Abstract
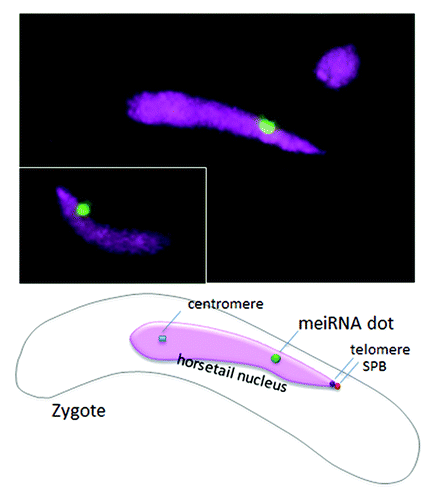
Pairing and recombination of homologous chromosomes are essential for ensuring correct segregation of chromosomes in meiosis. In S. pombe, chromosomes are first bundled at the telomeres (forming a telomere bouquet) and then aligned by oscillatory movement of the elongated “horsetail” nucleus ().Citation1,Citation2 Telomere clustering and subsequent chromosome alignment promote pairing of homologous chromosomes.Citation3-Citation5 However, this telomere-bundled alignment of chromosomes cannot be responsible for the specificity of chromosome pairing. Thus, there must be some mechanism to facilitate recognition of homologous partners after telomere clustering. Recent studies in S. pombe have shown that RNA transcripts retained on the chromosome, or RNA bodies, may play a role in recognition of homologous chromosomes for pairing ().Citation6 Acting as fiducial markers of homologous loci they would abrogate the need for direct DNA sequence homology searching.
DSB-Independent Search for Homologous Chromosomes
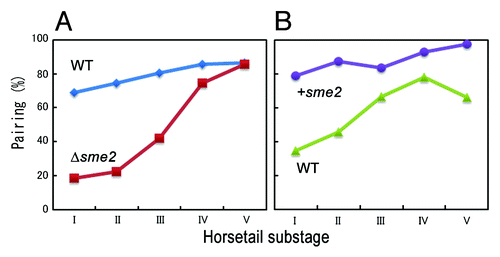
It has been proposed that in the yeast Saccharomyces cerevisiae, interactions between homologous DNA sequences with DNA double-strand breaks (DSB) and the consequent recombination are involved in homology searching for pairing.Citation7,Citation8 However, this issue remains debatable as it has been reported that pairing can be achieved through DSB-dependent or -independent pathways in other organisms.Citation7-Citation9 We recently demonstrated that the sme2 locus, which is located on chromosome II, shows robust pairing in the early stage of the horsetail movement and that the robust pairing occurs independently of DSB formation and, hence, independently of recombination.Citation6
The sme2 gene encodes a meiosis-specific polyadenylated non-coding RNA, named MeiRNA, that is essential for entry into meiosis in S. pombe.Citation10,Citation11 We reported that the sme2 RNA mediates robust pairing of homologous chromosomes in meiosis: deletion of the sme2 sequence eliminates this robust pairing and transposition to other chromosomal sites confers the robust pairing at those sites ().Citation6 Remarkably, the sme2 RNA transcripts accumulate at their respective gene loci and their retention at the homozygous sme2 loci is essential for robust pairing at the sme2 locus. Thus, the chromosomally-retained sme2 RNA seems to act as an “identifier” of homologous chromosomes.
Figure 2. Robust pairing is mediated by the sme2 sequence. (A) Pairing frequency at the sme2 locus during progression through the horsetail stage decreases with deletion of the sme2 sequence (Δsme2) compared with the wild-type (WT). (B) Pairing frequency at the ade8 locus increases with insertion of the sme2 sequence (+sme2) compared with the wild-type (WT).
The 3'-Region of meiRNA-L is Crucial for Robust Pairing
MeiRNA was initially identified as a 0.5 kb sme2 transcript that complements the defect of meiosis progression in the sme2Δ mutant.Citation10 On the other hand, we identified a longer transcript (1.5 kb), extended at the 3′ end, as a major transcript during meiosis.Citation6 This discrepancy was explained by the finding that MeiRNA is a major target of Mmi1, which selectively eliminates meiosis-specific transcripts in vegetative cells.Citation11 In an mmi1 temperature-sensitive mutant background, both the 1.5 kb and the 0.5 kb transcript were detected, suggesting that the short transcript is the product of Mmi1-mediated digestion.Citation11 Thus, the short (0.5 kb) and the long (1.5 kb) transcripts were named MeiRNA-S and MeiRNA-L, respectively.Citation6,Citation11
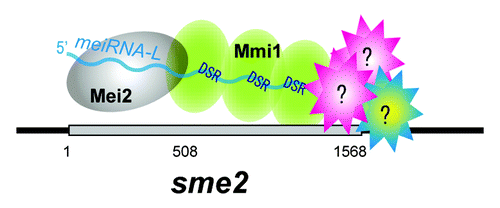
The sme2 transcript has two functional domains as shown in . The Mei2 protein, a key regulator of meiosis in S. pombe, binds to the sme2 transcript to form a distinct dot at the sme2 locus in the horsetail nucleus.Citation12-Citation14 The 5′-region of the sme2 transcript, corresponding to meiRNA-S, is necessary for Mei2 binding, but unnecessary for robust pairing. In contrast, meiRNA-L is essential for robust pairing of the sme2 loci.Citation6 Deletion of polyadenylation sites at the 3′-end of meiRNA-L results in the loss of meiRNA retention at the sme2 locus and of robust pairing. The 3′-extended region specific to meiRNA-L (that is meiRNA-L with the meiRNA-S sequence deleted) is sufficient for its chromosomal retention and for robust pairing. Thus, we expect that chromosomal retention of meiRNA-L through its 3′-region has a role in robust pairing, as described below.
