Abstract
In eukaryotes, genomic DNA is hierarchically packaged into chromatin by histones. A defined organization of the genome into chromatin with specific patterns of epigenetic modifications is crucial for transcriptional regulation, cell fate determination, and maintenance, in which the histone variant incorporation has been characterized as one of the most key players. The diversity of histone variants results in structural plasticity of chromatin and highlights functionally distinct chromosomal domains. Here we focus on the role of histone variant H3.3 and its coregulation with H2A.Z in chromatin dynamics at enhancers and promoters and transcriptional regulation.
Introduction
The accessibility of genomic DNA in eukaryotes is believed to be highly regulated by chromatin dynamics. Establishment of a proper chromatin landscape is critically important for genome function and integrity. The cell-type specific pattern of gene expression is characterized not only by the primary genomic DNA sequence but also by the distinct spatial organization of genome into chromatin with distinct structures that are modified by epigenetic factors including DNA methylations, histone variants and/or modifications, and other chromatin binding factors.Citation1,Citation2 In particular, the chromatin dynamics around the promoter and enhancer regions—which are highly regulated to accommodate binding of transcriptional factors and RNA polymerases machineries during the process of gene transcription—are of critical importance. Besides the bulk packaging role of canonical histones, different histone variants have evolved distinct regulatory mechanisms for their expression, deposition, and functional implications in chromatin dynamics and transcriptional activities.Citation3 Recent studies have demonstrated that two major histone variants, H3.3 and H2A.Z, are highly enriched at these regulatory regions and involved in transcriptional regulations.Citation4-Citation9 It is of great interest to investigate how the variants are incorporated into regions of genome, how they decorate specific chromatin states for gene regulation, and how they are involved in cell fate determination and maintenance. In this review we focus on the recent studies of epigenetic regulation and functional implications of the histone variant H3.3 and its coregulation with H2A.Z at enhancers and promoters.
Histone Variant H3.3 vs. Canonical H3
The histone variant H3.3 is one of the most conserved variants in all eukaryotes. In most metazoans, H3.3 displays only four amino acid differences compared with canonical H3, including residue 31 (Ser vs. Ala) in the N-terminal tail and residues 87 (Ala vs. Ser), 89 (Ile vs. Val), and 90 (Gly vs. Met) near the beginning of α2 helix. The canonical H3 protein is encoded by a gene cluster with more than a dozen genes, expressed in S phase and incorporated into chromatin during DNA replication by its chaperone CAF-1—while H3.3 protein, encoded by two distinct genes H3f3a and H3f3b, is expressed throughout the whole cell cycle and deposited into distinct chromatin regions in a DNA replication-independent manner by different mechanisms.Citation10 Histone variant H3.3 has been largely considered as a marker of transcriptionally active genes but also involved in the formation of telomere and pericentric heterochromatin in mouse embryonic stem (mES) cells.Citation11-Citation14 To date, a number of factors have been identified for proper incorporation of H3.3 at specific genomic regions. Different H3.3 chaperones are likely responsible for the deposition and localization of H3.3 at distinct genomic regions to accomplish different biological functions. The HIRA (HIR histone cell cycle regulation defective homolog A) complex has been found to account for the replication-independent assembly of H3.3 in transcribed chromatin regions.Citation15 It has been observed to physically associate with both initiation (Ser5 phosphorylation) and elongation (Ser2 phosphorylation) forms of RNA Pol II, which may account for the distinct H3.3 density profile in the highly transcribed genes; while the DNA binding ability of the HIRA complex without any sequence specificity may provide another potential nucleosome gap-filling mechanism for HIRA-dependent H3.3 deposition.Citation16 In contrast, DAXX (death-domain associated protein), another H3.3 specific chaperone, has been identified to be responsible for the deposition of H3.3 at telomeric heterochromatin in mES cells with the aid of ATRX (α-thalassemia X-linked mental retardation protein),Citation12,Citation14 which has been shown to prefer sequences capable of forming G-quadruplexes and contributed to the specific targeting H3.3 to tolemeres and pericentric heterochromatin.Citation9,Citation17 However, the precise mechanisms of H3.3 incorporation remain obscure.
