Abstract
Nuclear compartmentalization is achieved through the enclosure of the genome by the nuclear envelope; the nuclear envelope is perforated by nuclear pore complexes (NPCs), which form portals that control molecular exchange between the nucleus and cytoplasm. The number of NPCs per nucleus establishes a limit to the flux of molecules across the nuclear envelope and might directly impact genome organization and gene expression in a cell type specific manner. Mechanisms that control NPC number remain ill defined. Our recent study implicates a cytoplasmic pool of the nucleoporin Nsp1 as a factor that controls NPC number during the asymmetric division of budding yeast; Nsp1 acts to ensure that daughters inherit NPCs. We place our data within an emerging model of NPC inheritance in yeast and consider potential analogous mechanisms in multicellular eukaryotes, including the functional conservation of a cytoplasmic pool of Nsp1.
Introduction
Nuclear pore complexes (NPCs) form gateways in the nuclear envelope that control the exchange of molecules between the nucleus and cytoplasm in all eukaryotes. They are constructed from ~30 subunits termed nucleoporins or nups that form a ring-like scaffold and central transport channel. The number of NPCs within the nuclear envelope establishes a limit to the total flux of molecules that can permeate the nucleus at a given time. Further, the growing functional and physical connections between NPCs and chromatin support the concept that NPC number could directly influence gene expression, perhaps in a cell type specific fashion.Citation1,Citation2 Consistent with this idea, NPC number and distribution has been shown to vary between different cell typesCitation3-Citation6 and has even been correlated with cancer cell multidrug resistance.Citation7 The inputs and pathways that regulate NPC number remain to be fully uncovered.
Since there is no evidence suggesting that NPCs can be removed from the nuclear envelope, NPC number is likely controlled either by up or downregulation of the de novo NPC assembly pathway, or, as our recent work in yeast suggests, by ensuring inheritance of NPCs by daughter cells during cell division.Citation8 There has been considerable interest in understanding NPC inheritance in Saccharomyces cerevisiaeCitation8-Citation13 beyond mechanisms of NPC number control; there is also a potential relationship between NPCs and aging.Citation10,Citation14 In part, this is due to the remarkable lifetime—up to several years—of some of the NPC scaffold nups and the propensity for long lived proteins to accumulate damage with age.Citation14-Citation17 This posits that cells like yeast, which often restrict the passage of damaged or misfolded proteins to daughters in order to ensure their lifespan,Citation18 might have evolved mechanisms to restrict the transmission of NPCs as well.Citation9,Citation10 Since NPCs remain intact during mitosis in budding yeast, it provides a facile experimental system to investigate mechanisms that promote or restrict NPC transmission to daughters.
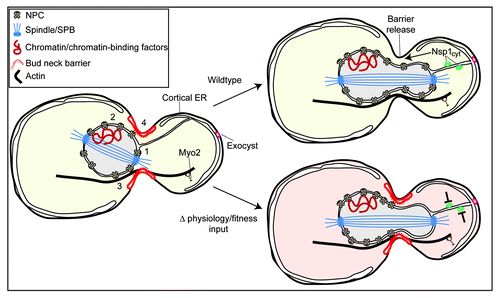
Our recent study helps to define an emerging consensus by which NPCs are transmitted to daughter cells through a mechanism that depends on four factors: (1) the dynamics of individual NPCs imposed by the asymmetric distribution of NPC binding partners; (2) a cytoplasmic pool of the nup, Nsp1; (3) a myosin motor (Myo2) and intact actin cytoskeleton; and (4) the modulation of a bud neck barrier (). In the following, we will interpret our data in the context of several additional studies that have investigated NPC inheritance and discuss the broader themes applicable to organisms that undergo an open mitosis in which NPCs break down during mitosis.
Figure 1. Mechanism of NPC inheritance in budding yeast. NPC dynamics and their segregation between mother and daughter cells are influenced by binding to cytoplasmic and nuclear structures including SPBs (1), and chromatin and/or chromatin binding partners like the LEM proteins (2). At least a subset of NPCs are likely attached to either a microtubule or actin-based cytoskeleton (3). There is a barrier at the bud neck (4) that impedes the passage of NPCs and other organelles, which likely responds to changes in cellular physiology. Under wild type conditions, Nsp1CYT is translocated into the bud through a mechanism that requires an actin cytoskeleton and Myo2. Nsp1CYT moves with ER tubules that extend from the mother nuclear envelope and contact the bud cortex; this ER might be connected to the cortex through the exocyst complex. The passage of Nsp1CYT licenses NPC passage by contributing to the dissolution of the barrier (arrow). Under conditions in which there are disruptions to bud physiology and/or fitness, we propose that Nsp1CYT function is inhibited, the barrier remains intact and NPCs are not transmitted.
