Abstract
The degree to which nucleosome positioning regulates transcription is an ongoing debate. To address this question, we recently followed dynamic changes in nucleosome occupancy, transcription factor binding and gene expression in yeast cells responding to oxidative stress. Integrating across these dynamic processes revealed new insights into the functions of nucleosome reorganization. Here, we used our data to address the extent to which upstream promoter architecture is a static feature inherent to specific genes vs. a dynamic platform that changes across conditions. Our results argue that, while some aspects of promoter architecture are fixed across environments, the level to which promoters are “open” or “covered” by nucleosomes depends on the conditions investigated.
Introduction
The role of chromatin structure in mediating genome biology is of great interest, particularly its role in regulating transcription. Genomic studies have characterized nucleosome positioning in a variety of organisms, collectively showing that most genes harbor nucleosome depleted regions (NDRs) immediately upstream of their transcription start site (TSS) and within which many cis-regulatory elements reside.Citation1,Citation2 However, a major open question is to what extent nucleosome positioning acts as a “gate keeper” to regulate access to the underlying DNA and thus control gene transcription.
Several recent studies have attempted to address this issue by investigating genome-wide changes in nucleosome occupancy in cells responding to a changing environment.Citation3-Citation7 Collectively, these studies showed that while the majority of nucleosomes do not change in position or occupancy after environmental change, those that do are correlated with downstream gene-expression changes: transcriptional induction is correlated with decreased nucleosome occupancy in the gene’s immediate upstream region, while gene repression is associated with increased occupancy of upstream nucleosomes.Citation4-Citation6 However, whether the nucleosome rearrangements are a regulatory prerequisite for transcription factor access remains an open question.
An important missing component from many studies is that of dynamics. To address this, we recently characterized dynamic changes in nucleosome occupancy in yeast responding to an acute dose of hydrogen peroxide (H2O2).Citation8 We purified and sequenced nucleosome-bound regions before and at five time points after H2O2 treatment, up to 60 min after treatment. Consistent with other reports, most nucleosome-bound regions in the cell population did not change appreciably in their association with nucleosomes (referred to as “nucleosome occupancy”). However, 25% of nucleosome-bound regions showed a statistically significant change in nucleosome binding at one or more time points. Decreased nucleosome occupancy generally occurred upstream of induced genes, whereas increased nucleosome occupancy was typically observed upstream of repressed genes. Most genes harbored only one upstream nucleosome with altered occupancy. These changes in nucleosome occupancy occurred with a wide variety of temporal patterns, with some regions changing occupancy almost immediately after stress treatment and others responding late in the time course (≥ 40 min after H2O2 addition).
To better understand the function of these changes, we also followed dynamic alterations in gene expression and binding of the stress-activated transcription factor Msn2p during the same response. Integrating the dynamics of these physiological processes allowed us to test several models regarding the role of chromatin change in transcription. Somewhat surprisingly, there was little correlation between the timing of upstream nucleosome reorganization and the timing of downstream gene expression (comparing either the peak changes or earliest changes in nucleosome occupancy and transcript abundance), at least on a global scale. Instead, dynamic differences in nucleosome occupancy were associated with distinct nucleosome positions. For example, nucleosomes lost transiently after H2O2 treatment were enriched at sites near the TSS (the so-called “+1” nucleosome) of induced genes. Their loss occurred after upstream Msn2p binding, but before the peak increase in transcript abundance of neighboring genes. Thus, transient loss of +1 nucleosomes appeared coincident with the transient burst of transcript induction from those promoters, suggesting that the loss was coupled with transcription initiation.
In contrast, nucleosomes lost late in the time course were strongly associated with Msn2p binding and were often positioned directly over Msn2p binding sites. Surprisingly, these nucleosomes were lost after the transcription factor was bound to those promoters and after the peak expression changes of downstream genes. These data strongly suggested that the late nucleosome loss was driven by transcription factor binding, rather than regulating transcription factor access. Indeed, we showed that Msn2p is required for subsequent nucleosome loss, since a strain lacking MSN2 showed defective H2O2-dependent nucleosome depletion at those sites. Msn2p may co-occupy its short binding sequence with a nucleosome, as found for other factors such as the mammalian glucocorticoid receptor and the yeast Pho4p transcription factor,Citation9-Citation12 and then recruit chromatin remodeling enzymes to evict or reposition nucleosomes.Citation13 Regardless of the precise mechanism, our results strongly suggest that, although many Msn2p binding sites are covered by nucleosomes before stress, those nucleosomes do not restrict Msn2p access to the underlying DNA. There was enrichment for genes with late upstream nucleosome loss and earlier nucleosome depletion coupled to transcription, but the number of genes with multiple nucleosomes was relatively small.