As mentioned above, H3.3 displays only several amino acid differences to canonical H3. How are these subtle differences translated into a loci-specific deposition of H3.3 with distinct functional properties? Recently, several mutational studies have been developed to specify the important roles of H3.3 specific residues in H3.3 deposition and function. The X-ray crystal structure of H3.3-H4 and DAXX revealed that the two H3.3 unique residues Ala87 and Gly90 are critical for the specific recognition of H3.3 by DAXX.Citation18,Citation19 In Arabidopsis, the four unique amino acids have been shown to serve as a guide for the dynamic deposition of H3.3 in plant chromatins, in which His87 and Leu90 in the core domain guide nucleosome assembly, while Thr31 and Tyr41 in the N-terminal tail guide nucleosome disassembly in nucleolar rDNA.Citation20 In Drosophila, the Ala87, Ile89, and Gly90 residues has been shown to be necessary for replication-independent incorporation of H3.3 into nucleosomes.Citation11 Although a single mutation at the residue Ser31 in H3.3 results no effect on the deposition of H3.3, the specific residue Ser31 in H3.3 can be phosphorylated, which has been found to be enriched at the telomeres in mES cells and at pericentric heterochromatin in differentiated cells during mitosis.Citation21,Citation22 But how these amino acid differences in H3.3 affect the properties of chromatin structures still remains to be addressed. Recently, we have conducted in vitro biophysical experiments to address this problem.Citation23 We found H3.3 greatly impairs higher-ordered chromatin folding, although it has no significant effect on the stability of mononucleosomes. Point-mutational analysis showed that all the four unique residues contribute to hinder the compaction of chromatin arrays. In addition, the incorporation of H3.3 can counteract the H2A.Z-mediated chromatin compaction, which is mainly caused by the two residues Ile89 and Gly90. Furthermore, H3.3 has also been found to counteract the association of the linker histone H1 in Drosophila; RNAi knockdown of H3.3 substantially increased the genomic binding of H1 and resulted an increase of nucleosome length.Citation24 The opening of higher-ordered chromatin structure caused by H3.3 incorporation may play an important role in connecting H3.3 with active transcription.
Dynamics of H3.3 at Enhancers and Promoters
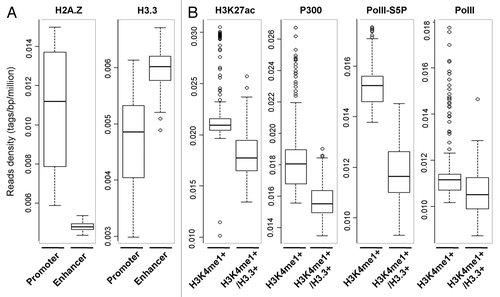
In mammalian cells the nucleosomes present at promoters and enhancers are dynamically regulated to accommodate binding of transcriptional factors and RNA polymerases machineries during the process of gene transcription. The deposition of histone variants into these regions will critically impact the properties of nucleosomes and/or chromatin and regulate transcriptional activity. Genome-wide analysis has shown that H3.3 is largely enriched in the actively transcribed genes, transcriptional factor binding sites, and telomeres in mammalian cells.Citation9,Citation25 Interestingly, our genomic analysis show that in mES cells the binding of H3.3 at enhancer regions is relatively higher than that at promoter regions, while the enrichment of H2A.Z is quite the reverse (). Furthermore, enhancer regions are bisected by H3.3 enrichment or not. The enhancers containing high level of H3.3 display lower level of H3K27ac, p300, and Pol II-Ser5p, which are mostly believed as “active enhancer” markers (). Our result indicates that the enhancers enriched with H3.3 exhibit distinct features compared with that without H3.3 enrichment, which suggests that H3.3 might be an epigenetic marker for the “poised enhancer.” Consistently, a recent study has shown that in hela cells, H3.3 largely colocalizes with HIRA complex at poised enhancers, the regions enriched of H3K4me1 but lack of H3K27ac and