end of S/G2 phase.
We found dramatic elevation of Daxx association with CEN/periCEN in response to HS (). HS-induced Daxx accumulation does not significantly affect H3.3 incorporation to CEN/periCEN, suggesting an alternative function of Daxx-containing complex at this region of heterochromatin during stress/recovery. In our search for this function, we turned to the epigenetic modifications of heterochromatin. Based on the currently accepted model,Citation28 CEN/periCEN regions are associated with a specific set of epigenetic modifications: centromeric CENP-A-containing nucleosomes (centromere-specific variant of histone H3) are interspersed with transcriptionally-prompted H3K4Me2 nucleosomes that participate in recruitment of CENP-A chaperone HJURP to centromere and maintenance of CENP-A balance.Citation4 Flanking heterochromatic TRs (periCEN) are mostly lacking H3K4Me2, but are enriched in repressive H3K9Me3 modification. This “epigenetic signature” is necessary for genome integrity and proper CEN/periCEN function; forced changes in these modifications lead to the loss of centromere structure/function, as shown recently for the human artificial chromosome.Citation4 We found that depletion of Daxx results in decrease of both modifications at CEN/periCEN in normal conditions; whether these changes are associated with reduced incorporation of histone H3.3 can be an appealing model that is awaiting further confirmation. TRs transcription participates in RNAi-mediated attraction of Clr4/Su(var)3–9 to CEN/periCEN,Citation32 that in turn methylates H3K9 thus recruiting HP1 for heterochromatin establishing. Thus, decrease of H3K9Me3 at CEN/periCEN upon Daxx depletion can be explained by reduced transcription from corresponding TRs and may potentially reduce hererochromatin formation.
What happens with “epigenetic signature” of heterochromatin in response to stress, including HS, is not well characterized, though burst of periCEN transcription may suggest some adaptive changes in heterochromatin epigenetics. In this regard, sequential waves of epigenetic modifications were documented at heterochromatin after HS;Citation33 it was proposed that transcription from periCEN is necessary for the post-stress reconstitution of heterochromatin structure.Citation34 In agreement with this model, depletion of Daxx induces robust changes in the epigenetic modifications at periCEN regions upon stress recovery. Specifically, we found elevation of H3K4Me2 that, in turn, may perturb the epigenetic balance important for centromere/kinetochore structure and function.Citation35 As H3K4Me2 at CEN is necessary to maintain CENP-A balance,Citation4 depletion of Daxx may also result in post-stress reduction in CENP-A loading.
In summary, we found that Daxx is important for loading of H3.3 on human CEN/periCEN TRs by transient association with these genomic regions in normal conditions. H3.3 loading is associated with elevated transcription at both TRs: at CEN, in normal conditions, and at periCEN, in stress-induced conditions. After stress, “epigenetic signature” is protected via Daxx-containing complex assembly at CEN/periCEN. Based on this model, repression or mutations of Daxx suppose to: (1) reduce incorporation of H3.3 on TRs; (2) reduce CEN/periCEN transcription; (3) change “epigenetic signature” of CEN/periCEN chromatin. Combination of these changes may compromise essential heterochromatin functions, therefore potentially increasing genomic instability and initiating malignant transformation of cells. In this context, the recent finding that mutations in Daxx and its H3.3 chaperone partner ATRX are elevated in pancreatic cancerCitation36 and glioblastomaCitation37 may suggest function of Daxx/ATRX H3.3 chaperone in tumorigenesis.
Several questions associated with this model remained unresolved: (1) What is the mechanism of Daxx association with CEN/periCEN in normal conditions and upon stress application? (2) How can Daxx protect “epigenetic signature” at CEN/periCEN after stress? (3) Finally, what is the function of CEN and periCEN transcription and/or transcripts in normal conditions, upon stress application and during cell transformation, given elevated TR expression recently described in epithelial cancers?Citation38,Citation39 Future studies are needed to elucidate mechanism of propagation and maintenance of heterochromatin, which may clarify functions of this part of the genome in normal and pathological conditions.