Open Vs. Closed Promoter Architecture: Static States or Dynamic Platforms?
Studies of both single genes and genomic profiles have reported two basic promoter architectures (and many variations spanning the two).Citation14-Citation17 Tirosh and Barkai defined yeast genes whose promoters were relatively ‘depleted’ of (DPN) or ‘occupied’ by (OPN) upstream nucleosomes, by focusing on the nucleosome occupancy 100 bp upstream of the TSS relative to the distal upstream region.Citation14 A similar distinction was made by Field et al., who defined classes based on the presence or absence of TATA regulatory elements, nucleosome coverage in the “typical” NDR position and whether or not transcription factor binding sites were obscured by nucleosomes.Citation15 Together, these studies and other single-gene analyses suggested that relatively “open” promoters (e.g., of DPN genes) are associated with constitutive transcription from TATA-less genes, while “closed” or “covered” promoters (e.g., of OPN genes) are linked to TATA-regulated genes and may drive high transcriptional noise, expression plasticity across environments and rates of expression evolution.Citation14-Citation17
These classes of promoter architecture have been distinguished based on actively dividing cells growing continuously in stress-free media, and therefore a major question is to what extent these architectures are inherent to specific genes or variable across conditions. Our data afforded the opportunity to revisit this question. We analyzed our data in terms of DPN and OPN genes classifications.Citation14 As expected, DPN genes had a clear NDR ~100 bp upstream of the TSS (). This group was enriched for H2O2-repressed genes (p = 5x10−6, hypergeometric distribution) but showed no significant enrichment beyond chance for genes with changes in upstream nucleosome occupancy.
Figure 1. Promoter architecture is influenced by NDR placement. Nucleosome occupancy 400 bp upstream to the TSS, in unstressed cells (left) or 60 min after treatment with 0.4 mM H2O2 (right) from.Citation8 Upstream nucleosome occupancy is shown for 493 DPN genes (A) or 543 OPN genes (B), as defined by Tirosh and Barkai;Citation14 genes were organized by hierarchical clustering of the upstream regions. Occupancy is shown on the same scale for both gene groups, according to the key.
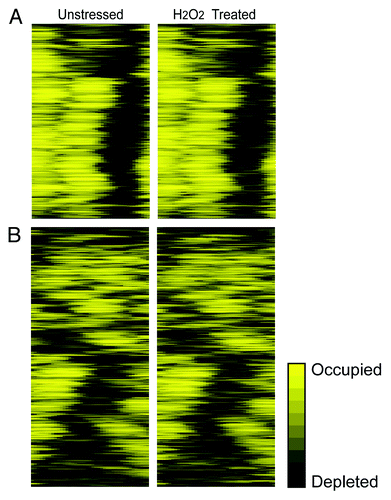
The OPN class was also enriched for genes repressed by H2O2 but, unlike DPN genes, showed an enrichment of genes with upstream nucleosome loss (p = 1 × 10−9). Somewhat surprisingly, however, we observed that most of the previously defined OPN genes in fact had very robust NDRs in our data set (). We scored the most likely NDR as the 100 bp upstream window with the minimum nucleosome occupancy over the 400 bp upstream region; the NDR boundaries were expanded until the nucleosome signal increased. The NDRs of OPN genes were very similar to the NDRs of DPN genes in terms of average nucleosome signal, NDR length and fraction of the gene’s transcription-factor binding sites contained withinCitation8—however, the NDRs of OPN genes were significantly farther and more variably distributed from the TSS (-253 ± 105 bp vs. -140 ± 46 bp for OPN and DPN genes, respectively, p = 1 × 10−45, T-test). This is partly explained by the preponderance of TATA-containing genes,Citation18 whose NDRs are significantly farther from the TSS compared with TATA-less genes (-211 ± 110 bp vs. -197 ± 96 bp, respectively, p = 3 × 10−13, T-test). The NDRs of OPN genes that we observed in our data set were also found in other data sets from unstressed cellsCitation4,Citation19 (data not shown). Thus, while the architectures of the previously defined DPN and OPN promoters are indeed different from one another, the distinction appears to be where the NDR is placed with regard to the TSS, rather than to what extent the NDR is occupied or depleted.
To further investigate the effects of promoter accessibility, we classified genes based on the level of nucleosome occupancy in the likely NDR, as described above. We first focused on 1,069 genes with zero sequencing counts in the likely NDR region before stress, regardless of where the NDR was positioned (). Over 50% of predicted transcription factor-binding sites in each upstream regionCitation8 fell within the NDR, compared with only 30% for genes outside this class (p = 1 × 10−70, T-test). The genes were heavily enriched for highly transcribed yeast genes, including those encoding ribosomal proteins, ribosome biogenesis and splicing proteins and transcription and translation factors. Although these processes are often thought of as essential ‘housekeeping’ functions, the genes are by no means constitutively expressed—most are strongly repressed upon diverse stress treatments.Citation20 Indeed, H2O2-repressed genes made up over half of the 1,069 genes with the most depleted NDRs before stress (p = 2 × 10−16). Not surprisingly, the group also showed a significant enrichment for genes with upstream nucleosome gain after stress treatment (p = 1 × 10−4).
