Abstract
The transitions between the successive cell cycle stages depend on reversible protein phosphorylation events. The phosphorylation state of every protein within a cell is strictly determined by spatiotemporally controlled kinase and phosphatase activities. Nuclear disassembly and reassembly during open mitosis in higher eukaryotic cells is one such process that is tightly regulated by the reversible phosphorylation of key proteins. However, little is known about the regulation of these mitotic events. In particular, although kinase function during entry into mitosis is better studied, very little is known about how proteins are dephosphorylated to allow nuclear reformation at the end of mitosis. We have identified LEM‑4, a conserved protein of the nuclear envelope, as an essential coordinator of kinase and phosphatase activities during mitotic exit. Inhibition of VRK‑1 kinase and promotion of a PP2A phosphatase complex by LEM‑4 tightly regulate the phosphorylation state of BAF, an essential player of nuclear reformation at the end of mitosis. Here I offer extended comments on the contribution of LEM‑4 in the regulation of protein phosphorylation and nuclear reformation.
The nuclear envelope (NE) is a sub-domain of the endoplasmic reticulum (ER) double membrane system, which is specialized to passively separate chromatin from the cytoplasm and to actively regulate different nuclear events.Citation1-Citation3 The diverse functions of the NE are mediated by membrane-associated and integral membrane proteins of the NE. Some of these proteins are part of networks of interactions that reach from chromatin and lamin filaments in the nucleus to the cytoskeleton in the cytoplasm.Citation4-Citation6 Particularly, barrier-to-autointegration factor (BAF), an essential and highly conserved metazoan protein, is required to link the NE with the chromatin.Citation7,Citation8 BAF is known to bind specifically as a dimer to one LEM (LAP2/emerin/MAN1) domainCitation9,Citation10 of the inner nuclear membrane (INM) proteins and to two DNA helixes in a sequence-independent manner.Citation11,Citation12 Through these interactions, BAF contributes to the integrity of the NE (). Other linking proteins include HP-1, which in a similar manner, but through distinct interactions, also links the NE with the chromatin.Citation1 This mechanism is however not conserved in all metazoa. Additionally, several integral INM proteins possess long basic domains that can mediate direct interactions between the NE and DNA.Citation13
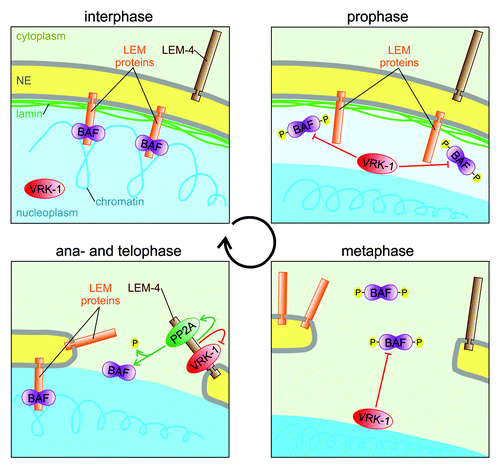
Figure 1. Model of the evolutionarily conserved regulatory mechanism controlling the function of BAF during cell cycle. During interphase BAF dimers (purple) bind LEM domain proteins (orange) of the INM and DNA to link the NE with the chromatin (blue). During mitotic entry the NE breaks down. At the nuclear periphery (prophase) and later on the condensed chromatin surface (prophase and metaphase) VRK-1 (red) phosphorylates BAF to keep it away from its binding partners. During mitotic exit (ana- and telophase) NE reforms around the decondensing chromatin. LEM-4 binds to VRK-1 and inhibits its enzymatic activity. Furthermore, LEM-4 also binds to a PP2A complex (green) and promotes its activity to dephosphorylate BAF. Consequently, BAF is recruited to the chromatin surface where it can bind LEM domain proteins to reform the NE.
