Abstract
A conserved organizational feature of eukaryotic nuclei is the peripheral heterochromatin compartment, which provides a protected area for epigenetically silent genes and gene-poor DNA. In metazoan cells this compartment is associated with the nuclear lamina, the protein meshwork at the inner edge of the nucleus. Heterochromatin-nuclear lamina interactions promote epigenetic gene silencing, which may drive many normal and diseased biological processes. We recently obtained evidence that a previously unstudied human protein, PRR14, participates in the tethering of heterochromatin to the inner nuclear periphery. PRR14 associates with the nuclear lamina and attaches to heterochromatin through its binding partner, heterochromatin protein 1 (HP1). After disassembly early in mitosis, PRR14 reassembles in two steps, first binding to anaphase chromosomes through HP1, followed by association with the nuclear lamina in telophase. PRR14 may thereby play a role in specifying HP1-bound heterochromatin for reattachment to the nuclear lamina at mitotic exit. Here we review the relevant literature, summarize our initial work, and provide additional comments and findings.
The three-dimensional organization of chromatin within the nucleus is a key facet of epigenetic control of gene expression and genome stability.Citation1-Citation3 One such organizational feature is the positioning of heterochromatin domains derived from different chromosomes within a compartment at the inner periphery of eukaryotic nuclei.Citation3-Citation10 This peripheral heterochromatin compartment is attached to the nuclear envelope and houses epigenetically silent genes and gene-poor DNA. There is evidence that the attachment of genes to the nuclear periphery promotes epigenetic gene silencing in both engineered and biological settings.Citation7,Citation11 The peripheral heterochromatin compartment is visualized as associating with the nuclear envelope, but the nature, breadth, and redundancies of the biochemical attachment mechanisms remain to be fully revealed. How these attachments might be faithfully restored during cell division is also largely unknown.
In metazoan cells, the peripheral heterochromatin compartment attaches to the nuclear lamina, the protein framework that lies under the inner nuclear membrane. The nuclear lamina is a meshwork composed of intermediate filament proteins, the A- and B-type lamins: Lamin A/C, Lamin B1, and Lamin B2.Citation12-Citation14 Important biological roles for the nuclear lamina as a docking site for peripheral heterochromatin continue to emerge within many normal and disease processes, including development,Citation11 cancer,Citation15,Citation16 aging,Citation17 and cell senescence.Citation18-Citation20 In diseases of the nuclear lamina, the laminopathies,Citation13,Citation21-Citation23 as well as in cancer, nuclear deformities are observed.Citation16 There are several non-mutually exclusive models regarding how these nuclear defects may contribute to disease, including effects on gene expression.Citation16 In particular, it will be important to explore nuclear lamina-chromatin attachment mechanisms in normal and diseased cells, as differential interactions may affect gene expression and thereby underlie disease phenotypes.Citation24 In view of the broad biological impact of nuclear lamina-heterochromatin attachment, it will be critical to obtain a comprehensive view of the attachment mechanisms.
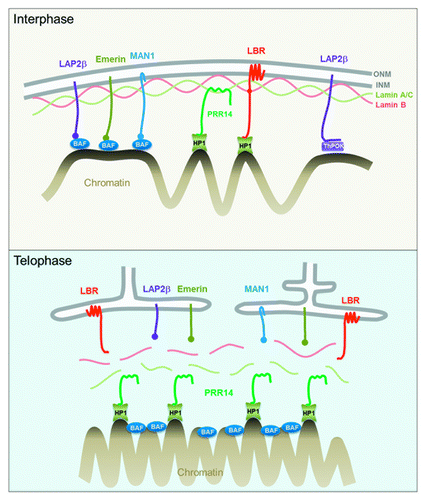
There are several known mechanisms by which heterochromatin, or generic chromatin, can attach to the nuclear lamina (, top). A group of inner nuclear membrane proteins that serve to anchor the nuclear lamina also participate in chromatin attachment through BAF, a chromatin-binding adaptor protein.Citation25 The Lamin B receptor (LBR), a multifunctional inner nuclear membrane protein, binds the Lamin B component of the lamina and also attaches to heterochromatin.Citation26 Recent studies have identified a mechanism through which the transcription factor ThPOK attaches DNA to the nuclear periphery through GAGA-enriched DNA sequences.Citation27 Collectively, these attachment modes have been referred to as “tethering,” indicating a direct or indirect linking of chromatin to the nuclear lamina. An emerging theme in the literature is that there are likely redundancies and specialization of tethering mechanisms.