Figure 3. Distinct functional domains of the sme2 transcript. The sme2 RNA transcript can be divided into two distinct domains: the 5′-portion of meiRNA-L, corresponding to meiRNA-S, and the 3′-extended region specific to meiRNA-L. Mei2 protein binds to the 5′-portion of meiRNA-L (meiRNA-S). Mmi1protein binds to DSR motifs, which are concentrated in the 3′-extended region of meiRNA-L. We speculate that as yet unknown proteins may bind to meiRNA-L and play a role in pairing.
Roles for RNA in Homologous Pairing
Chromosomally-accumulated meiRNA-L at the sme2 locus may play a role in recruiting specific RNA-binding proteins essential for pairing or recognition. On the other hand, even when the sme2 locus was removed from chromosome II, no obvious defects in chromosome segregation or spore formation were detected. These observations raise the possibility that other as yet undiscovered pairing sites may also exist on chromosome II. An array of recognition complexes at multiple sites on each chromosome might act as chromosome-specific identifiers. Such recognition complexes could be arrays of RNA-protein complexes, transcription factories or specialized chromatin structures.
Although Mei2 forms a dot at the sme2 locus in the horsetail stage, it has been demonstrated that Mei2 is unnecessary for robust pairing ().Citation6 Mmi1 also binds to MeiRNA-L and forms a dot at the sme2 locus. However, in a hypomorphic mmi1-48 mutant,Citation15 the robust pairing was enhanced, suggesting that Mmi1 negatively regulates robust pairing by degrading meiRNA-L.Citation6 Protein components that interact with the 3′-extended region specific to meiRNA-L may be involved in the robust pairing (), but such critical components are yet to be identified.
Alternatively, specific components may not be involved at all. Instead, RNA bodies formed at defined loci along the chromosome may act as a barcode for agglutination of homologous chromosomes. Recent reports demonstrate that RNA transcripts function as a scaffold in the assembly of many nuclear bodies at their respective gene loci.Citation16-Citation18 The most striking example of RNA bodies is in the nucleolus formed around the rRNA genes. S. pombe rRNA gene clusters locate on both ends of chromosome III, and occupy a defined position within the nucleolus.Citation19,Citation20 These findings provide an insight into the role of RNA nuclear bodies in the functional organization of chromosomes within the nucleus.
A previous model also proposed a role for transcription in homologous chromosome recognition and pairingCitation21-Citation23: this model depicts a chromosome as a linear array of transcription factories, providing a possible mechanism for how transcription results in recognition and pairing of homologous chromosomes when chromosomes are pre-aligned in a chromosome bouquet. A similar model, in which meiosis-specific polyadenylated RNA transcripts initiate the pairing process, has been previously proposed in the lily.Citation24 Other studies have reported a similar link between pairing and transcription: in Drosophila embryos, somatic pairing is initiated at the histone gene cluster at the time when zygotic expression begins;Citation25 in Drosophila male meiosis, the pairing sites correspond to the highly transcribed rDNA loci and histone genes.Citation26 Another more direct link has been reported: in mammals non-coding RNA accumulated at their respective gene loci has been shown to mediate transient X-chromosome pairing and to mark the onset of X inactivation.Citation27,Citation28 These findings imply that RNA transcripts retained at gene loci may act as chromosome identifiers for their homologous partner.
Precise molecular mechanisms for recognition between RNA bodies remain to be elucidated. Recognition can be achieved by an active mechanism based on DNA-RNA, RNA-RNA or protein-protein interactions, or by a passive mechanism based on agglutination of transcription factories, as discussed in recent literatures.Citation29,Citation30 These questions can be answered once critical factors for recognition are identified in the sme2 RNA body or similar pairing mechanisms are identified in other chromosomal loci of S. pombe or in other organisms.