Many of the molecular interactions at the NE are disrupted during mitotic entry to allow cytoplasmic spindle microtubules to unperturbedly reach and segregate the duplicated sister chromatids. At the end of mitosis these interactions should be reset in a controlled sequence of mitotic events to rebuild the interphase nuclear structures.Citation1,Citation7,Citation14 Many previous studies, including ours, have suggested that mitotic nuclear disassembly and reassembly are controlled by reversible phosphorylation events.Citation14 Cyclin-dependent kinase 1 (CDK1) and other kinases that function downstream of CDK1 activation, such as protein kinase C (PKC), Aurora A, polo-like kinase 1 (PLK1) and vaccinia-related kinase 1 (VRK1) have all been implicated in NE breakdown.Citation14-Citation17 They can phosphorylate different NE proteins leading to disassembly of the interphase nuclear structures. Consequently, soluble proteins are dispersed into the cytosol while trans-membrane proteins are absorbed into the ER, which serves as a reservoir for the NE membranes and membrane proteins during mitosis.Citation18 Upon mitotic exit and CDK1 inactivation all these proteins must be dephosphorylated in order to rebuild the interphase nuclear structures. It is still not known which phosphatases counteract CDK1 and other mitotic kinases during mitotic exit, and even less is known about how these phosphatases are regulated or how the opposing activities of kinases and phosphatases are integrated to allow orderly mitotic progression. It is known that NE reformation begins with the attachment of ER membranes to the anaphase chromatin surface and this step is mediated by direct interactions between DNA and the basic domains of a group of INM proteins.Citation13,Citation19,Citation20 Subsequently, BAF plays a central and evolutionarily conserved role in organizing and shaping these membranes around the decondensing chromatin into a closed NE rim.Citation21-Citation23
To reveal how BAF is regulated during mitosis, we have screened a collection of temperature sensitive embryonic lethal Caenorhabditis elegans mutants.Citation24 We identified a mutation in lem-4 gene, also known as ankle2, that encodes an evolutionarily conserved protein associated with the NE. Inactivation of lem-4 resulted in nuclear shape and NE membrane organization defects very similar to those caused by inactivation of the baf-1 gene. In both mutants the nuclei were multilobed and large parts of their chromatin were not covered with NE membranes.Citation21,Citation24 These phenotypic similarities prompted us to speculate that these two proteins might either function together or that one might regulate the other. While we found that BAF had no effect on LEM‑4, we found that LEM‑4 was an essential regulator of BAF‑1 localization during mitotic exit in worm and human cells.Citation24
BAF shows a very dynamic localization pattern throughout the cell cycle, and this is conserved from wormsCitation21 to humans.Citation22 During interphase, BAF is mainly enriched at the INM due to its specific interactions with the LEM domains of LEM‑2, EmerinCitation21,Citation25 and other proteins of the NE.Citation8 Inactivation of these proteins consequently results in the loss of BAF from the INM. During mitotic entry BAF is released from the NE and the chromatin and is uniformly distributed throughout the cytoplasm. During mitotic exit however, BAF is very rapidly recruited to the segregated chromatids. It is strongly enriched in the transient dense structures around the anaphase chromatin called “core” regions. The appearance of these structures coincides with the reformation of the closed NE and is hypothesized to be essential for the organization of the membranes around the chromatin.Citation21,Citation22 Inactivation of LEM‑4, but not other LEM domain proteins, completely abolished the recruitment of BAF to the chromatin surface and to the “core” region in worm and human cells.Citation24 Consequently, this resulted in abnormal nuclear structure and defects in NE membrane organization. We hypothesized that LEM‑4 localizes BAF during mitosis in a way other than via direct interaction. We based this assumption on the facts that; surprisingly, BAF cannot bind to the LEM domain of the human LEM-4 protein as found by immunoprecipitation experiments and GST pulldown assays and its worm ortholog does not possess a recognizable LEM domain. Second, because BAF is recruited to the “core” region prior to any LEM domain protein.Citation21,Citation22 The question was then; how does LEM‑4 regulate BAF localization and function during mitosis?
To address this question we turned to genetics and performed a suppressor screen on the temperature sensitive lem-4 mutant worm line. We identified a suppressor mutation in the vrk-1 gene, which suppressed not only the embryonic lethality, but also the nuclear defects seen in temperature sensitive lem-4 mutant worms at restrictive temperature. vrk-1 is an essential gene; its inactivation by dsRNA-interference (RNAi) or by the suppressor mutation results in embryonic lethality, but not when it is combined with the co-inactivation of lem-4. What is the explanation for this suppression and how might such information bring us closer to understanding the role of LEM-4 in BAF localization?