Figure 2. Two classes of promoters based on NDR accessibility. Data are shown as described in . (A) 1,069 genes with no nucleosome signal in unstressed cells, before (left) and 60 min after treatment with 0.4 mM H2O2 (right) from.Citation8 (B) 288 genes whose promoters rank in the top 5% based on nucleosome signal in the NDR. Genes in both classes were organized by position of the NDR. The contrast was increased for genes in (B) to distinguish the NDR.
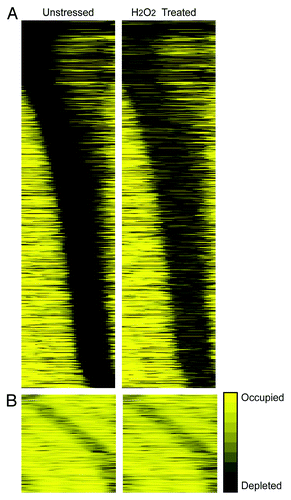
We repeated the promoter classification based on NDR nucleosome occupancy in cells acclimated for 60 min after H2O2 treatment. Although many of the 1,069 genes remained among those with the most depleted NDRs, over a third no longer ranked in this extreme in cells acclimated to H2O2 stress. Thus, for a substantial subset of genes with the most accessible NDRs before stress, the degree to which their promoters are “open” or “covered” depends on the conditions being investigated.
We also investigated the 288 genes (5th percentile) whose promoters were most covered by nucleosomes before stress, based on the signal in the NDR (). This group of genes was enriched for induced genes with uncharacterized functions, those regulated by TATA elements (p = 1 × 10−4) and genes with higher transcriptional noise (p = 2 × 10−7, T-test).Citation21 Although there was a clear enrichment for genes with upstream nucleosome depletion, most promoters (65%, p = 2 × 10−7, hypergeometric distribution) showed no significant change in upstream nucleosome occupancy after stress and remained ranked among the 5% of genes with the least accessible promoters. Transcriptional induction of these genes could occur if very rapid nucleosome exchangeCitation22-Citation24 or partial unwindingCitation25,Citation26 facilitates transient transcription factor binding, without apparent and constitutive nucleosome loss. While this group of genes was not strongly enriched for genes with rapidly exchanging (so-called “hot”) upstream nucleosomes (as measured under standard conditionsCitation24) it is possible that nucleosome exchange increases upon stress treatments to allow transcriptional induction without constitutive nucleosome loss.
Together, these results suggest important details about promoter architecture and the degree to which it is a defining feature of the downstream gene. With the exception of subtle nucleosome sliding in specific promoters,Citation27 the position of the NDR is largely fixed regardless of the conditions, consistent with the contribution of underlying sequence.Citation15,Citation28-Citation30 But the extent to which the NDR is covered by nucleosomes is, at least partly, a feature of the environment. Over a third of the most accessible NDRs before stress attained significant nucleosome coverage in cells acclimated to H2O2 stress (). It is interesting to note the gene classes represented by this group (including ribosomal proteins and transcription/translation factors) are reported to show low transcriptional noise and slower rates of expression evolution.Citation16,Citation21,Citation31,Citation32 Once again, we argue that some of these trends are specific to the conditions investigated, since other results from our lab demonstrate that environmental stress reveals cryptic variationCitation33 and transcriptional noiseCitation34 in these gene classes.
In contrast to many genes with open promoters, we find that genes with the most covered promoters retain this distinction in multiple environments, even when apparently induced. It will be interesting to uncover the extent to which this is due to rapid nucleosome exchange, co-occupancy with the relevant transcription factors, or post-transcriptional changes in gene expression. Regardless, it is clear that a full picture of genome biology will only be revealed as more studies incorporate a dynamic view of the response to changing environments.
Acknowledgments
This work was funded by NIH grants NIGMS R01GM083989 to A.P.G. and NHGRI P50HG004952.
