Abstract
We have raised antibodies against the profilin of Chironomus tentans to study the location of profilin relative to chromatin and to active genes in salivary gland polytene chromosomes. We show that a fraction of profilin is located in the nucleus, where profilin is highly concentrated in the nucleoplasm and at the nuclear periphery. Moreover, profilin is associated with multiple bands in the polytene chromosomes. By staining salivary glands with propidium iodide, we show that profilin does not co-localize with dense chromatin. Profilin associates instead with protein-coding genes that are transcriptionally active, as revealed by co-localization with hnRNP and snRNP proteins. We have performed experiments of transcription inhibition with actinomycin D and we show that the association of profilin with the chromosomes requires ongoing transcription. However, the interaction of profilin with the gene loci does not depend on RNA. Our results are compatible with profilin regulating actin polymerization in the cell nucleus. However, the association of actin with the polytene chromosomes of C. tentans is sensitive to RNase, whereas the association of profilin is not, and we propose therefore that the chromosomal location of profilin is independent of actin.
Introduction
β-actin is an abundant cellular protein of molecular weight 42 kDa and a major component of the cytoskeleton in all eukaryotic cells. β-actin is involved in essential biological processes such as maintaining cell shape, vesicle trafficking and cell motility. β-actin exists in two major forms: a globular, monomeric form referred to as “G-actin,” and a filamentous polymer named “F-actin.” The structure and activities of the actin cytoskeleton are regulated by the polymerization dynamics of actin, which in turn is modulated by a large number of proteins. Among them, ADF/cofilins and profilins are major regulators of actin polymerization.Citation1
β-actin is not only a major cytoskeletal component but also a nuclear protein that plays important roles in gene expression. β-actin is part of chromatin remodeling complexes, it is associated with the transcription machineries, it becomes incorporated into newly synthesized ribonucleoproteins, and it influences long-range chromatin organization (reviewed in ref. Citation2). Immuno-electron microscopy experiments in the salivary glands of the dipteran Chironomus tentans (C. tentans) have shown that actin is associated with active transcription units.Citation3 The association of actin with active chromatin was has been demonstrated also in mammalian cells. Chromatin immunoprecipitation experiments using anti-actin antibodies have shown that actin is associated with Pol I and Pol II genes.Citation4-Citation6
Several heterogeneous nuclear ribonucleoproteins (hnRNPs) bind actin and mediate its association with nascent mRNA. In C. tentans, actin is associated with the mRNA-binding proteins hrp36 Citation3 and hrp65.Citation7 In mammalian cells, actin associates with several hnRNP proteins, including hnRNP A2, hnRNP A3, CArG-box binding factor (CBF-A) and hnRNP U.Citation8,Citation9 Both in insect and mammalian cells, the actin-hnRNP complexes participate in the recruitment of co-activators to active genes.Citation6,Citation10,Citation11
Nuclear β-actin works in conjunction with different types of actin-binding proteins that regulate actin function and bridge interactions between actin and other nuclear components. NM1, a short-tailed myosin that acts as an actin-dependent ATPase, is found in the cell nucleus associated with actin.Citation12 Studies of Pol-I transcription have shown that an actin-NM1 interaction is required for Pol-I transcription, which has led to the proposal that an actomyosin motor helps the transcription machinery slide along the rDNA.Citation5,Citation13,Citation14 Studies of gene repositioning and chromatin organization point to the possibility that actin also plays a role in the large-scale organization of the genome.Citation15,Citation16 Altogether, these observations suggest that the dynamics of actin polymerization has important implications for chromatin organization and gene expression.
Other actin-binding proteins, including β-actinin, filamin, p190RhoGAP, paxillin, spectrin, tropomyosin, cofilin and profilin, have also been detected in the nucleus of human cells.Citation17,Citation18 Profilins are small proteins with masses in the 15–17 kDa range. They are evolutionarily conserved and play a key role in the regulation of F-actin formation. Profilins promote actin polymerization by favoring the exchange of ADP for ATP in actin. Localization studies using profilin fused to green fluorescent protein (GFP) showed that, in the cytoplasm of mammalian cells, profilin localizes to areas with high actin dynamics, such as leading lamellae and ruffles,Citation19 and participates in the formation Golgi vesicles.Citation20
Four profilin genes have been identified in mammals: Pfn1, Pfn2, Pfn3 and Pfn4, of which Pfn1 codes for the ubiquitous profilin I.Citation21 There is a single gene encoding profilin in the genome of Drosophila melanogaster, chickadee/CG9553.
The use of anti-profilin antibodies has revealed that profilin is located in several types of intranuclear structures in mammalian cells: splicing speckles, Cajal bodies and gems.Citation22,Citation23 Profilin is also involved in transporting actin out the nucleus via exportin 6.Citation24 The architecture of the mammalian cell nucleus in interphase is complex, and localization studies are limited by the co-existence of transcriptionally active chromatin intermingled with patches of dense, inactive chromatin. In order to learn more about the location of profilin and its role in the nucleus, we raised antibodies against the profilin of C. tentans and we performed immunolocalization studies in the polytene nuclei of the C. tentans salivary gland cells. The salivary gland cells of the fourth instar larvae have large polytene chromosomes and the interchromatin space is free of chromatin. The salivary gland cells are therefore an excellent material to study the location of profilin relative to chromatin and to active genes.
Results
A nucleotide sequence with high homology to profilins was found in a EST database of C. tentans cDNA.Citation25 This cDNA sequence, called p0825.218, contains an open reading frame that codes for a protein of 126 amino acids and 13.6 kDa. BLAST analyses showed that the predicted amino acid sequence encoded by the p0825.218 cDNA was 83% and 91% identical to the profilins of D. melanogaster and Anopheles gambiae, respectively (). The high degree of sequence homology to profilins from other diptera and the fact that the known insect genomes contain a single profilin gene support the conclusion that the p0825.218 cDNA codes for the profilin protein of C. tentans, Ct-Pfn.