Potential Models of NPC Transmission to Daughter Cells
There are at least three plausible mechanisms that could contribute to the transmission of NPCs to daughter cells during the asymmetric division of budding yeast: first, NPCs could freely diffuse along the nuclear envelope with NPC inheritance being a stochastic process largely dependent on the nuclear surface area inherited after nuclear division. Second, NPCs could be tethered to cytoplasmic or nuclear structures. In this scenario, NPC inheritance would be secondary to mechanisms that either retain or segregate the tethering factor. Third, in analogy to how several organelles are transported to daughters, NPCs could be moved by direct connections between motor proteins and NPCs. Importantly, any of the above mechanisms would be influenced by a barrier or restriction at the bud neck that could physically impede NPC transmission to the daughter ().
NPC diffusion
While individual NPCs may freely diffuse along the nuclear envelope, such a mechanism cannot explain the observation that there is an increased density of NPCs and a bias of “old” nups in daughters after anaphase.Citation8,Citation11 While an increased NPC density in daughters could also suggest enhanced daughter-specific NPC assembly, our data argue that this is not the case, as inhibiting NPC assembly does not influence NPC transmission. We also failed to detect a bias in the accumulation of newly synthesized NPC protomers in daughter cells. Perhaps most importantly, we are able to decouple nuclear envelope inheritance from NPC transmission by inhibiting the nup, Nsp1, or the myosin-V motor, Myo2.Citation8 Cumulatively, these results support that diffusion alone cannot explain NPC transmission and another mechanism is required to enrich NPCs in daughter cells.
NPC dynamics mediated by binding to cellular structures
NPCs in budding yeast are mobile within the nuclear envelopeCitation19,Citation20 suggesting that, as a population, they are not tethered to a stationary network analogous to the mammalian nuclear lamina.Citation21 However, individual NPCs interact with several cellular factors, both in the cytoplasm and nucleus that would directly influence their dynamics (). Should one of these factors be retained in mothers or segregated to daughters, this could impact the ultimate distribution of associated NPCs. A potential candidate for driving such asymmetry is the yeast centrosome (spindle pole body, SPB); NPCs have been observed clustered around SPBs,Citation22 potentially through an interaction with the nuclear basket protein, Mlp2.Citation23 Centrosome interactions might also contribute to the polarization of NPCs at the nuclear envelope observed in several organisms including PlasmodiumCitation5 and Chlamydomonas.Citation24
NPCs also interact with chromatin binding factors including the integral inner nuclear membrane proteins of the Lap2-emerin-MAN1 (LEM) family (Heh1 and Heh2),Citation25 and others reviewed in references Citation26, Citation27. Interestingly, in Schizosaccharomyces japonicus where there is a partial nuclear envelope break down during mitosis, the LEM proteins play a role in equally partitioning the nucleus and NPCs between daughters, perhaps by directly connecting NPCs to segregating chromosomes.Citation28 More insight into how interactions between NPCs and these binding factors are controlled and whether they are asymmetrically segregated between mother and daughter cells is required to fully understand their contribution to NPC transmission.
Motor-driven movement of NPCs
Several motor proteins have been shown to interact with NPCs in yeast and in multicellular eukaryotes.Citation29-Citation32 Consistent with the idea that NPCs can be coupled directly to the cytoskeleton through motor proteins, ATP-dependent movement of NPCs has been visualized in several yeasts including S. cerevisiae, where this movement also requires an intact actin cytoskeleton.Citation33 This dependence on actin might help to explain our observation that the myosin motor Myo2 is uniquely required for NPC density in daughter cells, although a direct connection between Myo2 and NPCs has not been established.Citation8 The directional movement of Nup2 foci between mother and daughter nuclei observed in budding yeastCitation11 is also consistent with an active motor-directed movement of NPCs, although since Nup2 is mobileCitation34 these results could benefit from visualizing a more stable component of the NPC. To clearly establish whether motors are required in NPC transmission will require the use of several nup markers and likely super-resolution approaches to resolve individual NPCs and monitor their movement during anaphase.