VRK-1 is the main mitotic kinase acting on BAF.Citation21,Citation26 During interphase VRK-1 is uniformly distributed in the nucleoplasm and its enzymatic activity is suppressed by macroH2A1.Citation27 During mitotic entry the decay of macroH2A1, probably together with other mitotic mechanisms, contributes to the reactivation of VRK‑1. VRK‑1 is then relocalized to the nuclear periphery and to the mitotic chromosomes where it can phosphorylate BAF and thus release BAF from its binding partners ().Citation21 Both, in vivo and in vitro evidence demonstrate that phosphorylation of BAF by VRK‑1 weakens its interactions with DNA and the LEM domains of its INM interaction partners.Citation21,Citation26,Citation28 Consistent with this, inactivation of vrk‑1 results in robust hyper-accumulation of BAF on the mitotic chromosomes and the NE membrane remnants throughout the entire mitosis.Citation21 This effect of VRK‑1 on BAF localization is the precise opposite of the effect of LEM‑4.Citation24 While VRK‑1 is required to release BAF from its binding partners during mitosis, LEM‑4 is required to recruit BAF to the chromatin surface during mitotic exit. Given the opposing effects of VRK‑1 and LEM‑4 on BAF localization and given the fact that VRK‑1 regulates BAF localization through phosphorylation; we hypothesized that LEM‑4 might also regulate BAF localization through regulating its phosphorylation state. Indeed, analyses of BAF phosphoisoforms in worms that were either mutant for these components or had been treated with RNAi to reduce their concentration revealed that, while inactivation of vrk‑1 resulted in hypo-phosphorylation of BAF, inactivation of lem‑4 resulted in its hyper-phosphorylation. The phosphorylation balance changed back to normal in the suppressor conditions, also suggesting that neither vrk‑1 nor lem‑4 was completely inactivated in these mutant conditions. The subcellular localization of BAF is thus determined by its phosphorylation state, where LEM‑4 is responsible for dephosphorylation of BAF and for its recruitment to the chromatin surface at the end of mitosis.
The intriguing question arising from this result is; how does LEM‑4 regulate the dephosphorylation of BAF? One possible explanation might be that LEM‑4 is a new phosphatase. However, we found that, at least in vitro, LEM‑4 cannot dephosphorylate BAF, and thus hypothesized that LEM‑4 might either inhibit the kinase activity of VRK‑1 or activate a so far unidentified phosphatase to dephosphorylate BAF. Our experiments testing the first idea showed that worm and human LEM‑4 can indeed directly bind to VRK‑1 and completely inhibit its kinase activity ().Citation24 The inhibitory effect of LEM‑4 was specific to VRK‑1, since the enzymatic activities of other mitotic kinases were not affected by the LEM‑4 proteins. We assumed that the inhibition of VRK‑1 by LEM‑4 in living cells might occur during mitotic exit, since inactivation of the worm temperature sensitive lem‑4 mutation specifically during mitosis, but not during interphase, resulted in abnormal NE structure. However, if the only effect of LEM‑4 were to inhibit the continued phosphorylation of BAF during mitotic exit, BAF would still be phosphorylated, i.e., the effect of LEM‑4 on VRK‑1 still does not explain how BAF is dephosphorylated in order that it may bind to chromatin and support the NE reformation around it.
Members of the protein phosphatase 1 (PP1) and 2A (PP2A) families are believed to counteract CDK1 and some other kinases during mitotic exit although, direct in vivo evidence is still lacking.Citation15,Citation29 PP2A functions in a heterotrimeric complex composed of catalytic, scaffold and regulatory subunits.Citation30 The regulatory subunit determines the substrate specificity and regulates the diverse functions of the PP2A complexes. Recently, a particular PP2A complex composed of the B55α regulatory subunit was shown to be the only phosphatase essential for mitotic exit.Citation31 The same PP2A‑B55α complex was also found to regulate centriole duplicationCitation32,Citation33 and interestingly, we found that the same complex directly binds to LEM‑4.Citation24 This finding prompted us to assume that PP2A might dephosphorylate BAF during mitotic exit and that LEM‑4 might promote this function (). Indeed, we confirmed that PP2A can dephosphorylate BAF in vitro, and systematic RNAi-mediated inactivation of worm phosphatases revealed that PP2A was the main phosphatase acting on BAF in vivo. Consequently, inactivation of PP2A resulted in hyper-phosphorylation of BAF and in the complete loss of BAF from the chromatin surface during mitotic exit in worm and human cells. Hence, the nuclear structure and the organization of the NE membranes in PP2A inactivated cells were very similarly disrupted as in the LEM‑4 inactivated cells.