Figure 1. The profilin protein of C. tentans. (A) The amino acid sequences of the profilins of C. tentans, D. melanogaster and H. sapiens were aligned using ClustalW (workbench.sdsc.edu/). Asterisk, colon and period indicate fully conserved residues, conservation of strong groups and conservation of weak groups, respectively. The amino acid sequence of the peptide used for antibody production is underlined. (B) Total protein extracts prepared from C. tentans tissue culture cells and from D. melanogaster S2 cells were probed by western blot with anti-profilin antibodies. Ab1 and ab2 were affinity purified from two rabbits immunized with the same peptide. Molecular mass standards are shown to the left in kDa. (C) A cytoplasmic extract (C), a nuclear pellet (NP) and a nuclear soluble extract (NS) were prepared from C. tentans tissue culture cells and probed with antibodies against profilin and actin, as indicated.
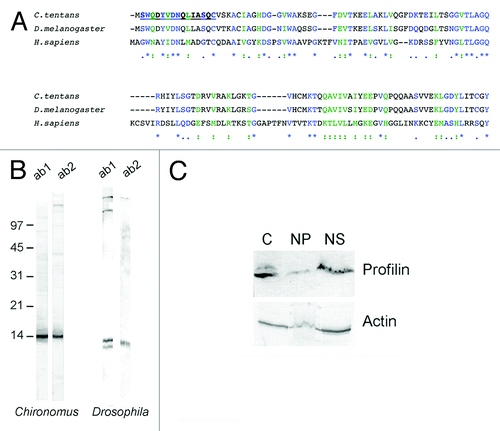
We immunized two rabbits with a synthetic peptide corresponding to amino acids 2–15 of Ct-Pfn based on the prediction of a highly immunogenic site near the N-terminus using the Protean-DNA LaserGene software. The affinity-purified antibodies detected a major band of the expected size when probed by western blotting against total protein extracts of C. tentans (). The anti-profilin antibodies, and in particular the antibody ab1, were specific and suitable for immunolocalization studies in C. tentans. D. melanogaster profilin, the product of the chickadee gene, was also detected by the anti-profilin antibodies.
Previous studies had shown the existence of profilin in the nucleus of mammalian cells.Citation23 To determine whether profilin was also a nuclear protein in C. tentans, we performed fractionation experiments using C. tentans cultured cells. The cells were homogenized in a detergent-containing buffer and the nuclei were collected by centrifugation. The supernatant was the cytoplasmic fraction (“C” in ). The nuclear pellet was resuspended in PBS, mildly sonicated to lysate the nuclei, and centrifuged again. The supernatant of this second centrifugation was the soluble fraction (“NS” in ) containing soluble proteins and mRNPs. The pellet was the nuclear insoluble fraction (“NP” in ) and contained the nuclear envelope, the nucleolus and the chromatin, including the nascent transcripts. We analyzed the presence of actin and profilin in these fractions by western blotting. The fractionation patterns of actin and profilin were very similar. Both proteins were predominantly found in the cytoplasm and in the nuclear soluble fraction. Profilin was much less abundant in the insoluble nuclear fraction.
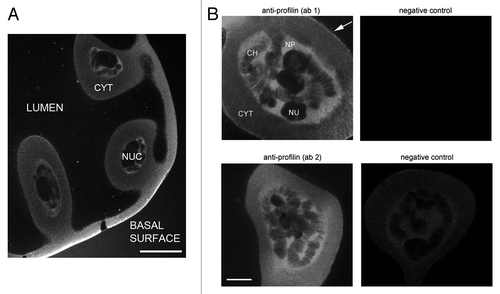
We next used the anti-profilin antibodies to stain the salivary gland cells of C. tentans by immunofluorescence. Salivary glands were dissected from fourth instar larvae, fixed, permeabilized and stained, and then imaged by laser confocal microscopy. shows that the anti-profilin antibodies gave a very strong cytoplasmic staining in the basal region of the cells. A thin rim was often also observed at the apical surface (arrow in ), and a faint but significant staining was observed throughout the cytoplasm. Also the nuclei of the salivary gland cells were stained. Profilin was concentrated in the nucleoplasm and associated with multiple bands in the polytene chromosomes, but was excluded from the nucleolus (). The Balbiani ring (BR) puffs were stained to a certain degree, but the levels of fluorescence in BR puffs were lower than that in the nucleoplasm (see Figs. 5 and 6). Both affinity-purified antibodies, ab1 and ab2, gave the same pattern of staining. Ab1 was chosen for subsequent studies.
Figure 2. Profilin in the salivary gland cells of C. tentans. Salivary glands were fixed and stained with ab1 and ab2 against profilin. (A) Overview showing the architecture of the salivary gland cells and the distribution of profilin. NUC: nucleus, CYT: cytoplasm. The bar represents 60 μm. (B) The nuclear localization of profilin in the salivary gland cells revealed with antibodies ab1 and ab2, as indicated. Control glands incubated in parallel with negative control antibodies are shown to the right. CH, chromosome; NU, nucleolus; NP, nucleoplasm. The bar represents 20 μm. The arrow points at the apical surface of the cell facing the lumen.
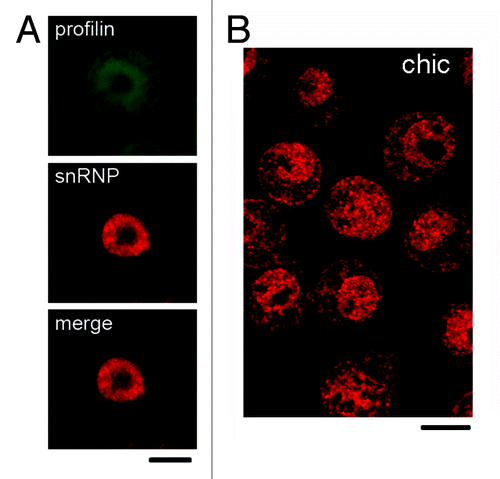
As shown in , the anti-profilin antibody also recognized the product of the D. melanogaster gene chickadee. We immunostained S2 cells with antibodies against profilin to determine whether profilin was also a nuclear protein in D. melanogaster. We used an anti-snRNP antibody as a nuclear marker. The anti-profilin ab1 antibody stained both the cytoplasm and the nucleus of the S2 cells, whereas the anti-snRNP antibody gave a predominantly nuclear staining (). We also stained S2 cells with a mAb raised against the profilin of D. melanogaster.Citation26 The pattern of staining observed with this mAb was very similar to the one obtained with our peptide-specific antibodies (compare A and B in ).
