Abstract
Posttranslational modification of proteins with SUMO and the RanGTP/GDP cycle are two essential cellular mechanisms contributing directly or indirectly to almost every cellular event. The SUMO E3 ligase RanBP2 (Nup358) and the Ran GTPase activating protein (RanGAP1) are known to form a stable complex throughout the cell cycle suggesting a link between sumoylation of proteins and RanGTP hydrolysis. In a recent study we demonstrated that the stable complex of RanBP2, sumoylated RanGAP1 and Ubc9 (and not RanBP2 by itself) represents the physiologically relevant form of the SUMO ligase. Characterization of the interactions reveals an intricate proximity of two catalytic activities, sumoylation and RanGTP hydrolysis. In this ExtraView we summarize our results and discuss some ideas about a potential coupling of both processes.
The regulation and fine-tuning of all cellular processes relies on a number of reversible posttranslational modifications of proteins, among them the attachment of proteins of the ubiquitin-like modifier family. The most intensely studied member in this family besides ubiquitin itself is SUMO, and in almost all eukaryotes the SUMO pathway is essential. All eukaryotic organisms express at least one SUMO, many organisms express several SUMO paralogs with distinct characteristics in respect to target specificity, abundance, the ability to form SUMO chains and involvement in stress responses.Citation1,Citation2
Following a maturation step to expose a C-terminal Gly-Gly motif, SUMO is conjugated to targets via an enzymatic cascade consisting of a unique E1 enzyme that activates SUMO in an ATP-dependent manner and the single E2 conjugating enzyme Ubc9, which transfers SUMO to a lysine side chain in the target protein with the help of one of several E3 ligases. Like other regulatory modifications, sumoylation is reversible due to specific isopeptidases that remove SUMO from the targets.Citation3
While the functional outcomes of sumoylation are manifold, the most common consequence at the molecular level is a change in protein interactions by either blocking or creating an (additional) binding surface. SUMO interaction motifs (SIM) that allow for noncovalent interaction with SUMO play a major role in SUMO-mediated assembly of protein complexes or subcellular structures like PML bodies,Citation4 but have also been shown to contribute to the efficiency and specificity of SUMO modification.Citation5,Citation6
The recent progress in experimental techniques and tools for the detection of sumoylation in cells provides us with a rapidly growing list of SUMO targets. However, our knowledge about enzymes mediating sumoylation, especially the E3 ligases, is still limited.
Among the few proteins that have unequivocally been demonstrated to act as SUMO E3 ligase in vitro and in vivo is the nucleoporin RanBP2 (Nup358).Citation7 This 358 kDa protein constitutes a major component of the cytoplasmic filaments of nuclear pore complexes in interphase;Citation8,Citation9 after nuclear envelope breakdown it is largely soluble but a pool of it is also found at the kinetochore and mitotic spindleCitation10 (). RanBP2 is an essential protein playing a role in various cellular processes, but only some of its functions have been linked to its SUMO ligase activity.Citation11-Citation13 The region in RanBP2 responsible for E3 ligase activity is contained within a C-terminal region harboring two 50 amino acid internal repeats (IR1 and IR2) separated by a 20 amino acid linker. Both repeats can interact with the E2 conjugating enzyme Ubc9, although with different affinities. Due to its higher activity as an isolated protein fragment in vitro, IR1 was assumed to be the RanBP2 region providing E3 ligase activity also in vivo.
Figure 1. Cellular localization and schematic domain structure of RanBP2 with leucine-rich N-terminus (LR), four RanGTP binding domains (RB), an 8-fold zinc finger (ZF), a kinesin binding domain (KBD), a cyclophilin domain (CY), several FG repeats (dashes) and the SUMO E3 ligase region (E3) that is shown in more detail in the lower part. The complex with sumoylated RanGAP1 and Ubc9 is an active SUMO E3 ligase. Two internal repeats (IR) accommodate the binding site for stable interaction with sumoylated RanGAP1 and the structural Ubc9 (IR1) as well as the site for transient interaction with the catalytic Ubc9 (IR2). IR2 is only active in the context of the complex, in free RanBP2 its E3 ligase activity is negligible. The “core” region of the complex is flanked on both sides by RanGTP binding domains (RB) and FG repeats (dashes) that serve as interaction sites for RanGTP and transport receptors.