A Bud Neck Barrier Controls NPC Inheritance
We envision that under different internal or external conditions, the contribution of any of the above mechanisms to NPC transmission could be altered to either promote or inhibit the numbers of NPCs inherited by daughters (). Furthermore, at least three studies, including our own, support the existence of a physical barrier at the bud neck that acts to prevent NPC transmission to daughters.Citation8-Citation10 Controversy over its existence is in large part due to an inability to reach a consensus as to the biochemical composition of the barrier, and the physiological conditions in which the barrier might be regulated. Indeed, as described above, we observe that NPCs are able to access daughter cells during anaphase suggesting that in a permissive laboratory setting and in vegetatively growing cells, the barrier is not overly active toward NPCs. In contrast, when Nsp1 is inhibited, we observe a striking reduction of the inheritance of NPCs.Citation8 While in principle, the inhibition of Nsp1 could influence any one of the three putative mechanisms of NPC transmission outlined above, by, for example, coupling NPCs to either cellular structures or motor proteins, our data support that it is a cytoplasmic pool of Nsp1 and not the NPC pool that impacts NPC inheritance. Therefore, the most plausible interpretation of our data is that Nsp1 acts by influencing the function of the bud neck barrier ().
Functional Conservation of Nsp1CYT
Perhaps one of the more surprising and interesting findings of our work is that a pool of Nsp1 that exists outside of NPCs, which we term “Nsp1CYT,” is responsible for ensuring NPC inheritance. To show this, we used a conditional “anchor-away” approach for inhibiting nup function,Citation35 which specifically inactivated cytoplasmic nups and not those bound to NPCs. The notion that Nsp1 can function outside of the NPC is not without precedent. Indeed, a quantitative proteomic analysis of fission yeast supports that Nsp1 is by far the most abundant nup. Whereas most nups were found in ~2000 to ~7000 copies per cell, Nsp1 was found in ~40 000 copies, an order of magnitude more.Citation36 Since there is no evidence that there is 10-fold more Nsp1 in NPCs compared with other nups,Citation37,Citation38 this excess Nsp1 likely exists outside of the NPC. Moreover, in HeLa cells, the Nsp1 ortholog Nup62 localizes to the leading edge of migrating cells and contributes to cell migration; this function is mediated by an interaction with the exocyst—a protein complex best known for its role in tethering secretory vesicles to the plasma membrane during exocytosis.Citation39 Further, Nup62 interacts with the endoplasmic reticulum (ER)-resident oxysterol binding protein, ORP8,Citation40 which might compete for binding with the exocyst subunit, Exo70.Citation41 Consistent with this, downregulation of ORP8 leads to the localization of Nup62 to the cell’s leading edge and a concomitant increase in cell migration.Citation41 Lastly, Nup62 has been found at the mid-body ring prior to cell abscission, an interaction that might be mediated by the actin-cap binding protein CapG.Citation42 Cumulatively, these studies point to a role for a cytoplasmic pool of Nup62 at the interface of membrane/actin-mediated processes at both the cell edge and mid-body ring. Interestingly, we observe analogous dynamics and interactions for Nsp1CYT in budding yeast including interactions with ER and the exocyst complex.Citation8
In analogy to the polarized delivery of Nup62 to the leading edge of migrating cells, Nsp1CYT is observed as a focus that moves from the mother cell to the bud tip directly preceding anaphase. During anaphase, Nsp1CYT is directed back toward the bud neck and the elongating anaphase nucleus, where it integrates into the nuclear envelope.Citation8 The dynamics of this movement are reminiscent of a battery of growth factors influenced by the polarization of the actin cytoskeleton, which is apically polarized during bud growth and then redirected to the bud neck during anaphase.Citation43 Indeed, actin depolymerization affects the localization of Nsp1CYT and the most abundant Nsp1CYT foci are observed in the context of extreme apical growth after overstimulation of the Wee1 kinase. Further, we observe a relationship between bud size and the abundance and bud-bias of the Nsp1CYT foci. Together, these data are suggestive of a link between Nsp1CYT function and actin-mediated processes that oversee bud growth and physiology.
Bud Fitness Modulates the Bud Neck Barrier
We hypothesize that Nsp1CYT is part of a network that couples bud physiology and/or fitness to modulation of the bud neck barrier in order to license NPC transmission (). Consistent with the sensitivity of the bud neck barrier to inputs from cellular physiology, it has been recently shown that ER stress is signaled through the MAP kinase Slt2, which regulates a bud neck barrier dependent on septin function.Citation44 It is possible that Nsp1CYT functions within this or an analogous pathway to either directly (or through another factor[s]) promote barrier dissolution. While a better understanding of Nsp1CYT’s direct binding partners is a necessity to evaluate this model, we are able to show that it interacts with ER, either through direct membrane bindingCitation45 or a yet to be identified ER protein.