Figure 3. Localization of profilin in S2 cells of D. melanogaster. (A) S2 cells were double-labeled with ab2 anti-profilin (green) and mAb Y12 against snRNPs (red). The anti-profilin antibody was positive in both nucleus and cytoplasm, whereas Y12 labeled preferentially the nucleus. (B) S2 cells were labeled with a mAb against the product of the chickadee gene, chic. This antibody also labeled both nucleus and cytoplasm. The bars represent 5 μm.
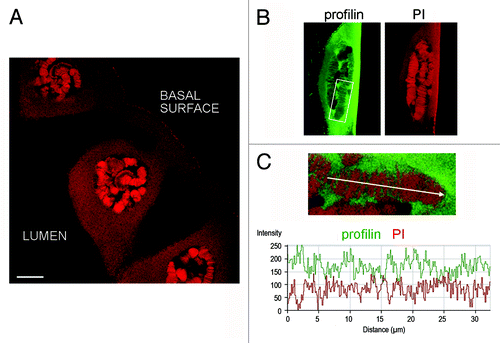
We performed co-localization experiments in the salivary glands of C. tentans to characterize the association of profilin with the chromosomes. In a first series of experiments, we immunostained salivary gland preparations with the anti-profilin ab1 antibody as in , and we counterstained the glands with propidium iodide (PI). PI binds to nucleic acids and stains predominantly dense chromatin in the polytene chromosomes. The rim of the nucleolus was also stained, and the cytoplasm to some extent, due to high concentrations of highly structured rRNA in these locations (). The extent of co-localization between profilin and PI in the chromosomes was very low. The PI-positive bands in the polytene chromosomes were negative for profilin and vice versa (). We concluded that profilin is excluded from dense chromatin.
Figure 4. Profilin is not associated with dense chromosome bands. Salivary glands were fixed and stained with ab1 against profilin and counterstained with propidium iodide to reveal dense chromatin bands in the polytene chromosomes. (A) Overview of salivary gland cells stained with propidium iodide (PI). The bar represents 25 μm. (B) The nucleus of a salivary gland cell stained with anti-profilin antibody (green) and PI (red). (C) The co-localization between profilin and PI was analyzed using the Profile function of the LSM 510 software. The graph shows the relative intensities for each channel, in arbitrary units, along the chromosome axis defined by the arrow. The analyzed chromosome segment is shown in the inset in (B). The opposite staining profiles for profilin and PI reveal that profilin is excluded from dense chromatin.
We performed double-staining experiments to analyze the localization of profilin relative to transcribed genes. We used antibodies against two different proteins that associate with the nascent mRNA: the hnRNP protein hrp23 and the core proteins of the snRNP complexes. Salivary glands stained with antibodies against profilin and hrp23 are shown in . Profilin and hrp23 co-localized to a large extent, and most profilin-positive bands in the polytene chromosomes were also stained with the anti-hrp23 antibody. However, the patterns of staining along the chromosomes were not identical. A few loci, such as the BR puffs, were predominantly stained by hrp23 with less profilin staining. Some loci were positive for profilin but negative for hrp23.
Figure 5. Co-localization of profilin with mRNA-binding proteins. Salivary glands were fixed and double stained with ab1 against profilin and against either hrp23. (A) or snRNPs (B). chrom, chromosomes; nu, nucleolus; cyt, cytoplasm. A large Balbiani ring puff (BR) strongly stained by the anti-hrp23 antibody is shown in the merged image in (A). The bars represent 20 μm. (C) The co-localization between profilin and snRNPs was analyzed using the Profile function of the LSM 510 software. The graph shows the relative intensities for each channel, in arbitrary units, along the chromosome axis defined by the arrow. Note the large extent of co-variation in the intensity profiles for profilin and snRNPs. (D) Isolated polytene chromosomes of C. tentans were double immuno-stained with anti-profilin and anti-snRNP antibodies. The image shows a chromosome IV, and the positions of the BR puffs are indicated. The white arrows point at loci that are positive for profilin but not for snRNPs or vice versa. The bar represents 10 μm.
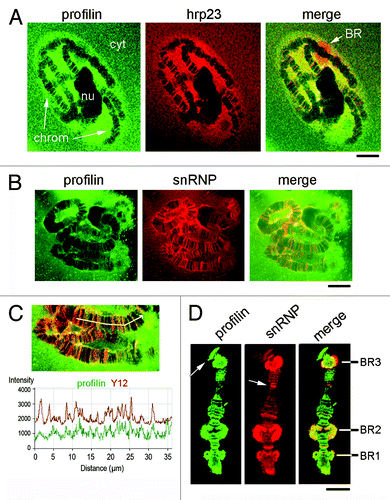
In another series of experiments, we stained salivary glands with antibodies against Ct-profilin and snRNPs (). Profilin co-localized to a large extent with snRNPs, although the patterns of staining were not identical, as in the case of hrp23. To better compare the distribution patterns of profilin and snRNPs, we immunostained isolated chromosomes. shows an example of chromosome IV immunostained with anti-profilin and anti-snRNP antibodies. The analysis of isolated chromosomes confirmed a partial co-localization. The BR puffs in chromosome IV were positively stained with the anti-profilin antibody in preparations of isolated chromosomes (). In whole-mount salivary glands, the BR puffs were positive but were not among the most prominently stained loci (see, for example, ). The difference in relative intensity can be explained by the different permeabilization and fixation conditions. In isolated chromosomes, profilin and snRNPs co-localized in the BRs. The snRNP staining was very intense in BR3, which is consistent with the fact the BR3 pre-mRNA contains a large number of introns, most of which are spliced co-transcriptionally.Citation27 Interestingly, the anti-snRNP antibody stained the entire BR3, whereas profilin had a more internal localization, near the body of the chromosome (see the merged image in ).