References
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 2009; 10:161 - 72; http://dx.doi.org/10.1038/nrg2522; PMID: 19204718
- Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter?. Dev Biol 2010; 339:258 - 66; http://dx.doi.org/10.1016/j.ydbio.2009.06.012; PMID: 19527704
- Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet 2006; 2:e158; http://dx.doi.org/10.1371/journal.pgen.0020158; PMID: 17002501
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 2008; 6:e65; http://dx.doi.org/10.1371/journal.pbio.0060065; PMID: 18351804
- Zawadzki KA, Morozov AV, Broach JR. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell 2009; 20:3503 - 13; http://dx.doi.org/10.1091/mbc.E09-02-0111; PMID: 19494041
- Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res 2011; 21:875 - 84; http://dx.doi.org/10.1101/gr.117465.110; PMID: 21515815
- Deal RB, Henikoff S. Capturing the dynamic epigenome. Genome Biol 2010; 11:218; http://dx.doi.org/10.1186/gb-2010-11-10-218; PMID: 20959022
- Huebert DJ, Kuan PF, Keleş S, Gasch AP. Dynamic changes in nucleosome occupancy are not predictive of gene expression dynamics but are linked to transcription and chromatin regulators. Mol Cell Biol 2012; 32:1645 - 53; http://dx.doi.org/10.1128/MCB.06170-11; PMID: 22354995
- Perlmann T, Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J 1988; 7:3073 - 9; PMID: 2846275
- Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol 1991; 11:688 - 98; PMID: 1846670
- Nagaich AK, Walker DA, Wolford R, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 2004; 14:163 - 74; http://dx.doi.org/10.1016/S1097-2765(04)00178-9; PMID: 15099516
- Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, et al. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 2009; 284:23461 - 71; http://dx.doi.org/10.1074/jbc.M109.019562; PMID: 19574230
- Erkina TY, Tschetter PA, Erkine AM. Different requirements of the SWI/SNF complex for robust nucleosome displacement at promoters of heat shock factor and Msn2- and Msn4-regulated heat shock genes. Mol Cell Biol 2008; 28:1207 - 17; http://dx.doi.org/10.1128/MCB.01069-07; PMID: 18070923
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res 2008; 18:1084 - 91; http://dx.doi.org/10.1101/gr.076059.108; PMID: 18448704
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 2008; 4:e1000216; http://dx.doi.org/10.1371/journal.pcbi.1000216; PMID: 18989395
- Tirosh I, Barkai N, Verstrepen KJ. Promoter architecture and the evolvability of gene expression. J Biol 2009; 8:95; http://dx.doi.org/10.1186/jbiol204; PMID: 20017897
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature 2009; 461:193 - 8; http://dx.doi.org/10.1038/nature08450; PMID: 19741699
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004; 116:699 - 709; http://dx.doi.org/10.1016/S0092-8674(04)00205-3; PMID: 15006352
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 2007; 39:1235 - 44; http://dx.doi.org/10.1038/ng2117; PMID: 17873876
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000; 11:4241 - 57; PMID: 11102521
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 2006; 441:840 - 6; http://dx.doi.org/10.1038/nature04785; PMID: 16699522
- Ahmad K, Henikoff S. Epigenetic consequences of nucleosome dynamics. Cell 2002; 111:281 - 4; http://dx.doi.org/10.1016/S0092-8674(02)01081-4; PMID: 12419239
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science 2007; 315:1408 - 11; http://dx.doi.org/10.1126/science.1134004; PMID: 17347439
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science 2007; 315:1405 - 8; http://dx.doi.org/10.1126/science.1134053; PMID: 17347438
- Anderson JD, Thåström A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol Cell Biol 2002; 22:7147 - 57; http://dx.doi.org/10.1128/MCB.22.20.7147-7157.2002; PMID: 12242292
- Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol 2008; 379:772 - 86; http://dx.doi.org/10.1016/j.jmb.2008.04.025; PMID: 18485363
- Becker PB. The chromatin accessibility complex: chromatin dynamics through nucleosome sliding. Cold Spring Harb Symp Quant Biol 2004; 69:281 - 7; http://dx.doi.org/10.1101/sqb.2004.69.281; PMID: 16117660
- Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys 2001; 34:269 - 324; http://dx.doi.org/10.1017/S0033583501003699; PMID: 11838235
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, et al. A genomic code for nucleosome positioning. Nature 2006; 442:772 - 8; http://dx.doi.org/10.1038/nature04979; PMID: 16862119
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009; 458:362 - 6; http://dx.doi.org/10.1038/nature07667; PMID: 19092803
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet 2005; 37:544 - 8; http://dx.doi.org/10.1038/ng1554; PMID: 15852004
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science 2007; 317:118 - 21; http://dx.doi.org/10.1126/science.1140247; PMID: 17525304
- Eng KH, Kvitek DJ, Keles S, Gasch AP. Transient genotype-by-environment interactions following environmental shock provide a source of expression variation for essential genes. Genetics 2010; 184:587 - 93; http://dx.doi.org/10.1534/genetics.109.107268; PMID: 19966067
- Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol 2011; 7:514; http://dx.doi.org/10.1038/msb.2011.48; PMID: 21772262