p300.Citation26 Further detailed analysis of the distribution of H3.3 at active and inactive genes has shown H3.3 enrichment at the promoters of transcribed gene but not of genes that are not expressed.Citation26,Citation27 In addition, the investigation on the dynamic incorporation of H3.3 in mouse embryonic fibroblasts (MEFs) also revealed that the H3.3-containing nucleosomes at enhancers and promoters undergo rapid turnover, which may remain chromatin at an “immature” uncondensed state and keep DNA highly accessible at these regions.Citation28 However, how and when H3.3 incorporates into these regions and whether the deposition of H3.3 drives or just reflects the transcriptional activity still remains to be elucidated. In order to address this problem, the dynamic incorporation of H3.3 and the related chromatin dynamics at regulatory elements during transcription activation have been investigated specifically for retinoid acid (RA)-induced genes.Citation23 Time-course ChIP analysis revealed that the incorporation of H3.3 at the enhancers occurs in the resting stages of transcription and prior to gene induction, with low level of H3.3 at the promoter.Citation23 In addition, EpiQ and genome-wide MNase-seq analysis demonstrated that the chromatins at enhancer regions enriched with H3.3 display a relative open conformation, which may facilitate the recognition and binding of transcriptional activators at this region.Citation23 Knockdown of H3.3 resulted in chromatin compaction and impaired the subsequent binding of RAR, TBP, and Pol II on the enhancers and greatly inhibited gene expression during tRA induction.Citation23 All the results indicated that the incorporation of H3.3 actively marks the enhancer region for gene activation. When the genes are activated by tRA, the H3.3-containing nucleosomes are rapidly displaced from enhancers; by contrast, the H3.3 is incorporated into the promoter regions in the meanwhile. The H3.3 deposition at the promoter is concomitant with gene activation. It is clear that the incorporation of H3.3 into enhancers and promoters occurs separately at different times during the process of transcription, but whether they are accomplished by different mechanisms still remains to be clear. It would also be interesting to investigate whether HIRA, DAXX, or other undetermined H3.3 specific chaperones account for these two different dynamic processes of H3.3 at enhancers and promoters during gene activation.
Figure 1. Genome-wide distributions of H2A.Z and H3.3 and their correlation with chromatin signatures. (A) Box-plots show the genome-wide binding pattern of H2A.Z and H3.3 between the promoters and enhancers in mES cells, in which the enhancers are defined as the Oct4/Sox2/Nanog cobound regions, and the promoters are from each enhancer-promoter units as described in reference Citation56. (B) Box-plots show the enrichments of H3K27ac, p300, Pol II, and Pol II-Ser5p at the region enriched of H3K4me1 with a high level of H3.3 (H3K4me1+/H3.3+) or not (H3K4me1+). H3K4me1 enriched region has been trimmed by subtracting TSS ± 2kb as described previously.Citation56 The y-axis value in panels A and B corresponds to reads density which is defined as the number of tags per bp per million. All ChIP-seq raw data comes from public databanks as follows: H2A.Z (GSE39237),Citation57 H3.3 (GSE16893), H3K4me1/3, H3K27ac, p300, and Pol II (GSE49847); Pol II-Ser5p (GSE20530).Citation58
Coregulation of H2A.Z with H3.3 in Gene Transcription
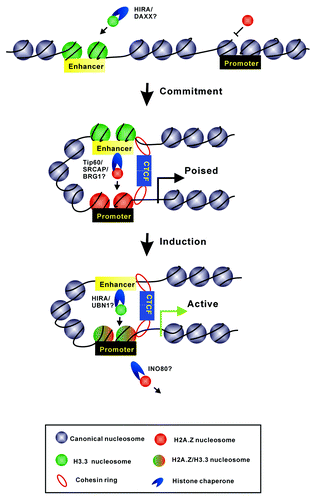