Materials and Methods
Cell lines and stress conditions
HEp2 cell lines stably expressing Daxx and control shRNACitation40 were additionally transfected with either Daxx siRNA or Non-Targeting siRNA correspondingly (Daxx siGENOME/SMARTpool M004420-01-0005; siGENOME Non-Targeting siRNA Pools D-001206-13, Dharmacon, Lafayette, CO) 48 h before experiments to achieve maximal Daxx depletion. HEp2 cells stably expressing Flag tagged H3.3 and H3.1 were created on the background of Daxx and control shRNA cells using pOZ plasmidCitation27 (Fig. S4). All cells were grown at 37°C in a humidified 5% CO2 atmosphere. For HS, cells were placed at 42°C in a humidified 5% CO2 atmosphere for 1 h; for recovery, cells were returned to 37°C in a humidified 5% CO2 atmosphere for indicated periods of time.
Quantitative RT-PCR
Total RNA was prepared using the Tri Reagent according to the manufacturer instructions (Sigma, St. Louis, MO). RNA was treated with the Turbo DNA-free kit (Applied Biosystems, Carlsbad, CA) to remove the potential traces of DNA. cDNA was produced with SuperScriptTM II Reverse Transcriptase (Invitrogen, Carlsbad, CA) using 1 ∝g of total RNA and random primers. Quantitative PCR was performed on the StepOne RT-PCR machine (Applied Biosystems, Foster City, CA) with either Fast SYBR Green Master Mix (HS3-9, HSPA1) or Power SYBR Green Master Mix (α-satellite) (Applied Biosystems, Foster City, CA); see Table S1 for primers and conditions. Expression was normalized to the GAPDH using the formulas: ΔCT = CT(HS3-9 or α-satellite) - CT(GAPDH), ΔΔCT = ΔCT - ΔCT (37°C), RQ = 2-ΔΔCT. RNA samples without RT reaction were included in qPCR to control for DNA contaminations. For each reaction, melting curves and agarose gel electrophoresis of PCR products were used to verify the identity of the amplification products (Fig. S3B). All experiments were repeated in triplicates.
Chromatin immunoprecipitation (ChIP)
Native ChIP was used to analyze association of H3.3 and H3.1 and histone H3 modifications at CEN and periCEN. Chromatin was prepared according to a published protocolCitation41 with some modifications. Briefly, cells were trypsinized, washed with DMEM/10% FBS to stop reaction, harvested by centrifugation at 500x g for 5 min and washed once with PBS, 1.5 mM MgCl2. The cells were lysed by incubation in PBS, 1.5 mM MgCl2 and 1% Triton X-100 for 5 min on ice with repeated vortexing. Nuclei were collected by centrifugation at 1,000x g for 5 min and resuspended in Chromatin Isolation Buffer (15 mM Tris pH 7.5, 15 mM NaCl, 60 mM KCl, 0.34 M sucrose) supplemented with 1 mM CaCl2. Nuclei were digested with 0.5 U MNase (N3755, Sigma) for 20 min on ice, and digestion was stopped by the addition of EDTA to 10 mM. Nuclei were collected by centrifugation at 16,000x g for 10 min, resuspended in IP buffer (10 mM TRIS-HCl pH 7.5, 500 mM NaCl, 0.1% Triton X-100), and incubated at 4°C for 1 h with rotation. Insoluble material was removed by centrifugation at 16,000x g for 15 min and soluble chromatin was recovered in the supernatant. Chromatin isolated from Flag-H3.1 or Flag-H3.3 expressing cell lines was immunoprecipitated with anti-Flag M2 Magnetic Beads (M8823, Sigma) overnight at 4°C with rotation. Chromatin isolated from cell line expressing empty pOZ-Flag vector was used as a negative control. To study post-transcriptional modifications, chromatin obtained from HEp2 cells was incubated overnight with anti-H3K4Me2 (ab32356, Abcam), anti-H3K9Me3 (#9754, Cell Signaling Technology, Danvers, MA) or without antibodies (negative control) at 4°C with rotation. Protein G dynabeads (Invitrogen) were added for 2 h. The beads were washed 6 times with IP buffer and chromatin was eluted with either 150 ng/ul 3xFlag peptide (from anti-Flag M2 Magnetic beads) or 1% SDS in IP buffer (from Dynabeads Protein G). DNA was purified by phenol/chloroform extraction and analyzed by quantitative PCR using StepOne RT-PCR machine (Applied Biosystems).