Figure 1. Top: Interphase diagram showing known chromatin-nuclear envelope tethering mechanisms, along with the role proposed for PRR14. Model depicts PRR14 as a non-membrane protein, tethering between Lamin A/C and heterochromatin-bound HP1. Bottom: Telophase model depicting a role for PRR14 in specifying heterochromatin destined for reassembly at the nuclear lamina at mitotic exit. Telophase model is based on the dynamic behavior of tagged PRR14.
An important achievement in this field was the development of methods to map DNA sequences in close contact with the nuclear lamina, lamina associated domains (LADs).Citation28,Citation29 Such studies have used both DNA modification (DamID)Citation28 and chromatin immunoprecipitation (ChIP) methods.Citation3,Citation19,Citation20,Citation30,Citation31 LAD mapping has allowed integration of LAD sequences with epigenome-wide features such as DNA methylation, histone modifications, and gene-specific transcriptional states. Remarkably, the LADs (0.1–10 Mb in length) have been found to constitute 35–40% of nuclear DNA. It appears that LADs represent epigenetically silent or gene-poor regions, and largely correspond to the peripheral heterochromatin compartment that can be visualized microscopically ().
Figure 2. Confocal imaging showing localization of GFP-tagged PRR14 at the nuclear lamina and the peripheral heterochromatin compartment as detected by anti-H3K9me3 staining.
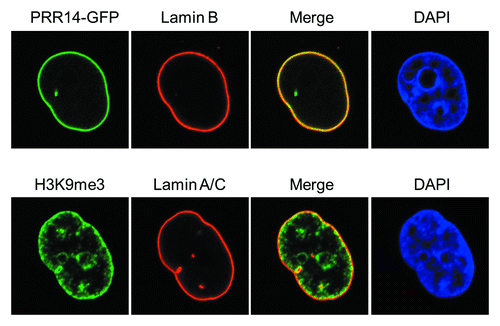
The repressive histone 3 lysine 9 di-, tri-methylation (H3K9me2/3) modification, a hallmark of epigenetically silent gene promoters and heterochromatin domains, is enriched at the inner nuclear periphery ().Citation10 The role, and maintenance, of H3K9 methylation at the nuclear periphery has recently been reviewed and we refer the reader to these excellent articles.Citation3,Citation32,Citation33 Recent work has indicated that H3K9 methylation is important for positioning of peripheral heterochromatin.Citation9,Citation34 Furthermore, H3K9me2/3-marked heterochromatin has been found to coincide with LADs,Citation34-Citation36 and the histone methyltransferase G9a that places the H3K9me2 mark was found to regulate contact of LADs with the nuclear lamina.Citation34 As H3K9 methylation appears to be important in positioning of peripheral heterochromatin, it might be expected that multiple tethering mechanisms utilize this modification. Early studies showed that the LBR tether binds to heterochromatin protein 1 (HP1), an H3K9me2/3 binding protein.Citation26,Citation37 Our recent studyCitation38 detected a new HP1-H3K9me3-dependent tether, as summarized below.
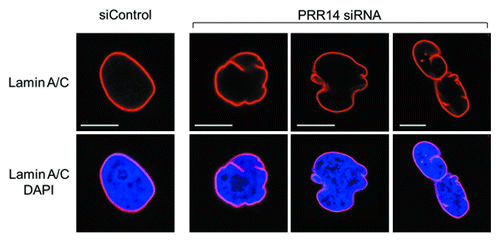
We identified a previously unstudied human protein, Proline Rich 14 (PRR14), during a genome-wide siRNA screen for epigenetic silencing factors (manuscript in preparation). The screening system utilizes reporter cells harboring chromosomally dispersed, epigenetically silent GFP transgenes that become reactivated in response to siRNA-based knockdown of epigenetic silencing factors.Citation39 The system was designed to be capable of detecting factors that have broad, rather than gene-specific roles in epigenetic silencing. The PRR14 gene is widely expressed and predicted to encode a 585 amino acid proline-rich protein. Bioinformatic analyses indicated that PRR14 lacks any identifiable functional domains.Citation38 We found that PRR14 localizes to the nuclear lamina in human interphase cells (), and its depletion results in cellular phenotypes reminiscent of laminopathies, including distorted nuclei ().Citation38 The mechanisms underlying the defects have not been fully investigated. We speculate that PRR14 may play a role in maintenance of the nuclear lamina or could also serve as part of a platform for reassembly of the nuclear lamina at mitotic exit. Notably, PRR14 has not been detected in protein-protein interaction studies using nuclear lamina or nuclear envelope components as baitCitation25,Citation40,Citation41 but has been detected in other protein-protein interaction screens (see below).