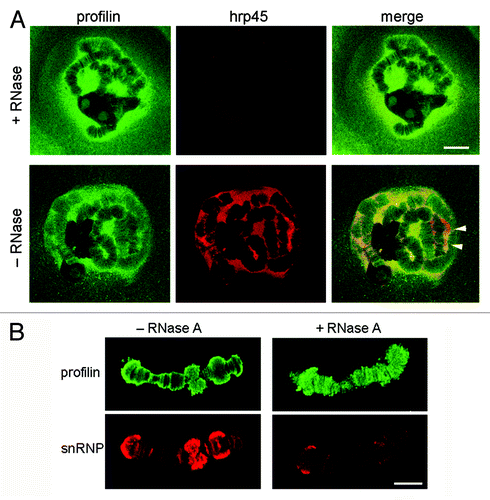
The results presented above suggest that profilin is associated with transcriptionally active loci in the polytene chromosomes. We next performed a series of experiments in which we digested the preparations with RNase A before the immunostaining, to determine whether the association of profilin with the chromosomes was mediated by RNA. The glands were double-stained with an antibody against the hnRNP protein hrp45 to monitor the effect of the RNase digestion. The nuclear localization of profilin was not affected by the RNase digestion, while the anti-hrp45 staining was drastically reduced (). We also performed experiments with RNase digestion of isolated polytene chromosomes. In this case, the preparations were counterstained with the anti-snRNP antibody (). The snRNP staining was drastically reduced by the digestion, but profilin was still associated with the chromosomes after digestion with RNase A. We concluded that the association of profilin with the chromosomes was not mediated by RNA.
Figure 6. The association of profilin with the chromosomes does not depend on RNA. (A) Salivary glands were digested with RNase A before fixation and immunostaining with antibodies against profilin (green) and hrp45 (red). The hnRNP protein hrp45 was used to monitor the efficiency of the enzymatic digestion. Note that the distribution of profilin in the nucleus was not affected by the digestion with RNase A. The arrowheads point at BR puffs intensely labeled by the anti-hrp45 antibody. The bar represents 20 μm. (B) RNase A digestion of isolated polytene chromosomes before immunostaining with antibodies against profilin (green) and snRNPs (red). The bar represents 10 μm.
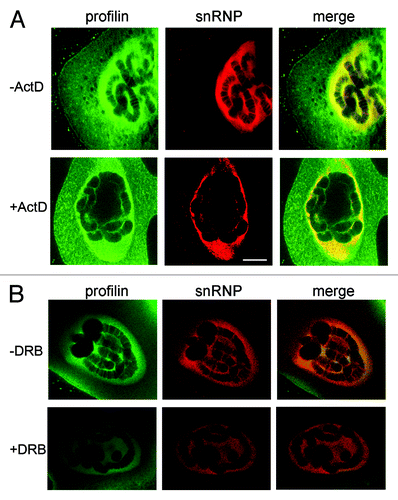
The results presented above suggest that profilin is associated with transcriptionally active loci, but that the association of profilin with the active loci does not depend on RNA. We next asked whether the chromosomal association of profilin depends on ongoing transcription. We incubated salivary glands in the presence of actinomycin D for one hour before fixation and immunostaining. Control glands were treated in parallel without actinomycin D. The glands were double-stained with antibodies against profilin and snRNPs. The effect of actinomycin D on the overall nuclear staining was not striking, but the chromosomes were devoid of snRNP staining after actinomycin D treatment, which indicated that the treatment was effective. The anti-profilin antibodies still stained the nucleoplasm, while the chromosomal staining was significantly reduced (). We concluded that the association of profilin with the chromosomes is transcription-dependent.
Figure 7. The association of profilin with the chromosomes requires ongoing transcription. (A) Salivary glands were incubated in the presence of actinomycin D (+Act) before fixation and immunostaining with antibodies against profilin (green) and snRNPs (red). Control glands were incubated in parallel without the drug, as indicated. (B) Fourth-instar larvae were treated with (+DRB) or without (-DRB) DRB before dissection of the glands and immunostaining. The bar represents 10 μm. Note that the banded labeling of the chromosomes is lost after actinomycin D or DRB treatment.
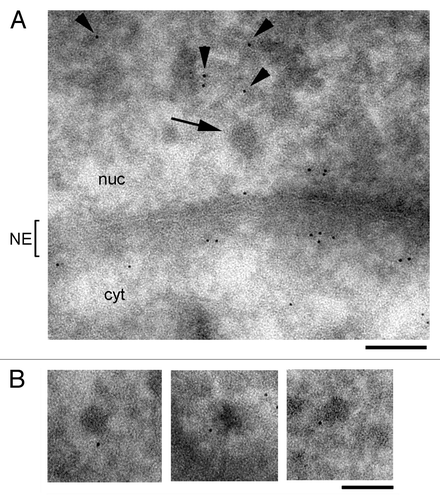
Actin becomes incorporated into nascent pre-mRNAs and remains associated with mRNP complexes during transport from the gene to the nuclear pores.Citation3 The results presented above suggested that profilin does not become incorporated into mRNP complexes. To further analyze this question, we used immuno-electron microscopy (immuno-EM) to analyze the association of profilin with BR mRNPs. The BR mRNPs are large ribonucleorotein particles that can be identified in the nucleoplasm of the salivary gland cells due to their large dimensions and distinctive morphology. Salivary glands were fixed, cryoprotected and cryosectioned. The thin cryosections were incubated with the anti-profilin antibody and with a secondary antibody conjugated to colloidal gold. The immuno-EM analysis failed to reveal any significant association of profilin with BR mRNP particles (). The density of gold labeling was relatively high in the nucleoplasm, where the labeling was associated with fibrillar material (arrowheads in ), but the BR mRNPs were devoid of labeling (arrow in ). A high density of labeling was observed near the nuclear envelope, on both the nuclear and the cytoplasmic sides. In some cases, profilin was found to decorate structures in the vicinity of the BR mRNPs (). These structures probably correspond to the connecting fibers described by Miralles et al.Citation28 In summary, the immuno-EM experiments confirmed that profilin is a nuclear protein preferentially located in the nucleoplasm and at the nuclear envelope, but not associated with nucleoplasmic BR mRNPs.
Figure 8. Immuno-EM analysis of profilin in the nucleoplasm of the salivary gland cells. Thin cryosections of salivary glands were stained with ab1 against profilin. A secondary antibody conjugated to 6-nm colloidal gold particles was used to visualize the immunolabelling. (A) Overview of a nucleoplasmic region near the nuclear envelope (NE). The arrow points to a BR mRNP particle. The arrowheads point at immuno-gold markers in the nucleoplasm. (B) Three examples of immuno-gold labeling in the vicinity of BR mRNP particles in the nucleoplasm of the salivary gland cells. The bars represent 100 nm.