Enrichment for Flag IP was calculated as follows: ΔCT = CT bound fraction - CT input fraction. ΔΔCT = ΔCT (pOZ-Flag-H3.1 or pOZ-Flag-H3.3) - ΔCT (pOZ-Flag, 37°C). RQ = 2-ΔΔCT.
Enrichment for epigenetic modifications was calculated as follows: ΔCT = CT bound fraction - CT input fraction, ΔΔCT = ΔCT (α-satellite or HS3-9) - ΔCT (HGB1), ΔΔΔCT = ΔΔCT - ΔΔCT (37°C), RQ = 2-ΔΔΔCT.
To analyze association of Daxx with CEN and periCEN, HEp2 cells were modified for stable expression of Flag tagged Daxx using pOZ plasmid.Citation27 Formaldehyde was added directly to cells media to final concentration 1%. The reaction was quenched after 10 min by adding Glycine to final concentration 0.125 M for 5 min. Cells were washed twice with PBS, resuspended in Lysis buffer (50 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton) and incubated for 10 min with rotation. Cells were centrifuged at 1,000 g for 5 min, supernatant was discarded and pellet was resuspended in IP buffer (50 mM Tris-Hcl, pH 7.4, 400 mM NaCl, 1 mM EDTA, 1% Triton) supplemented with 0.04% SDS, transferred into 5 ml Falcon round-bottom tube and sonicated using Misonix sonicator 3000 (6 cycles 10 sec on/50 sec off, power 2.5). Insoluble material was removed by centrifugation. Soluble chromatin fraction was diluted with equal volume of IP buffer to reduce SDS concentration to 0.02%. Chromatin was immunoprecipitated with anti-Flag M2 Magnetic Beads (Sigma) for 2 h with rotation. Beads were washed once with IP buffer supplemented with 0.02% SDS then three times with IP buffer and finally with TE buffer. Chromatin was eluted with a buffer containing 10 mM Tris pH 8.0, 1% SDS, 5 mM EDTA at 65°C for 30 min. Cross-link was reversed by addition of 5 M NaCl and incubation at 65°C for 4 h. The samples were diluted with equal volume of TE buffer and treated with proteinase K for 2 h. DNA was purified by Phenol/Chloroform extraction and ethanol precipitation. Enrichment for Flag IP was calculated as follows: ΔCT = CT bound fraction - CT input fraction, ΔΔCT = ΔCT (pOZ-Flag-Daxx) - ΔCT (pOZ-Flag), RQ = 2-ΔΔCT.
Immunofluorescence
Immunofluorescence analysis was completed on cells grown overnight on coverslips in 24 well plates (Corning Inc.) as described previously in reference Citation15. The following primary antibodies were used: Daxx 5.14 monoclonal,Citation15 PML 14 rabbit,Citation11 HSF1 rat polyclonal (Abcam), CREST human autoimmune antibodies (gift of Dr Gerd Maul, Wistar Institute). Cells were stained with Hoechst 33342 (Sigma) for DNA visualization and mounted on slides with Fluoromount G (Southern Biotech). Images were analyzed using Leica TCS SP5 confocal microscope. Colocalization analysis (Pearson’s correlation coefficientCitation42) on confocal images has been used to quantify the degree of association for Daxx/PML (for PML NBs) and Daxx/CREST (for centromeres). For calculation of the Pearson’s correlation coefficient, images were processed and analyzed using MetaMorph software, version 6.0. At least 50 cells were analyzed for each time points; experiments were repeated three times.