Figure 3. Confocal imaging showing nuclear lamina defects after PRR14 knockdown. Cells were transfected with the indicated siRNAs and monitored by confocal imaging 72h post-transfection. Scale bars, 10 μm.
Although full-length PRR14 localizes to the nuclear lamina, we found that the PRR14 N-terminal fragment colocalized with H3K9me3-marked heterochromatin throughout the nucleus.Citation38 This finding suggested that PRR14 was also a heterochromatin binding protein and could thereby function as a tether to mediate attachment of heterochromatin to the nuclear lamina. PRR14 had been identified as an HP1 binding partner in two independent protein-protein interaction screens,Citation42,Citation43 providing a mechanism by which PRR14 might bind heterochromatin. HP1 binds to histone H3K9me2/3 modifications on heterochromatin via its N-terminal chromodomain (CD), while the C-terminal chromoshadow domain (CSD) facilitates homo- and hetero-dimerization of HP1 family members HP1α, HP1β, and HP1γ.Citation44 HP1 functions as an adaptor protein to recruit binding partners that provide a repressive readout of H3K9 methyl marks. These HP1 binding partners frequently encode PxVxL motifs that interact with a pocket formed by dimerization of the HP1 CSDs.Citation45 HP1 proteins positioned on H3K9me2/3-marked heterochromatin thereby provide an extended platform for binding of effector proteins via the CSDs. In one of the aforementioned protein-protein interaction studies, PRR14 was detected as associating with HP1α dimers through the CSD PxVxL binding pocket.Citation42 Consistent with this study, we found that the PRR14 N-terminal fragment colocalized with HP1, and this binding was dependent on a PRR14 LAVVL motif, a variant of canonical PxVxL HP1-binding motif.Citation38 These findings provided a more detailed mechanism through which nuclear lamina-associated PRR14 could attach to, and possibly tether, heterochromatin. A role for PRR14 as a functional heterochromatin-nuclear lamina tether was supported by PRR14 siRNA-based knockdown experiments, which resulted in partial disruption of peripheral H3K9me3-marked heterochromatin.Citation38
As mentioned, LBR is also an HP1-binding nuclear lamina tether but, in apparent contrast to PRR14, its localization is membrane dependent. As we found that localization of PRR14 at the nuclear lamina required Lamina A/C, we hypothesized that PRR14 may attach heterochromatin to the nuclear lamina through Lamin A/C. Recent work has distinguished so-called “Lamin A/C-dependent” and “LBR-dependent” tethering mechanisms.Citation46 We found that, in HeLa cells, siRNA-based knockdown of LBR alone had no effect on the peripheral heterochromatin compartment, indicating the existence of redundant mechanisms.Citation38 However, knockdown of PRR14 produced significant disassociation of peripheral heterochromatin as compared with single knockdown of either Lamin A/C or LBR. As the effect of PRR14 knockdown on peripheral heterochromatin could only be reproduced by double knockdown of LBR and Lamin A/C, the tethering role of PRR14 may extend beyond attaching heterochromatin to Lamin A/C. We hypothesized that the more dramatic PRR14 knockdown phenotype might indicate a second, essential function for PRR14 during mitosis, as summarized below.Citation38
The proposal that PRR14 functions as a tether for peripheral heterochromatin suggested that its disassembly and reassembly during mitosis would be highly regulated. Indeed, the dynamic behavior of PRR14 during cell division indicated that it may serve to specify H3K9me3-HP1-marked heterochromatin for proper assembly at the nuclear lamina as cells exit mitosis, as follows (, bottom). Our working modelCitation38 encompasses the known mitotic behaviors of the HP1 protein and its ligand, the H3K9me2/3 modification.Citation47,Citation48 HP1 is bound to H3K9me2/3 in interphase to facilitate numerous processes, including heterochromatin-nuclear lamina tethering. In prophase, HP1 is ejected from H3K9me2/3 modifications through Aurora B kinase-mediated phosphorylation of the neighboring H3 serine 10 residue (H3S10P).Citation49 After ejection of HP1, the H3K9me2/3 modification is retained throughout mitosis as a stable chromatin mark.Citation48 During prophase and prometaphase, PRR14 is released from the nuclear lamina and appears to be largely excluded from chromatin.Citation38 Strikingly, PRR14 remains dispersed during metaphase, similar to the known and observed mitotic behavior of HP1 ( and ; Videos 2 and 3).