Nsp1CYT associates with ER tubules that extend from the mother nuclear envelope and contact the bud cortex (). We suspect that these tubules are identical to those seen being drawn into the bud from the mother nuclear envelope in tomographic reconstructions from electron microscopy of the yeast ERCitation46 and visualized by light microscopy by several groups.Citation47-Citation49 There is evidence that these tubules might be linked to the cortex through the exocyst.Citation47 Interestingly, acting through these ER-bud cortex connections, the exocyst plays a role in maintaining nuclear position at the bud neck prior to anaphase. This is achieved through the generation of forces that transiently pull the mother nuclear envelope into the bud and form so-called “nucleopodia.”Citation47 The dynamics of nucleopodia resemble “tugging”, suggesting that there is a dynamic fluctuation in the forces applied to the mother nuclear envelope. Since we show that nucleopodia are often directed toward Nsp1CYT foci, a key to unraveling the function of Nsp1CYT may be in understanding the nature of these forces and how they might translate information from the bud cortex to the mother nuclear envelope. We speculate that these forces might directly contribute to barrier dissolution, but testing this hypothesis awaits a more molecular understanding of this pathway.
Outlook to Multicellular Eukaryotes
The asymmetric division of budding yeast can serve as a model for mitoses in multicellular eukaryotes that result in daughter cells with distinct cell fates. While NPCs are thought to break down during mitosis into a small number of discrete subcomplexes, it seems feasible that the segregation of these building blocks might be biased to one daughter by diffusion barriers or the attachment to a polarity and/or motor-driven machinery, or to centrosomes. We suggest that this might be necessary because it is likely more efficient to reassemble NPCs from a small subset of preformed building blocks rather than from producing and assembling ~30 individual subunits. Moreover, the observation that interphase de novo NPC assembly is arrested in post-mitotic cells might place an additional burden on the inheritance of nups at the last mitosis before these cells enter quiescence.Citation14 How Nsp1/Nup62 might contribute to these putative mechanisms awaits further investigation and will require the capacity to visualize “new” and “old” nups through cell division and the completion of differentiation programs. The convergence of gene-editing technologies where fluorescent protein tags can be engineered into endogenous genes with super-resolution microscopy capable of visualizing the NPC substructure, promises to open the door for these experiments.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Megan King, Brant Webster, Jens Jäger, and Kristen Swithers for comments on the manuscript and Jordan Myers for help with the model. C.P.L. and P.C. are supported by a grant from the National Institutes of Health (RO1GM105672) to C.P.L.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet 2007; 8:507 - 17; http://dx.doi.org/10.1038/nrg2122; PMID: 17549064
- Köhler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell 2010; 38:6 - 15; http://dx.doi.org/10.1016/j.molcel.2010.01.040; PMID: 20385085
- Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J 2009; 59:243 - 55; http://dx.doi.org/10.1111/j.1365-313X.2009.03865.x; PMID: 19392704
- Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, Imamoto F, Imamoto N. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci 2006; 119:4442 - 51; http://dx.doi.org/10.1242/jcs.03207; PMID: 17074834
- Weiner A, Dahan-Pasternak N, Shimoni E, Shinder V, von Huth P, Elbaum M, Dzikowski R. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell Microbiol 2011; 13:967 - 77; http://dx.doi.org/10.1111/j.1462-5822.2011.01592.x; PMID: 21501361
- Maul GG, Deaven LL, Freed JJ, Campbell GL, Beçak W. Investigation of the determinants of nuclear pore number. Cytogenet Cell Genet 1980; 26:175 - 90; http://dx.doi.org/10.1159/000131439; PMID: 6966999