Discussion
We have identified the profilin of C. tentans and found that it is very similar to profilins from other diptera such as Drosophila and Anopheles. Previous studies have shown that profilins are not restricted to the cytoplasm but are also located in the cell nucleus, both in mammalian cells and in insects.Citation22,Citation23,Citation29-Citation31 Moreover, several nuclear proteins have been identified as ligands for profilin, including the survival motoneuron protein (SMN),Citation22 which is involved in snRNP biogenesis, and the Myb-related transcription factor p42POP.Citation29 However, previous studies showing that profilin is a nuclear protein have not addressed the detailed localization of profilin in relation to chromatin and active genes. The salivary glands of C. tentans offer unique advantages when analyzing the association of specific proteins, in this case profilin, with gene loci. The chromatin and interchromatin compartments in the polytene nuclei are easy to distinguish from each other, and the chromatin is confined to the large polytene chromosomes, which leaves the nucleoplasm free of chromatin. The chromosomes, visible using either light or electron microscopy, are composed of dense bands and interbands that represent different degrees of chromatin condensation. In spite of the fact that the general organization of the polytene nuclei differs from that of diploid nuclei, the basic processes of gene expression are the same in both types of cells (reviewed in refs. Citation32 and Citation33). We have used the advantages that polytene nuclei offer for in situ studies of gene expression to analyze the association of profilin with chromatin. For this purpose, we raised antibodies against the profilin of C. tentans.
Using immunofluorescence microscopy, we showed that a fraction of profilin is located in the nucleus of the salivary gland cells of C. tentans. Staining of D. melanogaster S2 cells also revealed a nuclear localization of profilin. From these results we conclude that profilin is a nuclear protein in insects as it is in vertebrates, and that the nuclear localization of profilin is characteristic of both polytene and diploid cells.
A more detailed analysis of the location of profilin in the nucleus of the salivary gland cells of C. tentans revealed that profilin is highly concentrated in the nucleoplasm and at the nuclear periphery. Immuno-EM experiments showed that profilin is located on both sides of the nuclear envelope. Moreover, a fraction of profilin is associated with the polytene chromosomes following a banded pattern, which indicates that profilin is not associated with all chromatin, but that it binds preferentially to certain chromatin domains or loci. We have performed experiments to characterize the type of loci that profilin associates with. By staining salivary glands with propidium iodide, we have shown that profilin does not co-localize with dense chromatin. Profilin associates instead with loci that are occupied by hnRNP and snRNP proteins. HnRNP and snRNP proteins bind co-transcriptionally to the nascent pre-mRNAs that are still bound to the gene. Therefore, the co-localization of profilin with pre-mRNA-binding factors strongly suggests that profilin associates with transcriptionally active genes. These experiments also show that profilin associates with genes that are transcribed by RNA polymerase II, although we cannot exclude the possibility that profilin also interacts with class III genes. The anti-profilin antibodies did not give any conspicuous staining of the nucleolus and we did not find any indication that profilin interacts with the rDNA genes.
We have performed experiments of transcription inhibition with actinomycin D and DRB, and we have shown that the association of profilin with the chromosomes requires ongoing transcription. These results agree well with previous studies in mammalian cells, in which treatment with actinomycin D led to similar reorganization of snRNPs and profilin.Citation23 However, digestion of the chromosomes with RNase A did not affect the chromosomal location of profilin in the polytene chromosomes of C. tentans, which indicates that the interaction of profilin with the gene loci is not mediated by RNA. Profilin must be associated with a chromosomal component other than RNA, either chromatin or the transcription machinery. Several studies have shown that actin interacts with the RNA polymerases,Citation4,Citation5,Citation11,Citation34 and actin is also a component of several chromatin remodelling complexes (reviewed in refs. Citation35 and Citation36). In principle, the association of profilin with the chromosomes could be explained by an interaction between profilin and actin molecules that are bound either to the RNA polymerase or to the chromatin factors located at active genes. However, the association of actin with the polytene chromosomes of C. tentans depends on RNA,Citation3 whereas profilin remains bound to chromosomes digested with RNase. This result suggests that the profilin located at sites of transcription is not bound to actin. Further studies will be necessary to characterize the molecular interactions that tether profilin to actively transcribed genes.
Several studies have supported the idea that nuclear profilin is involved in gene expression, but each of these studies ascribes profilin a different function. Skare et al. reported that an antibody against profilin has an inhibitory effect on pre-mRNA splicing in vitro and suggested a role for profilin in pre-mRNA processing. Lederer et al. showed that profilin binds to the Myb-related transcriptional repressor p42POP and regulates its activity, maybe by regulating its subcellular localization. Ye et al. proposed that an actomyosin motor is required for transcription by RNA polymerase I, which raises the possibility that profilin functions as a regulator of actin polymerization at transcription sites. Our immuno-localization studies are compatible with these suggested functions. Moreover, our immuno-EM experiments have revealed the existence of a fraction of profilin located at the nuclear envelope, which suggests a possible role for profilin in nucleo-cytoplasmic transport. Both actinCitation37 and NM1 Citation38 are involved in the nuclear export of different types of RNP complexes, and profilin enhances the interaction of actin with exportin 6, which in turn favors the nuclear export of actin.Citation24 It is unclear how these observations relate to each other and to profilin, but it is clear that actin and actin-binding proteins are implicated, directly or indirectly, in nucleo-cytoplasmic trafficking.
In summary, the work presented here and several previous reports link profilin with gene expression, but in quite divergent ways. It is clear that we are still far from understanding the roles of profilin in the cell nucleus. A key point will be to determine whether nuclear profilin has actin-independent functions or whether it acts primarily as an actin regulator. The associations of actin and profilin to the chromosomes differ in their RNA dependence, which suggests that the fraction of profilin that is bound to the chromosomes has a role that is independent of actin.