Figure 4. Further evidence for HP1-dependent localization of PRR14 on chromatin. For these experiments, HeLa cells stably expressing both GFP-PRR14 and RFP-HP1α were used. Left: Cells were treated with the indicated drugs for 3h, and images were acquired using confocal microscopy. Mock indicates untreated cells progressing to metaphase. Aurora B kinase inhibitor ZM447439 treatment resulted in prometaphase arrest and co-retention of PRR14 and HP1 on chromatin. Right: Cells were transfected with indicated siRNAs 72h prior to drug treatment. Cells were then treated with both nocodazole and ZM447439 for 3 h, and confocal images were acquired. As shown, only knockdown of all three HP1 isoforms resulted in PRR14 release from chromatin.
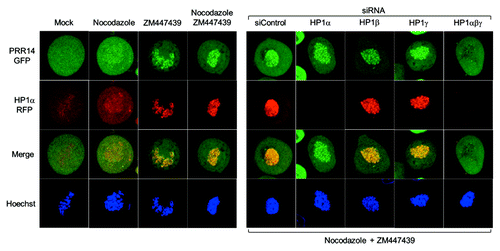
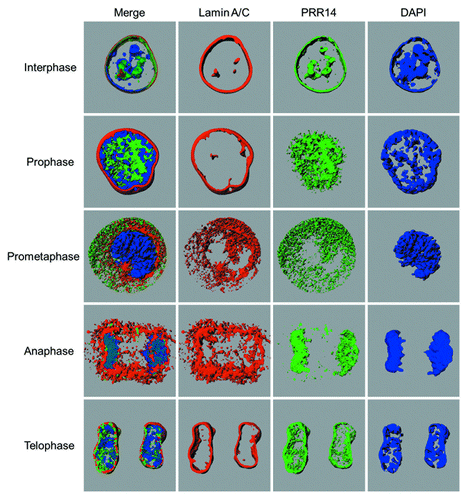
Figure 5. Snapshots from the 3D-renderings generated by Imaris software, showing localization of Lamin A/C, GFP-PRR14, and DAPI (DNA) in HeLa cells at various stages of mitosis. Videos 1–5 of 3D renderings correspond to interphase, prophase, prometaphase, anaphase, and telophase stages, respectively. The interphase nucleus shown was chosen to highlight the localization of PRR14 with nuclear channels.
To further investigate the mechanisms underlying PRR14 dynamics in early mitosis, we devised the following experiment (). Cells expressing GFP-tagged PRR14 and RFP-tagged HP1 were arrested in prometaphase using nocodazole, and, as expected, PRR14 and HP1 were largely absent from prometaphase chromatin. To test whether this pattern was related to ejection of HP1 though activity of Aurora B kinase, cells were treated with the Aurora B kinase inhibitor ZM447439. This treatment revealed binding of both PRR14 and HP1 to prometaphase chromatin. This co-retention of PRR14 and HP1 is consistent with our other finding that PRR14 binding to chromatin is HP1-dependent.Citation38 This experimental design also allowed us to address which HP1 isoforms (HP1α, HP1β, or HP1γ) are required for PRR14 chromatin binding. HP1 isoform knockdown was performed, followed by treatment with nocodazole plus ZM447439. Similar to our results with interphase cells,Citation38 knockdown of all three HP1 isoforms was required for PRR14 release from chromatin. Our work supports the role of PRR14 as a binding partner of HP1,Citation42 and it will be of interest to understand how HP1 isoforms or heterodimers contribute to PRR14 function.