- Lewin JM, Lwaleed BA, Cooper AJ, Birch BR. The direct effect of nuclear pores on nuclear chemotherapeutic concentration in multidrug resistant bladder cancer: the nuclear sparing phenomenon. J Urol 2007; 177:1526 - 30; http://dx.doi.org/10.1016/j.juro.2006.11.048; PMID: 17382772
- Colombi P, Webster BM, Fröhlich F, Lusk CP. The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. J Cell Biol 2013; 203:215 - 32; http://dx.doi.org/10.1083/jcb.201305115; PMID: 24165936
- Makio T, Lapetina DL, Wozniak RW. Inheritance of yeast nuclear pore complexes requires the Nsp1p subcomplex. J Cell Biol 2013; 203:187 - 96; http://dx.doi.org/10.1083/jcb.201304047; PMID: 24165935
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature 2008; 454:728 - 34; PMID: 18660802
- Khmelinskii A, Keller PJ, Bartosik A, Meurer M, Barry JD, Mardin BR, Kaufmann A, Trautmann S, Wachsmuth M, Pereira G, et al. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol 2012; 30:708 - 14; http://dx.doi.org/10.1038/nbt.2281; PMID: 22729030
- Khmelinskii A, Keller PJ, Lorenz H, Schiebel E, Knop M. Segregation of yeast nuclear pores. Nature 2010; 466:E1; http://dx.doi.org/10.1038/nature09255; PMID: 20651645
- Menendez-Benito V, van Deventer SJ, Jimenez-Garcia V, Roy-Luzarraga M, van Leeuwen F, Neefjes J. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc Natl Acad Sci U S A 2013; 110:175 - 80; http://dx.doi.org/10.1073/pnas.1207424110; PMID: 23248297
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009; 136:284 - 95; http://dx.doi.org/10.1016/j.cell.2008.11.037; PMID: 19167330
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013; 154:971 - 82; http://dx.doi.org/10.1016/j.cell.2013.07.037; PMID: 23993091
- Toyama BH, Hetzer MW. Protein homeostasis: live long, won’t prosper. Nat Rev Mol Cell Biol 2013; 14:55 - 61; http://dx.doi.org/10.1038/nrm3496; PMID: 23258296
- Savas JN, Toyama BH, Xu T, Yates JR 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science 2012; 335:942; http://dx.doi.org/10.1126/science.1217421; PMID: 22300851
- Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep 2012; 2:738 - 47; http://dx.doi.org/10.1016/j.celrep.2012.08.024; PMID: 23022486
- Bucci M, Wente SR. In vivo dynamics of nuclear pore complexes in yeast. J Cell Biol 1997; 136:1185 - 99; http://dx.doi.org/10.1083/jcb.136.6.1185; PMID: 9087436
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol 1997; 136:747 - 59; http://dx.doi.org/10.1083/jcb.136.4.747; PMID: 9049242
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol 2001; 154:71 - 84; http://dx.doi.org/10.1083/jcb.200101089; PMID: 11448991
- Winey M, Yarar D, Giddings TH Jr., Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell 1997; 8:2119 - 32; http://dx.doi.org/10.1091/mbc.8.11.2119; PMID: 9362057
- Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol 2005; 170:225 - 35; http://dx.doi.org/10.1083/jcb.200504140; PMID: 16027220
- Colón-Ramos DA, Salisbury JL, Sanders MA, Shenoy SM, Singer RH, García-Blanco MA. Asymmetric distribution of nuclear pore complexes and the cytoplasmic localization of beta2-tubulin mRNA in Chlamydomonas reinhardtii. Dev Cell 2003; 4:941 - 52; http://dx.doi.org/10.1016/S1534-5807(03)00163-1; PMID: 12791277
- Yewdell WT, Colombi P, Makhnevych T, Lusk CP. Lumenal interactions in nuclear pore complex assembly and stability. Mol Biol Cell 2011; 22:1375 - 88; http://dx.doi.org/10.1091/mbc.E10-06-0554; PMID: 21346187
- Taddei A, Gasser SM. Structure and function in the budding yeast nucleus. Genetics 2012; 192:107 - 29; http://dx.doi.org/10.1534/genetics.112.140608; PMID: 22964839
- Van de Vosse DW, Wan Y, Lapetina DL, Chen W-M, Chiang J-H, Aitchison JD, Wozniak RW. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 2013; 152:969 - 83; http://dx.doi.org/10.1016/j.cell.2013.01.049; PMID: 23452847
- Yam C, Gu Y, Oliferenko S. Partitioning and remodeling of the Schizosaccharomyces japonicus mitotic nucleus require chromosome tethers. [Internet] Curr Biol 2013; 23:2303 - 10; http://linkinghub.elsevier.com/retrieve/pii/S0960982213012451 http://dx.doi.org/10.1016/j.cub.2013.09.057; PMID: 24184107