The variant H2A.Z has been implicated in a wide range of DNA-mediated processes including transcription, DNA repair, and genomic stability. Numerous studies have shown that H2A.Z may play critical roles in heterochromatin formation; for instance, H2A.Z is shown to be required for proper centromere function by maintaining the integrity of pericentric heterochromatin, located in the boundary region to prevent the spread of heterochromatin into euchromatin, and co-localized with heterochromatin protein HP1α at various constitutive heterochromatic domains in different mammalian cell types.Citation29-Citation31 In addition, depletion of H2A.Z results in the disruption of constitutive heterochromatin and defects in chromosome segregation process.Citation32 However, H2A.Z has also been revealed to be involved in gene activation; H2A.Z antagonizes DNA methylation along the whole genome in plants and animals and is enriched at the promoters of inducible genes to poise genes for rapid transcriptional activation.Citation5,Citation33-Citation36 Genome-wide localization of H2A.Z shows that the variant flanks the nucleosome-free region at transcription start sites (TSSs) in a wide range of cell types.Citation8,Citation37,Citation38 In addition, the variant H2A.Z is also known as an interacting partner of H3.3; H2A.Z has been revealed to be colocalized with H3.3 at active promoters and many other regulatory regions.Citation8,Citation39 It is of particular interest to investigate how the two variants correlate with each other in the process of gene regulation. We have investigated the coregulation of H3.3 and H2A.Z at the enhancer and promoter regions during transcription activation of RA-induced genes.Citation23 We found that H2A.Z is actively recruited to promoters prior to gene induction and rapidly replaced during transcriptional activation. The deposition of H2A.Z into the promoter region prior to gene activation is dependent on the existence of H3.3 at the enhancer region. H3.3 knockdown greatly impairs the recruitment of the histone acetyltransferase complex and chromatin remodelers (such as Tip 60, SRCAP, and BRG1) to deposit H2A.Z at the promoter region. On the other hand, knockdown of H2A.Z does not affect the H3.3 incorporation at enhancers and promoters. It is of great interest to address how the deposition of H3.3 at enhancer region affects the incidences occurred at the promoter region, which is at least several kilobases far from the enhancer region. Notably, the two histone variants H2A.Z and H3.3 have also been found to mark CTCF (CCCTC-binding factor)-binding sites along the genome and might be important for the bindings of CTCF and cohesin to mediate higher-ordered chromatin organization.Citation40,Citation41 Interestingly, another recent study showed that HIRA/UBN1/ASF1a, which account for H3.3 incorporation, were found to biochemically interact with both CTCF and BRG1, a subunit of SWI/SNF which has been shown to function at the exchange of H2A.Z; in addition, genome-wide analysis showed that HIRA/UBN1/ASF1a colocalize with SWI/SNF.Citation26 All the studies give us a clue that the histone variants may provide specialized chromatin signatures for the loading and assembly of protein complexes (such as CTCF and cohesin complex) that mediate the form of chromatin loops to connect the enhancers and promoters ().
Figure 2. A model displays the dynamic regulation of H2A.Z and H3.3 at enhancer and promoter regions during gene transcriptional activation. The enrichment of H3.3 at enhancer regions facilitates the recruitment of H2A.Z into promoters prior to gene activation. Such variants’ incorporation might provide specialized chromatin signatures to mediate the formation of “chromatin loops” to connect the enhancers and promoters. During gene induction, H3.3-containing nucleosomes would be evicted from enhancers for the binding of transcriptional factors; meanwhile, H2A.Z at promoters would be replaced by H2A, which is accompanied by the deposition of H3.3. The model also depicts several histone chaperone candidates that might take part into this procedure.