After release of PRR14 from chromatin in early mitosis, PRR14 rapidly relocalizes to chromosomes at the onset of anaphase. PRR14 and HP1 were found to colocalize on anaphase chromosomes and such PRR14 binding required the PRR14 N-terminal HP1-binding motif.Citation38 Thus, anaphase chromatin binding of PRR14 appears to be HP1-dependent and likely corresponds with the H3S10P dephosphorylation that begins in anaphase to allow rebinding of HP1 to the mitotically stable H3K9me2/3 marks. These findings provide support for two mechanistically separable steps of PRR14 reassembly as a tether: a “specification step” whereby PRR14 binds to H3K9me3 on anaphase chromosomes via HP1 and an “association step” whereby PRR14 may track with H3K9me3-HP1-marked heterochromatin to the reassembling nuclear lamina at mitotic exit (, bottom). If this interpretation is correct, PRR14 may serve a “bookmarking” functionCitation50 to restore peripheral heterochromatin by guiding it to the nuclear lamina. The behavior of PRR14 throughout mitosis indicates that it is well suited to carry out this role, as it appears to remain soluble. This is in contrast to the BAF and LBR chromatin/heterochromatin binding proteins, which have membrane-associated functions during late mitosis ().Citation26,Citation51,Citation52 As opposed to the membrane-associated LBR heterochromatin binding protein, PRR14 assembles deep into chromosomes at the onset of anaphase. We speculate that the soluble nature of PRR14 allows the penetration of anaphase chromosomes for specification of HP1-bound heterochromatin. 3D-renderings of GFP-tagged PRR14 bound to anaphase chromosomes demonstrate such chromatin association (; Video S4).
A recent study used chromosome conformation capture methods to determine the three dimensional chromatin-folding states of metaphase chromosomes.Citation53 While interphase chromatin organization is locus-specific in different cell types, metaphase chromatin organization was found to be cell-type- and locus-independent; the topological associating domains (TADs) found in interphase were replaced with a linear chromatin organizational pattern. The rapid binding of PRR14 at the metaphase-anaphase transition may thereby reflect a very fundamental bookmarking role during the chromatin reorganization process from metaphase to interphase states.
Regarding a possible role for PRR14 in reassembly of peripheral heterochromatin, tracking of PRR14 and Lamin A/C during early telophase indicates that PRR14 is chromosome-associated at the time that the nuclear lamina begins to form, and only by late-telophase does PRR14 associate with the nuclear lamina ( and ). Here we provide detailed 3D renderings of this step (; Video 5). We note that our speculation and models regarding the mitotic functions of PRR14 are based largely on the dynamic behavior of tagged PRR14 proteins. Confirmation of this role will obviously require detailed functional studies.
The PRR14 gene appears to be conserved in mammals and is widely expressed in mammalian tissues, as well as early mouse development.Citation54,Citation55 The human paralogous gene, PRR14L, encodes a predicted protein of 2151 amino acids. This paralog is defined largely by strong amino acid homology within a segment near the C-termini. Interestingly, both PRR14 and PRR14L proteins were detected in proteomic studies designed to identify binding partners for the phosphatase PP2A.Citation56 This study also detected LEM-4 (ANKLE2) as a PP2A interactor. LEM-4 is a nuclear lamina-binding inner nuclear membrane protein that plays a role in PP2A recruitment for phosphoregulation of BAF.Citation57,Citation58 These findings place PRR14 as a PP2A interactor at the nuclear lamina. The assembly and disassembly of PRR14 are likely regulated by phosphorylation, consistent with this PP2A association.