Our experiments suggest a novel and evolutionarily conserved molecular mechanism where LEM‑4, by coordination of both kinase and phosphatase activities, controls the dephosphorylation of BAF to enable its essential function in NE reformation at the end of mitosis ().
Why is there a need for coordination of kinase and phosphatase activities during mitotic exit? The postmitotic reassembly of interphase structures is very rapid. For example, NE reformation in worm embryos is completed within ~2 minCitation21 and in human culture cells within ~10 min.Citation20 Therefore the switch between the inactive phosphorylated and the active dephosphorylated states of the implicated key molecules must occur rapidly. This likely requires mechanisms to avoid the futile cycles, which occur when kinases and their counteracting phosphatases are active simultaneously. Optimal protein dephosphorylation therefore requires two synchronous actions; efficient inhibition of a kinase and co-activation of the counteracting phosphatase. Proteins with functions similar to LEM‑4 might be the best candidates to ensure accurate spatial and temporal control of kinase and phosphatase activities. Since there are a multitude of proteins that form complexes with kinases and phosphatasesCitation34‑Citation36; I predict that this mechanism might be more generally employed to control mitotic exit and very probably other phosphorylation-dependent events.
Further work is needed to reveal the cell cycle cues that regulate the activity of LEM‑4 and its interactions with VRK‑1 and PP2A, as well as to understand, how other proteins are dephosphorylated in order that they can play their part in rebuilding the interphase nuclear structures such as nuclear pore complexes, lamin filaments or different nuclear bodies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I would like to thank Iain W. Mattaj for critically reading the manuscript. I was a Human Frontier Science Program Organization Fellow.
References
- Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 2005; 21:347 - 80; http://dx.doi.org/10.1146/annurev.cellbio.21.090704.151152; PMID: 16212499
- Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi JY, et al. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol 2003; 13:584 - 92; http://dx.doi.org/10.1016/j.tcb.2003.09.009; PMID: 14573352
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 2010; 11:317 - 28; http://dx.doi.org/10.1038/nrm2894; PMID: 20414256
- Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus 2011; 2:339 - 49; http://dx.doi.org/10.4161/nucl.2.5.17846; PMID: 21970986
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008; 22:832 - 53; http://dx.doi.org/10.1101/gad.1652708; PMID: 18381888
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 2010; 26:421 - 44; http://dx.doi.org/10.1146/annurev-cellbio-100109-104037; PMID: 20507227
- Gorjánácz M, Jaedicke A, Mattaj IW. What can Caenorhabditis elegans tell us about the nuclear envelope?. FEBS Lett 2007; 581:2794 - 801; http://dx.doi.org/10.1016/j.febslet.2007.03.052; PMID: 17418822
- Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y. Barrier-to-autointegration factor--a BAFfling little protein. Trends Cell Biol 2007; 17:202 - 8; http://dx.doi.org/10.1016/j.tcb.2007.02.004; PMID: 17320395
- Brachner A, Foisner R. Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem Soc Trans 2011; 39:1735 - 41; http://dx.doi.org/10.1042/BST20110724; PMID: 22103517
- Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol 2007; 261:1 - 46; http://dx.doi.org/10.1016/S0074-7696(07)61001-8; PMID: 17560279
- Bradley CM, Ronning DR, Ghirlando R, Craigie R, Dyda F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nat Struct Mol Biol 2005; 12:935 - 6; http://dx.doi.org/10.1038/nsmb989; PMID: 16155580
- Cai M, Huang Y, Suh JY, Louis JM, Ghirlando R, Craigie R, et al. Solution NMR structure of the barrier-to-autointegration factor-Emerin complex. J Biol Chem 2007; 282:14525 - 35; http://dx.doi.org/10.1074/jbc.M700576200; PMID: 17355960