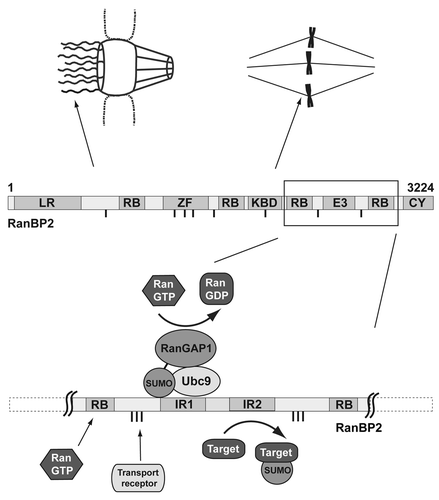
However, RanBP2 is not an isolated protein, but rather part of a protein complex: it stably associates with the RanGTPase activating protein RanGAP1 throughout the cell cycle, and a prerequisite for this stable interaction is the modification of RanGAP1 with SUMO and the presence of Ubc9 (for references see our original article). Notably, while RanGAP1 sumoylation does not depend on RanBP2, its association with RanBP2 requires RanBP2’s E3 ligase region; structural as well as biochemical analyses even suggested that it may encompass the same determinants as those required for E3 ligase activity and may therefore abrogate its E3 function.Citation14
In a recent study we provided insights into the questions whether all RanBP2 is integrated into such complexes and whether the RanBP2 complex is still capable to sumoylate other proteins.Citation15 While it had been evident that a significant pool of RanBP2 is stably engaged with RanGAP1*SUMO1, co-localization analyses combined with a co-depletion experiment revealed that most, if not all RanBP2 is stably incorporated into the complex in cells; RanBP2 in free form however could not be detected. Using biochemical reconstitution as well as competition experiments in cells we found that the stable complex with RanGAP1*SUMO1 required a Ubc9 molecule as molecular glue, and assembled as trimeric complex on the IR1 region of RanBP2. Of note, these observations showed that an existing crystal structure of a RanBP2 fragment in complex with the sumoylated RanGAP1 tail domain and Ubc9Citation14 describes the interactions between the three proteins in the stable complex.
The obvious question arising from these findings was how sumoylation can still be catalyzed by the RanBP2/RanGAP1*SUMO1/Ubc9 complex when the catalytically most active region in free RanBP2 is masked upon complex formation. A detailed analysis of in vitro reconstituted sumoylation assays revealed that the RanBP2/RanGAP1*SUMO1/Ubc9 complex is indeed active as a SUMO E3 ligase, even on its endogenous substrate Borealin.Citation16 In line with the existing crystal structure,Citation14 neither the structural Ubc9 nor RanBP2’s IR1 region was available for catalysis. Instead, our data indicated that upon complex formation a new catalytic center involving RanBP2’s IR2 region is formed that promotes sumoylation with catalytic properties distinct from those of RanBP2’s IR2 region alone. These findings have important implications for the question what is the functional SUMO E3 ligase: our data clearly showed that the RanBP2/RanGAP1*SUMO1/Ubc9 complex rather than free RanBP2 represents the functional entity.
What Makes the RanBP2/RanGAP1*SUMO1/Ubc9 Complex So Fascinating?
RanBP2 is a huge protein of 3224 amino acids (). Some of its domains are currently only poorly characterized: the N-terminus including a leucine-rich domain anchors RanBP2 at the nuclear pore complex and has been implicated in microtubule binding and organization,Citation17 the 8-fold zinc finger motif was described as interaction site for the export receptor Crm1,Citation18 a kinesin binding domain mediates interaction with the motor protein kinesin-1Citation19 and the motor adaptor BicD2,Citation20 and the C-terminal cyclophilin domain binds directly to the HIV-1 capsid.Citation21 Better characterized are the already mentioned SUMO E3 ligase region, four RanGTP binding domains and several FG repeats distributed over nearly the whole protein that serve as interaction sites for transport receptors.Citation8,Citation9 Considering the enormous size of RanBP2 and the variety of cellular processes that RanBP2 may be involved in it is intriguing that only 140 amino acids in the C-terminus are sufficient to accommodate the SUMO E3 ligase core as well as the platform for complex formation with sumoylated RanGAP1. In addition, this area is flanked by two out of the four Ran binding domains. These serve together with RanGAP1 as key regulators of the small GTPase Ran by stimulating its low intrinsic GTPase activity. This places two distinct enzymatic activities, namely the SUMO E3 ligase and the RanGTP hydrolyzing activities, in close proximity within the same protein complex suggesting an intricate link between sumoylation and the Ran GTPase cycle.
This however does not seem to be an evolutionarily conserved feature: plant RanGAP is targeted to the nuclear pore and the mitotic cell plate by a plant specific WWP domain, a mechanism independent of RanBP2 and sumoylationCitation22 suggesting that the precise RanGAP localization is of special importance in higher eukaryotes, yet not necessarily linked to sumoylation. Interestingly, Drosophila dmRanGAP has acquired the capacity to interact with dmRanBP2,Citation23 however it is unclear whether this interaction involves sumoylation of dmRanGAP and whether dmRanBP2 is a SUMO E3 ligase as the described active site is not discernable by sequence comparison. Since the E3 ligase area is recognizable in Xenopus, one may wonder whether the capacity of the RanBP2-RanGAP complex to also regulate SUMO modification has been acquired to serve vertebrate-specific functions.