During the course of our work we observed that, in addition to localization to the nuclear lamina in interphase, GFP tagged-PRR14 surrounds intranuclear channels (; Video 1). Such channels have been reported to share components with the nuclear lamina, including A- and B-type lamins.Citation59,Citation60 As reported,Citation60 we found that these channels are also associated with heterochromatin. Thus, PRR14 may also participate in tethering heterochromatin to such internal structures in addition to the peripheral nuclear lamina.Citation3
We know very little about the PRR14 protein and many fundamental questions remain regarding its functional importance. The high proline content suggests that it may form a disordered, extended structure, with the predicted folded N-terminal region attaching to HP1-marked heterochromatin.Citation38 An extended structure may fit with a role as being tightly associated with the filamentous nuclear lamina or other nucleoskeletal components. PRR14 also has a highly basic isoelectric point and is relatively arginine-serine-rich. Whether these features reflect a direct bivalent tethering mechanism, or if PRR14 is part of a more extensive complex remains to be determined. Another fundamental question relates to the nature of the heterochromatin that may be bound by PRR14 through HP1 at the nuclear periphery. Is it comprised of genes, gene promoters, or gene-poor sequences? Lastly, the behavior of PRR14 during mitosis implicates a possible role in HP1-dependent “bookmarking” of heterochromatin for return to the nuclear lamina. Although reshuffling of LADs during cell division has been reported,Citation34 it has been argued that faithful post-mitotic restoration of peripheral heterochromatin is a fundamental process.Citation3 But how might heterochromatin that is destined for the nuclear periphery at mitotic exit be selected among the collective heterochromatin? Might HP1 isoforms be involved in such specificity?
Using a functional screen, we detected a role in epigenetic gene silencing for the human PRR14 protein. From our initial study, PRR14 appears to participate in several static and dynamic processes, from heterochromatin tethering to nuclear lamina structure. Confirmation of some of these roles will require additional rigorous experimentation. Furthermore, there have been no biological functions described for PRR14 as yet, in health or disease. As knockdown of PRR14 causes misshapen nuclei as seen in cancer and laminopathies, it will be of interest to investigate possible disease connections and develop genetic models.
Additional material
Download Zip (26.8 MB)Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was supported in whole or in part by National Institutes of Health grants Roadmap Epigenomics DK082498 (R.A.K.), CA071515, and CA006927. This project is also funded in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. This work was also supported by the Fox Chase Cancer Center Keystone Program in Epigenetics and Progenitor Cells. A.P. is a recipient of an AACR Centennial Predoctoral Fellowship in Cancer Research. We thank Fabrice Roegiers and Hong Yen for critical comments and Marie Estes for assistance in preparing this manuscript. We also thank Martijn Luijsterburg for providing the mRFP-HP1α expression plasmid. The Fox Chase Cancer Center Bioimaging and Cell Sorting Facilities supported this work. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or any other sponsoring organization.
References
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 2013; 152:1270 - 84; http://dx.doi.org/10.1016/j.cell.2013.02.001; PMID: 23498936
- Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol 2013; 20:290 - 9; http://dx.doi.org/10.1038/nsmb.2474; PMID: 23463314
- Collas P, Lund EG, Oldenburg AR. Closing the (nuclear) envelope on the genome: how nuclear lamins interact with promoters and modulate gene expression. Bioessays 2014; 36:75 - 83; http://dx.doi.org/10.1002/bies.201300138; PMID: 24272858
- Bank EM, Gruenbaum Y. The nuclear lamina and heterochromatin: a complex relationship. Biochem Soc Trans 2011; 39:1705 - 9; http://dx.doi.org/10.1042/BST20110603; PMID: 22103511
- Geyer PK, Vitalini MW, Wallrath LL. Nuclear organization: taking a position on gene expression. Curr Opin Cell Biol 2011; 23:354 - 9; http://dx.doi.org/10.1016/j.ceb.2011.03.002; PMID: 21450447
- Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Curr Opin Genet Dev 2011; 21:167 - 74; http://dx.doi.org/10.1016/j.gde.2011.01.023; PMID: 21345665
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008; 452:243 - 7; http://dx.doi.org/10.1038/nature06727; PMID: 18272965
- Shevelyov YY, Nurminsky DI. The nuclear lamina as a gene-silencing hub. Curr Issues Mol Biol 2012; 14:27 - 38; PMID: 21795760
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 2012; 150:934 - 47; http://dx.doi.org/10.1016/j.cell.2012.06.051; PMID: 22939621
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 2011; 12:7 - 18; http://dx.doi.org/10.1038/nrg2905; PMID: 21116306
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 2010; 38:603 - 13; http://dx.doi.org/10.1016/j.molcel.2010.03.016; PMID: 20513434
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol 2010; 2:a000547; http://dx.doi.org/10.1101/cshperspect.a000547; PMID: 20826548
- Dittmer TA, Misteli T. The lamin protein family. Genome Biol 2011; 12:222; http://dx.doi.org/10.1186/gb-2011-12-5-222; PMID: 21639948
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol 2011; 12:695 - 708; http://dx.doi.org/10.1038/nrm3207; PMID: 21971041
- Reddy KL, Feinberg AP. Higher order chromatin organization in cancer. Semin Cancer Biol 2013; 23:109 - 15; http://dx.doi.org/10.1016/j.semcancer.2012.12.001; PMID: 23266653
- Davidson PM, Lammerding J. Broken nuclei - lamins, nuclear mechanics, and disease. Trends Cell Biol 2013; http://dx.doi.org/10.1016/j.tcb.2013.11.004; PMID: 24309562
- Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection?. Nat Rev Mol Cell Biol 2010; 11:567 - 78; http://dx.doi.org/10.1038/nrm2944; PMID: 20651707
- Cruickshanks HA, McBryan T, Nelson DM, Vanderkraats ND, Shah PP, van Tuyn J, Singh Rai T, Brock C, Donahue G, Dunican DS, et al. Senescent cells harbour features of the cancer epigenome. Nat Cell Biol 2013; 15:1495 - 506; http://dx.doi.org/10.1038/ncb2879; PMID: 24270890
- Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young AR, Narita M, Pérez-Mancera PA, Bennett DC, Chong H, et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 2013; 27:1800 - 8; http://dx.doi.org/10.1101/gad.217281.113; PMID: 23964094
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 2013; 27:1787 - 99; http://dx.doi.org/10.1101/gad.223834.113; PMID: 23934658
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet 2012; 28:464 - 71; http://dx.doi.org/10.1016/j.tig.2012.06.001; PMID: 22795640
- Gordon LB, Cao K, Collins FS. Progeria: translational insights from cell biology. J Cell Biol 2012; 199:9 - 13; http://dx.doi.org/10.1083/jcb.201207072; PMID: 23027899
- Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000760; http://dx.doi.org/10.1101/cshperspect.a000760; PMID: 20182615
- Kubben N, Adriaens M, Meuleman W, Voncken JW, van Steensel B, Misteli T. Mapping of lamin A- and progerin-interacting genome regions. Chromosoma 2012; 121:447 - 64; http://dx.doi.org/10.1007/s00412-012-0376-7; PMID: 22610065
- Montes de Oca R, Shoemaker CJ, Gucek M, Cole RN, Wilson KL. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One 2009; 4:e7050; http://dx.doi.org/10.1371/journal.pone.0007050; PMID: 19759913
- Olins AL, Rhodes G, Welch DB, Zwerger M, Olins DE. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus 2010; 1:53 - 70; PMID: 21327105
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 2012; 149:1474 - 87; http://dx.doi.org/10.1016/j.cell.2012.04.035; PMID: 22726435
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948 - 51; http://dx.doi.org/10.1038/nature06947; PMID: 18463634
- Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 2010; 22:320 - 5; http://dx.doi.org/10.1016/j.ceb.2010.04.002; PMID: 20444586
- Lund E, Oldenburg AR, Delbarre E, Freberg CT, Duband-Goulet I, Eskeland R, Buendia B, Collas P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res 2013; 23:1580 - 9; http://dx.doi.org/10.1101/gr.159400.113; PMID: 23861385
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 2013; 23:260 - 9; http://dx.doi.org/10.1101/gr.138032.112; PMID: 23152449
- Meister P, Taddei A. Building silent compartments at the nuclear periphery: a recurrent theme. Curr Opin Genet Dev 2013; 23:96 - 103; http://dx.doi.org/10.1016/j.gde.2012.12.001; PMID: 23312840
- Towbin BD, Gonzalez-Sandoval A, Gasser SM. Mechanisms of heterochromatin subnuclear localization. Trends Biochem Sci 2013; 38:356 - 63; http://dx.doi.org/10.1016/j.tibs.2013.04.004; PMID: 23746617