- Ulbert S, Platani M, Boue S, Mattaj IW. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J Cell Biol 2006; 173:469 - 76; http://dx.doi.org/10.1083/jcb.200512078; PMID: 16717124
- Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 2009; 10:178 - 91; http://dx.doi.org/10.1038/nrm2641; PMID: 19234477
- Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 2011; 12:469 - 82; http://dx.doi.org/10.1038/nrm3149; PMID: 21750572
- Gorjánácz M, Mattaj IW. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci 2009; 122:1963 - 9; http://dx.doi.org/10.1242/jcs.044750; PMID: 19494125
- Mall M, Walter T, Gorjánácz M, Davidson IF, Nga Ly-Hartig TB, Ellenberg J, et al. Mitotic lamin disassembly is triggered by lipid-mediated signaling. J Cell Biol 2012; 198:981 - 90; http://dx.doi.org/10.1083/jcb.201205103; PMID: 22986494
- Walters AD, Bommakanti A, Cohen-Fix O. Shaping the nucleus: factors and forces. J Cell Biochem 2012; 113:2813 - 21; http://dx.doi.org/10.1002/jcb.24178; PMID: 22566057
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 2008; 182:911 - 24; http://dx.doi.org/10.1083/jcb.200805140; PMID: 18779370
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol 2009; 186:183 - 91; http://dx.doi.org/10.1083/jcb.200901106; PMID: 19620630
- Gorjánácz M, Klerkx EP, Galy V, Santarella R, López-Iglesias C, Askjaer P, et al. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J 2007; 26:132 - 43; http://dx.doi.org/10.1038/sj.emboj.7601470; PMID: 17170708
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci 2008; 121:2540 - 54; http://dx.doi.org/10.1242/jcs.033597; PMID: 18628300
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol 2007; 9:1160 - 6; http://dx.doi.org/10.1038/ncb1636; PMID: 17828249
- Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, et al. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 2012; 150:122 - 35; http://dx.doi.org/10.1016/j.cell.2012.04.043; PMID: 22770216
- Liu J, Lee KK, Segura-Totten M, Neufeld E, Wilson KL, Gruenbaum Y. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2003; 100:4598 - 603; http://dx.doi.org/10.1073/pnas.0730821100; PMID: 12684533
- Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell 2006; 17:2451 - 64; http://dx.doi.org/10.1091/mbc.E05-12-1179; PMID: 16495336
- Kim W, Chakraborty G, Kim S, Shin J, Park CH, Jeong MW, et al. Macro histone H2A1.2 (macroH2A1) protein suppresses mitotic kinase VRK1 during interphase. J Biol Chem 2012; 287:5278 - 89; http://dx.doi.org/10.1074/jbc.M111.281709; PMID: 22194607
- Bengtsson L, Wilson KL. Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol Biol Cell 2006; 17:1154 - 63; http://dx.doi.org/10.1091/mbc.E05-04-0356; PMID: 16371512
- Bollen M, Gerlich DW, Lesage B. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol 2009; 19:531 - 41; http://dx.doi.org/10.1016/j.tcb.2009.06.005; PMID: 19734049
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 2001; 353:417 - 39; http://dx.doi.org/10.1042/0264-6021:3530417; PMID: 11171037
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol 2010; 12:886 - 93; http://dx.doi.org/10.1038/ncb2092; PMID: 20711181
- Kitagawa D, Flückiger I, Polanowska J, Keller D, Reboul J, Gönczy P. PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev Cell 2011; 20:550 - 62; http://dx.doi.org/10.1016/j.devcel.2011.02.005; PMID: 21497765
- Song MH, Liu Y, Anderson DE, Jahng WJ, O’Connell KF. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell 2011; 20:563 - 71; http://dx.doi.org/10.1016/j.devcel.2011.03.007; PMID: 21497766
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, et al. A global protein kinase and phosphatase interaction network in yeast. Science 2010; 328:1043 - 6; http://dx.doi.org/10.1126/science.1176495; PMID: 20489023
- Colledge M, Scott JD. AKAPs: from structure to function. Trends Cell Biol 1999; 9:216 - 21; http://dx.doi.org/10.1016/S0962-8924(99)01558-5; PMID: 10354567
- Alto N, Carlisle Michel JJ, Dodge KL, Langeberg LK, Scott JD. Intracellular targeting of protein kinases and phosphatases. Diabetes 2002; 51:Suppl 3 S385 - 8; http://dx.doi.org/10.2337/diabetes.51.2007.S385; PMID: 12475780