Additional material
Download Zip (16.7 MB)Funding
This work was supported by NIH/NCI R01 CA127378-01A1 for V.M.M., E.V.G. and A.M.I., by MCB grant from Presidium of Russian Academy of Sciences for E.V.G. and by grants from “La Ligue Contre le Cancer” (9ADO1217/1B1-BIOCE) and the “Institut National du Cancer” (247343/1B1-BIOCE) to V.V.O.
Acknowledgments
We want to thank Dr Gerd Maul, Wistar Institute, Philadelphia, PA for inspiration of this work and dedicate this manuscript to his memory.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: landesbioscience.com/journals/nucleus/article/20180/
References
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays 1998; 20:532 - 5; http://dx.doi.org/10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M; PMID: 9723001
- Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF. Characterization of a chromosome-specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics 1996; 33:220 - 8; http://dx.doi.org/10.1006/geno.1996.0187; PMID: 8660971
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 2007; 17:1146 - 60; http://dx.doi.org/10.1101/gr.6022807; PMID: 17623812
- Bergmann JH, Rodríguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 2011; 30:328 - 40; http://dx.doi.org/10.1038/emboj.2010.329; PMID: 21157429
- Choo KH. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet 1997; 61:1225 - 33; http://dx.doi.org/10.1086/301657; PMID: 9399915
- Moyzis RK, Albright KL, Bartholdi MF, Cram LS, Deaven LL, Hildebrand CE, et al. Human chromosome-specific repetitive DNA sequences: novel markers for genetic analysis. Chromosoma 1987; 95:375 - 86; http://dx.doi.org/10.1007/BF00333988; PMID: 3677921
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, et al. Stress-induced transcription of satellite III repeats. J Cell Biol 2004; 164:25 - 33; http://dx.doi.org/10.1083/jcb.200306104; PMID: 14699086
- Eymery A, Souchier C, Vourc’h C, Jolly C. Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells. Exp Cell Res 2010; 316:1845 - 55; http://dx.doi.org/10.1016/j.yexcr.2010.02.002; PMID: 20152833
- Lindsay CR, Giovinazzi S, Ishov AM. Daxx is a predominately nuclear protein that does not translocate to the cytoplasm in response to cell stress. Cell Cycle 2009; 8:1544 - 51; http://dx.doi.org/10.4161/cc.8.10.8379; PMID: 19372739
- Lindsay CR, Morozov VM, Ishov AM. PML NBs (ND10) and Daxx: from nuclear structure to protein function. Front Biosci 2008; 13:7132 - 42; http://dx.doi.org/10.2741/3216; PMID: 18508722
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 1999; 147:221 - 34; http://dx.doi.org/10.1083/jcb.147.2.221; PMID: 10525530
- Maul GG, Negorev D, Bell P, Ishov AM. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol 2000; 129:278 - 87; http://dx.doi.org/10.1006/jsbi.2000.4239; PMID: 10806078
- Negorev D, Maul GG. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 2001; 20:7234 - 42; http://dx.doi.org/10.1038/sj.onc.1204764; PMID: 11704851
- Pluta AF, Earnshaw WC, Goldberg IG. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J Cell Sci 1998; 111:2029 - 41; PMID: 9645950
- Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci 2004; 117:3807 - 20; http://dx.doi.org/10.1242/jcs.01230; PMID: 15252119
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140:678 - 91; http://dx.doi.org/10.1016/j.cell.2010.01.003; PMID: 20211137
- Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 2010; 24:1253 - 65; http://dx.doi.org/10.1101/gad.566910; PMID: 20504901
- Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci U S A 2006; 103:6428 - 35; http://dx.doi.org/10.1073/pnas.0600803103; PMID: 16571659
- Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev 2010; 20:110 - 7; http://dx.doi.org/10.1016/j.gde.2010.01.003; PMID: 20153629
- Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res 2011; 21:421 - 34; http://dx.doi.org/10.1038/cr.2011.14; PMID: 21263457