As mentioned above, H2A.Z has been found to function both in gene activation and silencing. Many biophysical analyses have shown that H2A.Z itself can enhance the stability of histone octamers and nucleosomes reconstituted in vitro.Citation23,Citation42,Citation43 Moreover, the incorporation of H2A.Z can promote the folding of chromatin arrays, including H1- and HP1α-mediated chromatin folding, to form a compact higher-ordered structure.Citation23,Citation31,Citation44 It is reasonable to speculate that H2A.Z would repress or poise the gene transcription. Indeed, our FRET, magnetic tweezer, AUC, and EM studies showed the stabilization and condensation effect of H2A.Z on nucleosome and/or chromatin structures.Citation23 In addition, our in vitro transcription assay demonstrated that the incorporation of H2A.Z can inhibit transcriptional activity in the chromatin level.Citation23 Knockdown of H2A.Z in cells impaired the compact structure of chromatin at promoter regions and promoted the RA-induced expression of Cyp26A1 and HoxA1.Citation23 All the data agree well with the repressive role of H2A.Z in gene functions. Then, how can H2A.Z function in transcriptional activation? The apparently contradictory and complicated roles of H2A.Z might be caused by different combinations with other histone variants or specific histone modifications. H2A.Z has been found to colocalize with H3.3 in the nucleosomes located at TSS of active promoters and other regulatory regions, which exhibits a much more unstable structure and higher turnover rates in vivo than the canonical and single-variant containing nucleosomes.Citation8,Citation39,Citation45 Our biophysical analyses have shown that although the incorporation of double variant H2A.Z-H3.3 resulted in a more stable nucleosome compared with the canonical one, it impaired the compaction of higher-ordered chromatin structure and promoted the in vitro transcriptional activity at chromatin template.Citation23 In addition, our ChIP analysis showed that during the early stage of gene induction, the H2A.Z was rapidly replaced by H2A, which was accompanied by a significant increase of H3.3 at the promoter regions.Citation23 The dynamic displacement of histone variant H2A.Z may increase the ratio of heterotypic H2A.Z-H2A nucleosomes existing at the promoter regions. Recently, the unstable heterotypic H2A.Z-H2A nucleosomes have been observed to be enriched around TSSs for active genes in mES cells.Citation41 Based on the crystal structure of H2A.Z-containing nucleosome, the steric clashes between the L1 loop regions of H2A and H2A.Z have been observed to restrict the stability of hybrid H2A.Z-H2A nucleosomes.Citation46 In addition, the different combinations of H2A.Z with specific histone modifications may also contribute to the complicated functions of H2A.Z in gene regulation. H2A.Z-containing nucleosomes in mammalian cells have also been found to be enriched for H3K4 methylation with less H3K9 methylation compared with the H2A-containing nucleosomes; acetylation of H2A.Z N-terminus is a hallmark of active genes while ubiquitylation and sumolyation at H2A.Z C-terminus have been implicated in heterochromatin formation and DNA repair, including the inactive X chromosome of female cells.Citation47
H3.3 in Epigenetic Inheritance
Most recently, epigenetic regulations, including histone variants, have emerged as key players to control the cell fate by modulating the chromatin state and gene transcription during development and differentiation. Cell differentiation is believed to be mediated by lineage-determining transcription factors, which activate differentiation-specific genes by binding to the corresponding regulatory regions with appropriate chromatin features. Once a differentiated cell identity is established, the cell lineage remains stable and unlikely to change over many cell divisions. It is generally believed that the faithful inheritance of the epigenetic signature from mother to daughter cells, named as “epigenetic memory,” is crucial for the maintenance of a cell identity. It’s very interesting to investigate how a lineage-committed by undifferentiated cell maintains the ability to specifically activate the appropriate differentiation program upon differentiation signaling