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013; 153:178 - 92; http://dx.doi.org/10.1016/j.cell.2013.02.028; PMID: 23523135
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 2011; 43:768 - 75; http://dx.doi.org/10.1038/ng.865; PMID: 21706001
- Jørgensen HF, Fisher AG. LOCKing in Cellular Potential. Cell Stem Cell 2009; 4:192 - 4; http://dx.doi.org/10.1016/j.stem.2009.02.007; PMID: 19265653
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem 1996; 271:14653 - 6; http://dx.doi.org/10.1074/jbc.271.25.14653; PMID: 8663349
- Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, Katz RA. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep 2013; 5:292 - 301; http://dx.doi.org/10.1016/j.celrep.2013.09.024; PMID: 24209742
- Poleshko A, Einarson MB, Shalginskikh N, Zhang R, Adams PD, Skalka AM, Katz RA. Identification of a functional network of human epigenetic silencing factors. J Biol Chem 2010; 285:422 - 33; http://dx.doi.org/10.1074/jbc.M109.064667; PMID: 19880521
- Kubben N, Voncken JW, Demmers J, Calis C, van Almen G, Pinto Y, Misteli T. Identification of differential protein interactors of lamin A and progerin. Nucleus 2010; 1:513 - 25; PMID: 21327095
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 2012; 196:801 - 10; http://dx.doi.org/10.1083/jcb.201112098; PMID: 22412018
- Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, Kimura H, Obuse C. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol 2010; 12:719 - 27; http://dx.doi.org/10.1038/ncb2075; PMID: 20562864
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005; 437:1173 - 8; http://dx.doi.org/10.1038/nature04209; PMID: 16189514
- Zeng W, Ball AR Jr., Yokomori K. HP1: heterochromatin binding proteins working the genome. Epigenetics 2010; 5:287 - 92; http://dx.doi.org/10.4161/epi.5.4.11683; PMID: 20421743
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ 3rd. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun 2005; 331:929 - 37; http://dx.doi.org/10.1016/j.bbrc.2005.04.016; PMID: 15882967
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013; 152:584 - 98; http://dx.doi.org/10.1016/j.cell.2013.01.009; PMID: 23374351
- Sugimoto K, Tasaka H, Dotsu M. Molecular behavior in living mitotic cells of human centromere heterochromatin protein HPLalpha ectopically expressed as a fusion to red fluorescent protein. Cell Struct Funct 2001; 26:705 - 18; http://dx.doi.org/10.1247/csf.26.705; PMID: 11942629
- Wang F, Higgins JM. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 2013; 23:175 - 84; http://dx.doi.org/10.1016/j.tcb.2012.11.005; PMID: 23246430
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 2005; 438:1176 - 80; http://dx.doi.org/10.1038/nature04254; PMID: 16222244
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet 2010; 11:583 - 9; http://dx.doi.org/10.1038/nrg2827; PMID: 20628351
- Wandke C, Kutay U. Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell 2013; 152:1222 - 5; http://dx.doi.org/10.1016/j.cell.2013.02.046; PMID: 23498932
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, Yamamoto A, Hiraoka Y. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci 2008; 121:2540 - 54; http://dx.doi.org/10.1242/jcs.033597; PMID: 18628300
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science 2013; 342:948 - 53; http://dx.doi.org/10.1126/science.1236083; PMID: 24200812
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 2010; 18:675 - 85; http://dx.doi.org/10.1016/j.devcel.2010.02.012; PMID: 20412781
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 2010; 6:468 - 78; http://dx.doi.org/10.1016/j.stem.2010.03.015; PMID: 20452321
- Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 2012; 337:1348 - 52; http://dx.doi.org/10.1126/science.1221483; PMID: 22984071
- Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW, Gorjánácz M. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 2012; 150:122 - 35; http://dx.doi.org/10.1016/j.cell.2012.04.043; PMID: 22770216
- Gorjánácz M. LEM-4 promotes rapid dephosphorylation of BAF during mitotic exit. Nucleus 2013; 4:14 - 7; http://dx.doi.org/10.4161/nucl.22961; PMID: 23211644
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev 2006; 86:967 - 1008; http://dx.doi.org/10.1152/physrev.00047.2005; PMID: 16816143
- Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol 1997; 136:531 - 44; http://dx.doi.org/10.1083/jcb.136.3.531; PMID: 9024685