Materials and Methods
Culturing conditions
Chironomus tentans was cultured as described by Meyer et al.Citation39 Salivary glands were isolated from fourth instar larvae. C. tentans tissue culture cells were cultivated at 24°C as previously described in reference Citation40. Drosophila melanogaster S2 cells were cultured at 28°C in Schneider’s Drosophila medium (Invitrogen, 21720-024) supplemented with 10% heat-inactivated fetal calf serum, 50 μg/ml penicillin and 50 μg/ml streptomycin.
Antibodies
A synthetic peptide spanning amino acids 2 to15 (SWQ DYV DNQ LIA SQC) located in the N-terminal domain of Ct-profilin was conjugated to keyhole limpet hemocyanin and used to immunize two rabbits following standard procedures. Immunization was performed at AgriSera (Vännäs). The antibodies were affinity purified and eluted in two steps at pH 2.5 and pH 7.0. A mAb against the product of the chickadee gene of D. melanogaster was described by Verheyen and CooleyCitation26 and purchased from the Developmental Studies Hybridoma Bank (University of Iowa). The Y12 antibody is a mAb against the Sm antigen of snRNPs, and was originally produced and characterized by Lerner et al.Citation41 mAbs against hrp23 and hrp45 were produced and characterized by Sun et al.Citation42 and Alzhanova-Ericsson et al.Citation43 respectively.
SDS-PAGE and western blotting
For analysis of antibody specificity, whole cell extracts were prepared from C. tentans tissue culture cells or D. melanogaster S2 cells. The cells were boiled in 2x sample buffer containing 5% β-mercaptoethanol and separated by SDS-PAGE. After electrophoresis, the proteins in the gel were transferred to polyvinylidenefluoride (PVDF) membranes (Millipore, IPVH00010) in Tris-glycine buffer containing 0.02% SDS and 4 M urea using a semi-dry electrophoretic transfer cell (BioRad, 170-3940). The membranes were blocked with 10% non-fat dry milk in PBS, and probed with primary antibody at a final concentration of approximately 2 μg/ml, followed by a goat-anti-rabbit secondary antibody conjugated with alkaline phosphatase (DAKO, D0487). The antibodies were diluted in PBS containing 0.05% Tween-20 and 1% milk. The NBT/BCIP system was used for detection of the alkaline phosphatase activity.
Cytoplasmic and nuclear extracts from C. tentans tissue culture cells
C. tentans tissue culture cells were homogenized in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 2 mM NaH2PO4, pH 7.2) containing 0.2% Nonident P40 (NP-40) and protease inhibitors (Complete cocktail, Roche, 04693159001) using a glass homogenizer (tight pestle). The homogenate was centrifuged at 2,000 g for 10 min at 4°C. The supernatant was the cytoplasmic fraction. The pellet containing the nuclei was resuspended in PBS containing protease inhibitors and sonicated three times for 4–5 sec each time, and centrifuged as above. The resulting supernatant was the soluble nuclear extract and the pellet was the nuclear insoluble fraction. The proteins in the fractions were precipitated by the addition of 6 volumes of cold acetone and stored at -20°C. Prior to SDS-PAGE, the samples were centrifuged at 4°C at 11,000 g and the pellet air-dried and redissolved in 2× sample buffer.
Immunofluorescence of whole salivary glands
C. tentans salivary glands were isolated and fixed in 4% formaldehyde in PBS for 10 min on ice, and incubated 20 min on ice in PBS containing 4% formaldehyde and 5% Triton X-100. The glands were then further permeabilized in 10% Triton X-100 in PBS in a humid chamber for 10 min at room temperature, and washed three times 5 min each in PBS. Blocking was done in 5% new-born calf serum (NBCS) and 3% bovine serum albumin (BSA) in PBS for 30 min. Incubation with primary antibody was done for 2 h in a humid chamber. The primary antibodies were diluted to a final concentration of approximately 1–5 μg/ml in PBS containing 0.5% NBCS and 0.3% BSA. After primary antibody incubation, the glands were washed three times for 5 min each in PBS, followed by incubation with secondary antibodies, either FITC or Texas Red conjugated, for 1 h. After final washes, the samples were mounted in Vectashield mounting medium (Vector, H-1000) and examined in an LSM 510 laser confocal microscope (Carl Zeiss). In the negative controls, the primary antibody was replaced by an unrelated rabbit antibody against mouse immunoglobulins. Co-localization was analyzed using the “Profile” function of the LSM 510 software by drawing a test line along the region of interest and measuring the relative fluorescence intensity along the line in arbitrary units.
Immunofluorescence of isolated polytene chromosomes
C. tentans polytene chromosomes were isolated from the salivary glands essentially as described by Björkroth et al.Citation44 with some modifications. The salivary glands were permeabilized with 2% NP40 in TKM buffer (10 mM triethanolamine-HCl at pH 7.0, 100 mM KCl and 1 mM MgCl2) and the chromosomes were isolated by repeatedly pipetting through a siliconized glass micropipette. Isolated chromosomes were transferred to 8-well slides and fixed with 2% formaldehyde in TKM for 30 min. After fixation, the chromosomes were washed in TKM three times for 5 min each, and blocked in 3% BSA in TKM for 30 min. After blocking, the chromosomes were incubated with primary antibodies diluted to a final concentration of approximately 5–10 μg/ml in 0.5% BSA in TKM at 4°C overnight. The chromosomes were washed with 0.01% Tween-20 in TKM three times for 10 min each, followed by incubation with secondary antibodies either FITC or Texas Red conjugated. The chromosomes were mounted with Vectashield mounting medium (Vector, H-1000) and examined in an LSM 510 microscope.
RNase A digestion
For experiments of RNase A digestion, the glands were washed with TBS after dissection, then incubated in glycerol buffer (20 mM TRIS-HCl at pH 7.2, 5 mM MgCl2, 0.5 mM PMSF, 0.5 mM EGTA, 25% glycerol) for 3 min, in glycerol buffer containing 0.05% Triton X-100 for a further 3 min, and then washed three times 5 min in transcription buffer (5 mM TRIS-HCl at pH 7.4, 100 mM KCl, 0.5 mM EGTA, 25% glycerol). The glands were incubated in RNase A diluted to 100 μg/ml in TBS for 20 min at 37°C. Control samples were incubated in TBS without enzyme under the same conditions. The glands were then processed for immunofluorescence as described above.