and how the established epigenetic information could be propagated to the next cell generation. Intriguingly, H3.3 has been shown to play a key role in global changes in the transcriptome linked to cellular fate and has been implicated in cell differentiation; it was found at many developmental regulatory genes that are bivalent genes marked with transcription-repressing H3K27me3 and transcription-activating H3K4me3 in mES cells. HIRA-dependent deposition of H3.3 has been shown to be required for transcriptional reprogramming of mammalian nuclei transplanted to Xenopus oocytes;Citation48 which also has been found to facilitate the recruitment of PRC2 for proper establishment of H3K27me3 at the promoters of developmentally regulated genes in mES cells.Citation49 For skeletal muscle differentiation, MyoD, the myogenic transcription factor, recruits Chd2, a member of the SNF2 family of chromatin remodeling enzymes, specifically at myogenic gene promoters to mediate H3.3 deposition into the regulatory regions prior to the onset of myogenic gene expression.Citation50 Other than the faithful replication of DNA methylation by Dnmt1, which provides a plausible mechanism for the propagation of a silent gene state, the deposition of H3.3 has been found to be associated with the persistence of epigenetic memory of an active gene state in Xenopus nuclear transplant embryos.Citation51 In addition, our results show that H3.3 is actively incorporated at enhancers prior to gene activation and primes the transcription of RA-regulated genes, indicating that H3.3 might play an important role in “epigenetic memory” for the inducible genes including RA-regulated genes. However, the mechanism by which H3.3 is inherited at enhancer regions of RA-regulated genes still remains to be clear. The replication-independent assembly of H3.3 will dilute H3.3 nucleosomes by a factor of 2 during each cell cycle. Although the canonical H3-H4 tetramers rarely split into H3-H4 dimers during cell division, approximately 10% of H3.3-H4 tetramers do experience splitting events in each cell cycle, which are remarkably enriched at cell-type specific enhancers at genome-wide level,Citation52,Citation53 which may provide a possible mechanism for H3.3 inheritance at these regions.
Conclusion
Numerous studies have emphasized that various epigenetic mechanisms (such as DNA methylation, histone modifications, histone variants, and architectural proteins) are involved in the generation of stable and inheritable epigenetic complexity through organizing genome into functionally distinct higher-ordered chromatin domains. However, the mechanisms by which the featured chromatin signatures generated by distinct epigenetic factors are inherited from generation to generation during cell division or organism propagation in which new chromatin features need to be established, still remains unclear. In particular, whether H3.3 incorporation at enhancers is crucial for transcriptional memory or epigenetic inheritance during cell division or organism propagation will need further investigation. In this context it is of great interest to analyze whether H3.3 can cooperate with other histone variants (such as H2A.Z) or DNA and/or histone modifications to generate a transcriptionally permissive state for the differentiation-specific genes by modulating the chromatin dynamics at regulatory regions of the genome during development and differentiation. In future studies in order to illuminate the roles of histone variants in the regulation of higher-ordered chromatin organization, it will also be of great interest to analyze the dynamics of histone variants upon gene activation during development by ChIP-seq assays and CRISPR/Cas or TALEs imaging techniques developed recently.Citation54,Citation55
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
The work was supported by grants from the Ministry of Science and Technology (2011CB966300 to G.L.), the National Natural Science Foundation of China (Nos. 91219202, 91019007, and 31071147 to G.L. and 31000566 to P.C.), Chinese Academy of Sciences (CAS) Strategic Priority Research Program (XDA01010304 to G.L.) and Key Research Program (KJZD-EW-L05 to P.C), and the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry to P.C.