- Payne C, Rawe V, Ramalho-Santos J, Simerly C, Schatten G. Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. [Internet] J Cell Sci 2003; 116:4727 - 38; http://jcs.biologists.org/content/116/23/4727.full http://dx.doi.org/10.1242/jcs.00784; PMID: 14600259
- Stelter P, Kunze R, Flemming D, Höpfner D, Diepholz M, Philippsen P, Böttcher B, Hurt E. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol 2007; 9:788 - 96; http://dx.doi.org/10.1038/ncb1604; PMID: 17546040
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 2010; 8:e1000350; http://dx.doi.org/10.1371/journal.pbio.1000350; PMID: 20386726
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 2011; 192:855 - 71; http://dx.doi.org/10.1083/jcb.201007118; PMID: 21383080
- Steinberg G, Schuster M, Theisen U, Kilaru S, Forge A, Martin-Urdiroz M. Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J Cell Biol 2012; 198:343 - 55; http://dx.doi.org/10.1083/jcb.201201087; PMID: 22851316
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol 2001; 153:1465 - 78; http://dx.doi.org/10.1083/jcb.153.7.1465; PMID: 11425876
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 2008; 31:925 - 32; http://dx.doi.org/10.1016/j.molcel.2008.07.020; PMID: 18922474
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bähler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 2012; 151:671 - 83; http://dx.doi.org/10.1016/j.cell.2012.09.019; PMID: 23101633
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature 2007; 450:683 - 94; http://dx.doi.org/10.1038/nature06404; PMID: 18046405
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature 2007; 450:695 - 701; http://dx.doi.org/10.1038/nature06405; PMID: 18046406
- Hubert T, Vandekerckhove J, Gettemans J. Exo70-mediated recruitment of nucleoporin Nup62 at the leading edge of migrating cells is required for cell migration. Traffic 2009; 10:1257 - 71; http://dx.doi.org/10.1111/j.1600-0854.2009.00940.x; PMID: 19552648
- Zhou T, Li S, Zhong W, Vihervaara T, Béaslas O, Perttilä J, Luo W, Jiang Y, Lehto M, Olkkonen VM, et al. OSBP-related protein 8 (ORP8) regulates plasma and liver tissue lipid levels and interacts with the nucleoporin Nup62. PLoS One 2011; 6:e21078; http://dx.doi.org/10.1371/journal.pone.0021078; PMID: 21698267
- Béaslas O, Vihervaara T, Li J, Laurila P-P, Yan D, Olkkonen VM. Silencing of OSBP-related protein 8 (ORP8) modifies the macrophage transcriptome, nucleoporin p62 distribution, and migration capacity. Exp Cell Res 2012; 318:1933 - 45; http://dx.doi.org/10.1016/j.yexcr.2012.05.026; PMID: 22683860
- Hubert T, Van Impe K, Vandekerckhove J, Gettemans J. The actin-capping protein CapG localizes to microtubule-dependent organelles during the cell cycle. Biochem Biophys Res Commun 2009; 380:166 - 70; http://dx.doi.org/10.1016/j.bbrc.2009.01.064; PMID: 19166812
- Irazoqui JE, Lew DJ. Polarity establishment in yeast. J Cell Sci 2004; 117:2169 - 71; http://dx.doi.org/10.1242/jcs.00953; PMID: 15126618
- Babour A, Bicknell AA, Tourtellotte J, Niwa M. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell 2010; 142:256 - 69; http://dx.doi.org/10.1016/j.cell.2010.06.006; PMID: 20619447
- Patel SS, Rexach MF. Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol Cell Proteomics 2008; 7:121 - 31; http://dx.doi.org/10.1074/mcp.M700407-MCP200; PMID: 17897934
- West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol 2011; 193:333 - 46; http://dx.doi.org/10.1083/jcb.201011039; PMID: 21502358
- Kirchenbauer M, Liakopoulos D. An auxiliary, membrane-based mechanism for nuclear migration in budding yeast. Mol Biol Cell 2013; 24:1434 - 43; http://dx.doi.org/10.1091/mbc.E12-08-0602; PMID: 23447703
- Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol 2000; 150:461 - 74; http://dx.doi.org/10.1083/jcb.150.3.461; PMID: 10931860
- Fehrenbacher KL, Davis D, Wu M, Boldogh I, Pon LA. Endoplasmic reticulum dynamics, inheritance, and cytoskeletal interactions in budding yeast. Mol Biol Cell 2002; 13:854 - 65; http://dx.doi.org/10.1091/mbc.01-04-0184; PMID: 11907267