Isolated polytene chromosomes were incubated with 100 μg/ml RNase A for 30 min and washed with TKM before fixation and immunofluorescence as above.
Propidium iodide staining
The glands were processed for immunofluorescence as described above. After incubation with the secondary antibody, the glands were washed three times 5 min each in PBS and incubated in 500 mM propidium iodide in PBS for 10 min at room temperature. Then the glands were washed two times 10 min each in PBS and two times 10 min each in PBS supplemented with 0.001% Tween-20. The glands were finally mounted in Vectashield mounting medium (Vector, H-1000) and examined in an LSM 510 microscope.
Actinomycin D and DRB treatments
Salivary glands were incubated in 10 μg/ml actinomycin D in ZO medium for 1 h at room temperature before fixation and immunofluorescent staining as above. Negative controls (-Act) were incubated in ZO medium without the drug.
A 10 mg/ml stock solution of 5,6-Dichlorobenzimidazole 1-μ-D-ribofuranoside (DRB, Sigma-Aldrich, D1916) in DMSO was prepared. Fourth-instar larvae were kept in culture water containing 0.4 μM DRB for 45 min. The salivary glands were then dissected, fixed and immunostained as described above. Negative controls (-DRB) were incubated in parallel in culture water with DMSO alone.
Immunolabeling of S2 cells
S2 cells were allowed to adhere to glass slides coated with poly-l-lysine in a humid chamber at 28°C for 30 min. After a quick wash in PBS, the cells were fixed in 3.7% PBS for 10 min at room temperature, washed three times 5 min each in PBS, and permeabilized in 0.05% Triton-X100 in PBS for 13 min at room temperature. The cells were blocked in 5% milk and 5% BSA in PBS for 1 h. The primary antibody was diluted in 0.5% milk and 0.3% BSA to a concentration of 1–5 μg/ml. Antibody incubations were performed in a humid chamber.
Immuno-electron microscopy
Salivary glands were dissected from fourth instar larvae, fixed for 20–25 min in 4% paraformaldehyde and 0.1% glutaraldehyde, cryoprotected with 2.3 M sucrose, frozen by immersion in liquid nitrogen and cryosectioned. Thin cryosections were picked up on drops of 2.3 M sucrose and mounted onto nickel grids coated with formvar and carbon. The grids were floated onto drops of PBS containing 0.1 M glycine and 10% fetal calf serum before incubation with the antibody solutions. The primary antibody was affinity-purified anti-profilin diluted to 17 μg/ml in PBS containing 0.1 M glycine and 10% fetal calf serum. The secondary antibody, conjugated to 6 nm gold particles (Jackson ImmunoResearch Laboratories, 111-195-144), was diluted 1:50 in the same solution. After immunolabelling, the sections were stained with 2% aqueous uranyl acetate for 3–5 min and embedded in polyvinyl alcohol (9–10 kDa, Aldrich, 360627). The specimens were examined and photographed in a FEI Tecnai G2 electron microscope at 80 kV.
Acknowledgments
We thank R. Karlsson (Wenner-Gren Institute, Stockholm University) for fruitful discussions about profilin, Ann Kristin Östlund Farrants for critical reading of the manuscript and G.F. for English editing. This research has been financed by grants from the Swedish Research Council (Vetenskapsrådet) to N.V. E.S. was financed by the Research School for Genomics and Bioinformatics at the Stockholm University.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kardos R, Pozsonyi K, Nevalainen E, Lappalainen P, Nyitrai M, Hild G. The effects of ADF/cofilin and profilin on the conformation of the ATP-binding cleft of monomeric actin. Biophys J 2009; 96:2335 - 43; http://dx.doi.org/10.1016/j.bpj.2008.12.3906; PMID: 19289059
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol 2010; 2:a000620; http://dx.doi.org/10.1101/cshperspect.a000620; PMID: 20452941
- Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol 2001; 153:229 - 36; http://dx.doi.org/10.1083/jcb.153.1.229; PMID: 11285288
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol 2004; 6:1094 - 101; http://dx.doi.org/10.1038/ncb1182; PMID: 15502823
- Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 2004; 6:1165 - 72; http://dx.doi.org/10.1038/ncb1190; PMID: 15558034
- Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol 2008; 28:6342 - 57; http://dx.doi.org/10.1128/MCB.00766-08; PMID: 18710935
- Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, et al. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci U S A 2003; 100:6475 - 80; http://dx.doi.org/10.1073/pnas.1131933100; PMID: 12743363
- Percipalle P, Jonsson A, Nashchekin D, Karlsson C, Bergman T, Guialis A, et al. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res 2002; 30:1725 - 34; http://dx.doi.org/10.1093/nar/30.8.1725; PMID: 11937625
- Raju CS, Göritz C, Nord Y, Hermanson O, López-Iglesias C, Visa N, et al. In cultured oligodendrocytes the A/B-type hnRNP CBF-A accompanies MBP mRNA bound to mRNA trafficking sequences. Mol Biol Cell 2008; 19:3008 - 19; http://dx.doi.org/10.1091/mbc.E07-10-1083; PMID: 18480411
- Sjölinder M, Björk P, Söderberg E, Sabri N, Farrants AK, Visa N. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev 2005; 19:1871 - 84; http://dx.doi.org/10.1101/gad.339405; PMID: 16103215
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol 2005; 12:238 - 44; http://dx.doi.org/10.1038/nsmb904; PMID: 15711563
- Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, et al. A myosin I isoform in the nucleus. Science 2000; 290:337 - 41; http://dx.doi.org/10.1126/science.290.5490.337; PMID: 11030652
- Fomproix N, Percipalle P. An actin-myosin complex on actively transcribing genes. Exp Cell Res 2004; 294:140 - 8; http://dx.doi.org/10.1016/j.yexcr.2003.10.028; PMID: 14980509
- Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev 2008; 22:322 - 30; http://dx.doi.org/10.1101/gad.455908; PMID: 18230700