References
- Varga-Weisz PD, Becker PB. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 2006; 16:151 - 6; http://dx.doi.org/10.1016/j.gde.2006.02.006; PMID: 16503135
- Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 2011; 21:175 - 86; http://dx.doi.org/10.1016/j.gde.2011.01.022; PMID: 21342762
- Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 2010; 11:264 - 75; http://dx.doi.org/10.1038/nrm2861; PMID: 20197778
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 2005; 3:e384; http://dx.doi.org/10.1371/journal.pbio.0030384; PMID: 16248679
- Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, Milburn PJ, Parish CR, Tremethick DJ, Rao S. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol 2009; 29:1972 - 86; http://dx.doi.org/10.1128/MCB.01590-08; PMID: 19158270
- Dalvai M, Bellucci L, Fleury L, Lavigne AC, Moutahir F, Bystricky K. H2A.Z-dependent crosstalk between enhancer and promoter regulates cyclin D1 expression. Oncogene 2013; 32:4243 - 51; http://dx.doi.org/10.1038/onc.2012.442; PMID: 23108396
- Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 2005; 19:804 - 14; http://dx.doi.org/10.1101/gad.1259805; PMID: 15774717
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 2009; 41:941 - 5; http://dx.doi.org/10.1038/ng.409; PMID: 19633671
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140:678 - 91; http://dx.doi.org/10.1016/j.cell.2010.01.003; PMID: 20211137
- Frank D, Doenecke D, Albig W. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene 2003; 312:135 - 43; http://dx.doi.org/10.1016/S0378-1119(03)00609-7; PMID: 12909349
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 2002; 9:1191 - 200; http://dx.doi.org/10.1016/S1097-2765(02)00542-7; PMID: 12086617
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 2010; 20:351 - 60; http://dx.doi.org/10.1101/gr.101477.109; PMID: 20110566
- Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 2010; 12:853 - 62; http://dx.doi.org/10.1038/ncb2089; PMID: 20676102
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A 2010; 107:14075 - 80; http://dx.doi.org/10.1073/pnas.1008850107; PMID: 20651253
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004; 116:51 - 61; http://dx.doi.org/10.1016/S0092-8674(03)01064-X; PMID: 14718166
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 2011; 44:928 - 41; http://dx.doi.org/10.1016/j.molcel.2011.12.006; PMID: 22195966
- Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 2010; 24:1253 - 65; http://dx.doi.org/10.1101/gad.566910; PMID: 20504901
- Elsässer SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 2012; 491:560 - 5; http://dx.doi.org/10.1038/nature11608; PMID: 23075851
- Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, Zhang Z, Li G, Xu RM. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol 2012; 19:1287 - 92; http://dx.doi.org/10.1038/nsmb.2439; PMID: 23142979
- Shi L, Wang J, Hong F, Spector DL, Fang Y. Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes. Proc Natl Acad Sci U S A 2011; 108:10574 - 8; http://dx.doi.org/10.1073/pnas.1017882108; PMID: 21670303
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res 2009; 19:404 - 14; http://dx.doi.org/10.1101/gr.084947.108; PMID: 19196724
- Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, Allis CD. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci U S A 2005; 102:6344 - 9; http://dx.doi.org/10.1073/pnas.0502413102; PMID: 15851689
- Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, Huang L, Wen Z, Li W, Li X, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 2013; 27:2109 - 24; http://dx.doi.org/10.1101/gad.222174.113; PMID: 24065740
- Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J 2009; 28:3635 - 45; http://dx.doi.org/10.1038/emboj.2009.301; PMID: 19834459
- Jin C, Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc Natl Acad Sci U S A 2006; 103:574 - 9; http://dx.doi.org/10.1073/pnas.0509974103; PMID: 16407103
- Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, Adams PD. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep 2013; 3:1012 - 9; http://dx.doi.org/10.1016/j.celrep.2013.03.026; PMID: 23602572
- Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep 2005; 6:354 - 60; http://dx.doi.org/10.1038/sj.embor.7400366; PMID: 15776021
- Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol 2013; 14:R121; http://dx.doi.org/10.1186/gb-2013-14-10-r121; PMID: 24176123
- Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci U S A 2007; 104:525 - 30; http://dx.doi.org/10.1073/pnas.0607870104; PMID: 17194760
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 2003; 112:725 - 36; http://dx.doi.org/10.1016/S0092-8674(03)00123-5; PMID: 12628191
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell 2004; 16:655 - 61; http://dx.doi.org/10.1016/j.molcel.2004.10.023; PMID: 15546624
- Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol 2004; 11:650 - 5; http://dx.doi.org/10.1038/nsmb786; PMID: 15195148
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 2008; 456:125 - 9; http://dx.doi.org/10.1038/nature07324; PMID: 18815594
- Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res 2010; 20:1383 - 90; http://dx.doi.org/10.1101/gr.106542.110; PMID: 20709945
- Li B, Pattenden SG, Lee D, Gutiérrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A 2005; 102:18385 - 90; http://dx.doi.org/10.1073/pnas.0507975102; PMID: 16344463
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 2008; 29:611 - 24; http://dx.doi.org/10.1016/j.molcel.2008.02.010; PMID: 18342607
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 37; http://dx.doi.org/10.1016/j.cell.2007.05.009; PMID: 17512414
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell 2010; 39:901 - 11; http://dx.doi.org/10.1016/j.molcel.2010.08.026; PMID: 20864037
- Henikoff S. Labile H3.3+H2A.Z nucleosomes mark ‘nucleosome-free regions’. Nat Genet 2009; 41:865 - 6; http://dx.doi.org/10.1038/ng0809-865; PMID: 19639024
- Millau JF, Gaudreau L. CTCF, cohesin, and histone variants: connecting the genome. Biochem Cell Biol 2011; 89:505 - 13; http://dx.doi.org/10.1139/o11-052; PMID: 21970734
- Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol 2012; 19:1076 - 83; http://dx.doi.org/10.1038/nsmb.2424; PMID: 23085713
- Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem 2004; 279:24274 - 82; http://dx.doi.org/10.1074/jbc.M313152200; PMID: 15020582
- Hoch DA, Stratton JJ, Gloss LM. Protein-protein Förster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A.Z. J Mol Biol 2007; 371:971 - 88; http://dx.doi.org/10.1016/j.jmb.2007.05.075; PMID: 17597150
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 2002; 9:172 - 6; http://dx.doi.org/10.1038/nsb0402-316b; PMID: 11850638
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 2007; 21:1519 - 29; http://dx.doi.org/10.1101/gad.1547707; PMID: 17575053
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol 2000; 7:1121 - 4; http://dx.doi.org/10.1038/81971; PMID: 11101893
- Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol 2007; 27:6457 - 68; http://dx.doi.org/10.1128/MCB.00241-07; PMID: 17636032
- Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 2012; 5:17; http://dx.doi.org/10.1186/1756-8935-5-17; PMID: 23102146
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsässer SJ, Chapgier A, Goldberg AD, Canaani E, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 2013; 155:107 - 20; http://dx.doi.org/10.1016/j.cell.2013.08.061; PMID: 24074864
- Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S, Kumamaru H, Saiwai H, Tsubota T, Kurumizaka H, et al. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J 2012; 31:2994 - 3007; http://dx.doi.org/10.1038/emboj.2012.136; PMID: 22569126
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 2008; 10:102 - 9; http://dx.doi.org/10.1038/ncb1674; PMID: 18066050
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 2010; 328:94 - 8; http://dx.doi.org/10.1126/science.1178994; PMID: 20360108
- Huang C, Zhang Z, Xu M, Li Y, Li Z, Ma Y, Cai T, Zhu B. H3.3-H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet 2013; 9:e1003558; http://dx.doi.org/10.1371/journal.pgen.1003558; PMID: 23754967
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013; 155:1479 - 91; http://dx.doi.org/10.1016/j.cell.2013.12.001; PMID: 24360272
- Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol 2013; 20:1321 - 4; http://dx.doi.org/10.1038/nsmb.2680; PMID: 24096363
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153:307 - 19; http://dx.doi.org/10.1016/j.cell.2013.03.035; PMID: 23582322
- Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol 2012; 13:R85; http://dx.doi.org/10.1186/gb-2012-13-10-r85; PMID: 23034477
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell 2010; 141:432 - 45; http://dx.doi.org/10.1016/j.cell.2010.03.030; PMID: 20434984