References
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, et al. Telomere-led premeiotic chromosome movement in fission yeast. Science 1994; 264:270 - 3; http://dx.doi.org/10.1126/science.8146661; PMID: 8146661
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci 1998; 111:701 - 12; PMID: 9471999
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell 2009; 17:598 - 605; http://dx.doi.org/10.1016/j.devcel.2009.10.014; PMID: 19922865
- Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 2004; 6:329 - 41; http://dx.doi.org/10.1016/S1534-5807(04)00059-0; PMID: 15030757
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006; 125:59 - 69; http://dx.doi.org/10.1016/j.cell.2006.01.048; PMID: 16615890
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, et al. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science 2012; 336:732 - 6; http://dx.doi.org/10.1126/science.1219518; PMID: 22582262
- Gerton JL, Hawley RS. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat Rev Genet 2005; 6:477 - 87; http://dx.doi.org/10.1038/nrg1614; PMID: 15931171
- Barzel A, Kupiec M. Finding a match: how do homologous sequences get together for recombination?. Nat Rev Genet 2008; 9:27 - 37; http://dx.doi.org/10.1038/nrg2224; PMID: 18040271
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma 2006; 115:158 - 74; http://dx.doi.org/10.1007/s00412-006-0048-6; PMID: 16570189
- Watanabe Y, Yamamoto MS. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 1994; 78:487 - 98; http://dx.doi.org/10.1016/0092-8674(94)90426-X; PMID: 7520368
- Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat-Todeschini L, Vavasseur A, et al. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol 2012; 2:120014; http://dx.doi.org/10.1098/rsob.120014; PMID: 22645662
- Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 1997; 386:187 - 90; http://dx.doi.org/10.1038/386187a0; PMID: 9062192
- Yamashita A, Watanabe Y, Nukina N, Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 1998; 95:115 - 23; http://dx.doi.org/10.1016/S0092-8674(00)81787-0; PMID: 9778252
- Shimada T, Yamashita A, Yamamoto M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell 2003; 14:2461 - 9; http://dx.doi.org/10.1091/mbc.E02-11-0738; PMID: 12808043
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 2006; 442:45 - 50; http://dx.doi.org/10.1038/nature04881; PMID: 16823445
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95 - 101; http://dx.doi.org/10.1038/ncb2140; PMID: 21170033
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011; 13:167 - 73; http://dx.doi.org/10.1038/ncb2157; PMID: 21240286
- Carmo-Fonseca M, Rino J. RNA seeds nuclear bodies. Nat Cell Biol 2011; 13:110 - 2; http://dx.doi.org/10.1038/ncb0211-110; PMID: 21283118
- Uzawa S, Yanagida M. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J Cell Sci 1992; 101:267 - 75; PMID: 1629244
- Hiraoka Y. Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells 1998; 3:405 - 13; http://dx.doi.org/10.1046/j.1365-2443.1998.00205.x; PMID: 9753423
- Cook PR. The transcriptional basis of chromosome pairing. J Cell Sci 1997; 110:1033 - 40; PMID: 9175699
- Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta 2008; 1783:2155 - 60; http://dx.doi.org/10.1016/j.bbamcr.2008.07.013; PMID: 18706455
- Wilson PJ, Riggs CD, Hasenkampf CA. Plant chromosome homology: hypotheses relating rendezvous, recognition and reciprocal exchange. Cytogenet Genome Res 2005; 109:190 - 7; http://dx.doi.org/10.1159/000082399; PMID: 15753576
- Hotta Y, Tabata S, Stubbs L, Stern H. Meiosis-specific transcripts of a DNA component replicated during chromosome pairing: homology across the phylogenetic spectrum. Cell 1985; 40:785 - 93; http://dx.doi.org/10.1016/0092-8674(85)90338-1; PMID: 2580636
- Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J Cell Biol 1993; 120:591 - 600; http://dx.doi.org/10.1083/jcb.120.3.591; PMID: 8425892
- McKee BD. The license to pair: identification of meiotic pairing sites in Drosophila. Chromosoma 1996; 105:135 - 41; http://dx.doi.org/10.1007/BF02509494; PMID: 8781181
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science 2006; 311:1149 - 52; http://dx.doi.org/10.1126/science.1122984; PMID: 16424298
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 2009; 23:1831 - 42; http://dx.doi.org/10.1101/gad.1811209; PMID: 19684108
- Dernburg AF. Molecular biology. RNA plays meiotic matchmaker. Science 2012; 336:681 - 2; http://dx.doi.org/10.1126/science.1222647; PMID: 22582252
- Yamashita A, Kariyazono R, Watanabe Y. Meiotic pairing by non-coding RNA?. EMBO Rep 2012; 13:766; http://dx.doi.org/10.1038/embor.2012.124; PMID: 22898975