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007; 179:1095 - 103; http://dx.doi.org/10.1083/jcb.200710058; PMID: 18070915
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol 2006; 16:825 - 31; http://dx.doi.org/10.1016/j.cub.2006.03.059; PMID: 16631592
- Dingová H, Fukalová J, Maninová M, Philimonenko VV, Hozák P. Ultrastructural localization of actin and actin-binding proteins in the nucleus. Histochem Cell Biol 2009; 131:425 - 34; http://dx.doi.org/10.1007/s00418-008-0539-z; PMID: 19039601
- Obrdlik A, Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus 2011; 2:72 - 9; http://dx.doi.org/10.4161/nucl.2.1.14508; PMID: 21647301
- Wittenmayer N, Rothkegel M, Jockusch BM, Schlüter K. Functional characterization of green fluorescent protein-profilin fusion proteins. Eur J Biochem 2000; 267:5247 - 56; http://dx.doi.org/10.1046/j.1432-1327.2000.01600.x; PMID: 10931210
- Dong J, Radau B, Otto A, Müller E, Lindschau C, Westermann P. Profilin I attached to the Golgi is required for the formation of constitutive transport vesicles at the trans-Golgi network. Biochim Biophys Acta 2000; 1497:253 - 60; http://dx.doi.org/10.1016/S0167-4889(00)00056-2; PMID: 10903430
- Schlüter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta 1997; 1359:97 - 109; http://dx.doi.org/10.1016/S0167-4889(97)00100-6; PMID: 9409807
- Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem 1999; 274:37908 - 14; http://dx.doi.org/10.1074/jbc.274.53.37908; PMID: 10608857
- Skare P, Kreivi JP, Bergström Å, Karlsson R. Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp Cell Res 2003; 286:12 - 21; http://dx.doi.org/10.1016/S0014-4827(03)00102-2; PMID: 12729790
- Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 2003; 22:5928 - 40; http://dx.doi.org/10.1093/emboj/cdg565; PMID: 14592989
- Arvestad L, Visa N, Lundeberg J, Wieslander L, Savolainen P. Expressed sequence tags from the midgut and an epithelial cell line of Chironomus tentans: annotation, bioinformatic classification of unknown transcripts and analysis of expression levels. Insect Mol Biol 2005; 14:689 - 95; http://dx.doi.org/10.1111/j.1365-2583.2005.00600.x; PMID: 16313569
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 1994; 120:717 - 28; PMID: 7600952
- Wetterberg I, Baurén G, Wieslander L. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 1996; 2:641 - 51; PMID: 8756407
- Miralles F, Öfverstedt LG, Sabri N, Aissouni Y, Hellman U, Skoglund U, et al. Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J Cell Biol 2000; 148:271 - 82; http://dx.doi.org/10.1083/jcb.148.2.271; PMID: 10648560
- Lederer M, Jockusch BM, Rothkegel M. Profilin regulates the activity of p42POP, a novel Myb-related transcription factor. J Cell Sci 2005; 118:331 - 41; http://dx.doi.org/10.1242/jcs.01618; PMID: 15615774
- Birbach A, Verkuyl JM, Matus A. Reversible, activity-dependent targeting of profilin to neuronal nuclei. Exp Cell Res 2006; 312:2279 - 87; http://dx.doi.org/10.1016/j.yexcr.2006.03.026; PMID: 16716297
- Nie Z, Xu J, Chen J, Lv Z, Wang D, Sheng Q, et al. Expression analysis and characteristics of profilin gene from silkworm, Bombyx mori.. Appl Biochem Biotechnol 2009; 158:59 - 71; http://dx.doi.org/10.1007/s12010-008-8302-4; PMID: 18633732
- Wieslander L, Baurén G, Bernholm K, Jiang WQ, Wetterberg I. Processing of pre-mRNA in polytene nuclei of Chironomus tentans salivary gland cells. Exp Cell Res 1996; 229:240 - 6; http://dx.doi.org/10.1006/excr.1996.0366; PMID: 8986604
- Kiesler E, Visa N. Intranuclear pre-mRNA trafficking in an insect model system. Prog Mol Subcell Biol 2004; 35:99 - 118; http://dx.doi.org/10.1007/978-3-540-74266-1_5; PMID: 15113081
- Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev 2004; 18:3010 - 5; http://dx.doi.org/10.1101/gad.1250804; PMID: 15574586
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem 2002; 71:755 - 81; http://dx.doi.org/10.1146/annurev.biochem.71.110601.135507; PMID: 12045110
- Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol 2004; 5:410 - 5; http://dx.doi.org/10.1038/nrm1370; PMID: 15122354
- Hofmann W, Reichart B, Ewald A, Müller E, Schmitt I, Stauber RH, et al. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J Cell Biol 2001; 152:895 - 910; http://dx.doi.org/10.1083/jcb.152.5.895; PMID: 11238447
- Obrdlik A, Louvet E, Kukalev A, Naschekin D, Kiseleva E, Fahrenkrog B, et al. Nuclear myosin 1 is in complex with mature rRNA transcripts and associates with the nuclear pore basket. FASEB J 2010; 24:146 - 57; http://dx.doi.org/10.1096/fj.09-135863; PMID: 19729515
- Meyer B, Mähr R, Eppenberger HM, Lezzi M. The activity of Balbiani rings 1 and 2 in salivary glands of Chironomus tentans larvae under different modes of development and after pilocarpine treatment. Dev Biol 1983; 98:265 - 77; http://dx.doi.org/10.1016/0012-1606(83)90357-3; PMID: 6192024
- Wyss C. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp Cell Res 1982; 139:309 - 19; http://dx.doi.org/10.1016/0014-4827(82)90255-5; PMID: 7044809
- Lerner EA, Lerner MR, Janeway CA Jr., Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A 1981; 78:2737 - 41; http://dx.doi.org/10.1073/pnas.78.5.2737; PMID: 6789322
- Sun X, Alzhanova-Ericsson AT, Visa N, Aissouni Y, Zhao J, Daneholt B. The hrp23 protein in the balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J Cell Biol 1998; 142:1181 - 93; http://dx.doi.org/10.1083/jcb.142.5.1181; PMID: 9732280
- Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev 1996; 10:2881 - 93; http://dx.doi.org/10.1101/gad.10.22.2881; PMID: 8918889
- Björkroth B, Ericsson C, Lamb MM, Daneholt B. Structure of the chromatin axis during transcription. Chromosoma 1988; 96:333 - 40; http://dx.doi.org/10.1007/BF00330699