In which cellular processes could this play a role? Ran is a well-known key regulator with essential functions throughout the cell cycle, including nucleocytoplasmic transport, mitotic spindle assembly and nuclear envelope reformation.Citation24 The common molecular mechanism underlying its diverse functions is the localized assembly and disassembly of transport receptor/cargo complexes depending on its nucleotide-bound state: GTP hydrolysis leads to the disruption of export receptor/cargo/RanGTP complexes, and releases import receptors from inhibitory interactions with RanGTP. Considering that several FG repeats representing interaction sites for transport receptors flank RanBP2’s E3 ligase region in addition to the Ran binding domain, it is likely that transport complexes will encounter the RanBP2 E3 ligase complex. This allows envisioning various scenarios for a dual role of the RanBP2/RanGAP1*SUMO1/Ubc9 complex: (1) Sumoylation may serve to fine-tune the transport machinery; (2) The transport machinery may recruit (cargo) proteins as substrates for sumoylation; (3) Components of the transport machinery may regulate the E3 ligase activity. In the following, we would like to discuss some of these speculative functions in context of RanBP2 biology.
The cellular process for which a direct link between sumoylation and RanGTP hydrolysis has long been anticipated is nucleocytoplasmic transport, due to the identification of RanGAP1 as the first and up to now most prominent SUMO target, and the requirement of sumoylation for RanGAP1 localization at the nuclear pore (see ref. Citation25 and references therein). While RanBP2’s most essential transport functions have been mapped to the N-terminal half of the protein excluding the E3 ligase region,Citation13,Citation26 RanBP2-associated RanGAP1 has been found to be important for the efficiency of some nucleocytoplasmic transport processes.Citation27
In a recent study, Rothenbusch and collegues reported that sumoylation serves to regulate the efficiency of a specific transport pathway in yeast: Mms21 dependent sumoylation of the Kap114 import receptor is required to assist RanGTP-mediated cargo release in the nucleus.Citation28 In analogy, RanBP2 complex-dependent sumoylation could influence the stability of nuclear export complexes or may assist in the recycling of import receptors at the nuclear pore.
While a direct involvement of RanBP2’s E3 ligase region in nucleocytoplasmic transport has not yet been demonstrated, it has been found to be important in mitosis: both known SUMO targets of RanBP2, Topoisomerase 2 α and Borealin, are sumoylated in mitosis,Citation12,Citation16 and the E3 ligase region of RanBP2 is sufficient to alleviate chromosome segregation defects observed upon RanBP2 depletionCitation13 that have been linked to Topoisomerase 2 α sumoylation.Citation12 Interestingly, a fraction of the RanBP2/RanGAP1*SUMO1/Ubc9 complex localizes to kinetochores in dependence of Crm1 and RanGTP,Citation29 raising the possibility that associations with export-like complexes may tether the E3 ligase complex at kinetochores. While there is currently no evidence supporting the notion that kinetochore associations are important for SUMO substrate selection, several centromere/kinetochore proteins can be modified with SUMO,Citation30 and both endogenous RanBP2 substrates can be found at the centromere/kinetochore region. Strikingly, for centromere localization of Survivin, a component of the chromosomal passenger complex also comprising Borealin, Incenp and AuroraB, an interaction with Crm1 is essential.Citation31 Whether Crm1 influences Borealin sumoylation remains to be seen.
Finally, while it is tempting to speculate about potential links between sumoylation and the Ran system, the multidomain protein RanBP2 may obviously participate in many more processes than sumoylation and RanGTP hydrolysis. Deciphering the puzzle of RanBP2’s complex functions will be a challenging, but fascinating subject for future studies involving various aspects of cellular biology.
Acknowledgments
We thank Frauke Melchior for continuous support and critical reading of the manuscript and all lab members for sharing advice.