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004; 116:51 - 61; http://dx.doi.org/10.1016/S0092-8674(03)01064-X; PMID: 14718166
- Maul GG, Yu E, Ishov AM, Epstein AL. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem 1995; 59:498 - 513; http://dx.doi.org/10.1002/jcb.240590410; PMID: 8749719
- Nefkens I, Negorev DG, Ishov AM, Michaelson JS, Yeh ET, Tanguay RM, et al. Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J Cell Sci 2003; 116:513 - 24; http://dx.doi.org/10.1242/jcs.00253; PMID: 12508112
- Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J 1999; 18:3702 - 11; http://dx.doi.org/10.1093/emboj/18.13.3702; PMID: 10393185
- Morozov VM, Massoll NA, Vladimirova OV, Maul GG, Ishov AM. Regulation of c-met expression by transcription repressor Daxx. Oncogene 2008; 27:2177 - 86; http://dx.doi.org/10.1038/sj.onc.1210865; PMID: 17952115
- Boellmann F, Guettouche T, Guo Y, Fenna M, Mnayer L, Voellmy R. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc Natl Acad Sci U S A 2004; 101:4100 - 5; http://dx.doi.org/10.1073/pnas.0304768101; PMID: 15016915
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 2003; 370:430 - 44; http://dx.doi.org/10.1016/S0076-6879(03)70037-8; PMID: 14712665
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 2004; 11:1076 - 83; http://dx.doi.org/10.1038/nsmb845; PMID: 15475964
- Velichko AK, Kantidze OL, Razin SV. HP1α is not necessary for the structural maintenance of centromeric heterochromatin. Epigenetics 2011; 6:380 - 7; http://dx.doi.org/10.4161/epi.6.3.13866; PMID: 20962594
- Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol 2009; 53:259 - 68; http://dx.doi.org/10.1387/ijdb.082673ae; PMID: 19412885
- Campos EI, Reinberg D. New chaps in the histone chaperone arena. Genes Dev 2010; 24:1334 - 8; http://dx.doi.org/10.1101/gad.1946810; PMID: 20595228
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 2010; 20:134 - 41; http://dx.doi.org/10.1016/j.gde.2010.02.003; PMID: 20207534
- Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell 2004; 15:543 - 51; http://dx.doi.org/10.1091/mbc.E03-07-0487; PMID: 14617804
- Biamonti G, Vourc’h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol 2010; 2:a000695; http://dx.doi.org/10.1101/cshperspect.a000695; PMID: 20516127
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 2008; 14:507 - 22; http://dx.doi.org/10.1016/j.devcel.2008.02.001; PMID: 18410728
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011; 331:1199 - 203; http://dx.doi.org/10.1126/science.1200609; PMID: 21252315
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012; 482:226 - 31; http://dx.doi.org/10.1038/nature10833; PMID: 22286061
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011; 331:593 - 6; http://dx.doi.org/10.1126/science.1200801; PMID: 21233348
- Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res 2009; 37:6340 - 54; http://dx.doi.org/10.1093/nar/gkp639; PMID: 19720732
- Lindsay CR, Scholz A, Morozov VM, Ishov AM. Daxx shortens mitotic arrest caused by paclitaxel. Cell Cycle 2007; 6:1200 - 4; http://dx.doi.org/10.4161/cc.6.10.4244; PMID: 17471023
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2002; 2:319 - 30; http://dx.doi.org/10.1016/S1534-5807(02)00135-1; PMID: 11879637
- Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem 2007; 40:101 - 11; http://dx.doi.org/10.1267/ahc.07002; PMID: 17898874
- Crusselle-Davis VJ, Zhou Z, Anantharaman A, Moghimi B, Dodev T, Huang S, et al. Recruitment of coregulator complexes to the beta-globin gene locus by TFII-I and upstream stimulatory factor. FEBS J 2007; 274:6065 - 73; http://dx.doi.org/10.1111/j.1742-4658.2007.06128.x; PMID: 17970752
- Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, et al. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp Cell Res 2002; 276:10 - 23; http://dx.doi.org/10.1006/excr.2002.5513; PMID: 11978004