References
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11:861 - 71; http://dx.doi.org/10.1038/nrm3011; PMID: 21102611
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8:947 - 56; http://dx.doi.org/10.1038/nrm2293; PMID: 18000527
- Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci 2009; 122:4249 - 52; http://dx.doi.org/10.1242/jcs.050542; PMID: 19923268
- Shen TH, Lin H-K, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell 2006; 24:331 - 9; http://dx.doi.org/10.1016/j.molcel.2006.09.013; PMID: 17081985
- Meulmeester E, Kunze M, Hsiao H-H, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell 2008; 30:610 - 9; http://dx.doi.org/10.1016/j.molcel.2008.03.021; PMID: 18538659
- Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell 2009; 35:669 - 82; http://dx.doi.org/10.1016/j.molcel.2009.07.013; PMID: 19748360
- Pichler A, Gast A, Seeler J-S, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 2002; 108:109 - 20; http://dx.doi.org/10.1016/S0092-8674(01)00633-X; PMID: 11792325
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem 1995; 270:14209 - 13; http://dx.doi.org/10.1074/jbc.270.23.14209; PMID: 7775481
- Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, et al. A giant nucleopore protein that binds Ran/TC4. Nature 1995; 376:184 - 8; http://dx.doi.org/10.1038/376184a0; PMID: 7603572
- Joseph J, Tan S-H, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol 2002; 156:595 - 602; http://dx.doi.org/10.1083/jcb.200110109; PMID: 11854305
- Aslanukov A, Bhowmick R, Guruju M, Oswald J, Raz D, Bush RA, et al. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet 2006; 2:e177; http://dx.doi.org/10.1371/journal.pgen.0020177; PMID: 17069463
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 2008; 133:103 - 15; http://dx.doi.org/10.1016/j.cell.2008.01.045; PMID: 18394993
- Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Wälde S, et al. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol 2011; 194:597 - 612; http://dx.doi.org/10.1083/jcb.201102018; PMID: 21859863
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 2005; 435:687 - 92; http://dx.doi.org/10.1038/nature03588; PMID: 15931224
- Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell 2012; 46:287 - 98; http://dx.doi.org/10.1016/j.molcel.2012.02.017; PMID: 22464730
- Klein UR, Haindl M, Nigg EA, Muller S. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell 2009; 20:410 - 8; http://dx.doi.org/10.1091/mbc.E08-05-0511; PMID: 18946085
- Joseph J, Dasso M. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett 2008; 582:190 - 6; http://dx.doi.org/10.1016/j.febslet.2007.11.087; PMID: 18070602
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 1999; 274:37370 - 8; http://dx.doi.org/10.1074/jbc.274.52.37370; PMID: 10601307
- Cai Y, Singh BB, Aslanukov A, Zhao H, Ferreira PA. The docking of kinesins, KIF5B and KIF5C, to Ran-binding protein 2 (RanBP2) is mediated via a novel RanBP2 domain. J Biol Chem 2001; 276:41594 - 602; http://dx.doi.org/10.1074/jbc.M104514200; PMID: 11553612
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 2010; 8:e1000350; http://dx.doi.org/10.1371/journal.pbio.1000350; PMID: 20386726
- Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 2011; 7:e1002439; http://dx.doi.org/10.1371/journal.ppat.1002439; PMID: 22174692
- Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci U S A 2001; 98:15377 - 82; http://dx.doi.org/10.1073/pnas.261459698; PMID: 11752475
- Forler D, Rabut G, Ciccarelli FD, Herold A, Köcher T, Niggeweg R, et al. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol Cell Biol 2004; 24:1155 - 67; http://dx.doi.org/10.1128/MCB.24.3.1155-1167.2004; PMID: 14729961
- Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci 2008; 121:1577 - 86; http://dx.doi.org/10.1242/jcs.005959; PMID: 18469014
- Palancade B, Doye V. Sumoylating and desumoylating enzymes at nuclear pores: underpinning their unexpected duties?. Trends Cell Biol 2008; 18:174 - 83; http://dx.doi.org/10.1016/j.tcb.2008.02.001; PMID: 18313922
- Wälde S, Thakar K, Hutten S, Spillner C, Nath A, Rothbauer U, et al. The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic 2012; 13:218 - 33; http://dx.doi.org/10.1111/j.1600-0854.2011.01302.x; PMID: 21995724
- Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell 2008; 19:2300 - 10; http://dx.doi.org/10.1091/mbc.E07-12-1279; PMID: 18305100
- Rothenbusch U, Sawatzki M, Chang Y, Caesar S, Schlenstedt G. Sumoylation regulates Kap114-mediated nuclear transport. EMBO J 2012; 31:2461 - 72; http://dx.doi.org/10.1038/emboj.2012.102; PMID: 22562154
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 2005; 7:626 - 32; http://dx.doi.org/10.1038/ncb1263; PMID: 15908946
- Zhang X-D, Goeres J, Zhang H, Yen TJ, Porter ACG, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell 2008; 29:729 - 41; http://dx.doi.org/10.1016/j.molcel.2008.01.013; PMID: 18374647
- Knauer SK, Bier C, Habtemichael N, Stauber RH. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep 2006; 7:1259 - 65; http://dx.doi.org/10.1038/sj.embor.7400824; PMID: 17099693