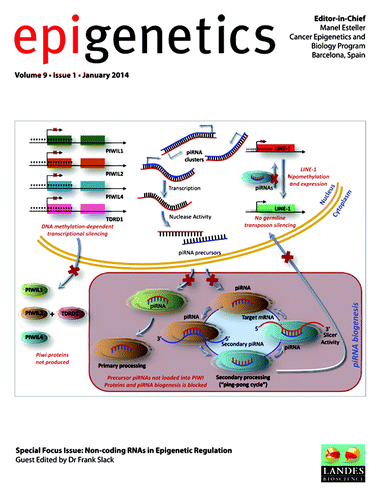
Chromatin remodeling by the small RNA machinery in mammalian cells
Chromatin states can impact fundamental cellular processes such as determination of cell identity and development of disease, but there are many open questions as to how these chromatin states are established and regulated. In several lower eukaryotes, the small RNA machinery comprised of small RNA and its partners, the Argonaute proteins, is known to play important roles in the establishment of heterochromatin and silencing of repetitive sequences. In mammalian cells, however, the nuclear function of the small RNA machinery is largely unknown. Emerging evidence suggests that components of the small RNA pathway interact with chromatin to regulate gene transcription and alternative splicing. In addition, these endogenous mechanisms are being exploited to target specific genomic loci for manipulation of gene expression and splicing events. In a recent review, Dr Long-Cheng Li summarizes the current understanding of chromatin remodeling by small RNAs in mammalian cells and highlights recent efforts to map genome-wide interactions between RNAi-related factors and chromatin ().
Nuclear actin polymerization critical for coordination of HoxB expression
Hox genes are co-regulated and expressed in the order of their location in the genome, a mechanism termed collinearity. The HoxB cluster expression is activated by retinoic acid (RA), and is accompanied by translocation of the cluster to regions outside the chromosomal territory and toward the center of the nucleus. The DNA-binding Pknox1-Pbx1 complex modulates Hox protein activity. A recent study by Dr Gabriela Naum-Onganía and colleagues used teratocarcinoma cells as a model of Hox gene expression, to show that upon retinoic acid induction, Pknox1 co-localizes with polymeric nuclear actin. They found that globular actin accumulates, and polymeric actin, the elongating RNA polymerase II, and THOC (a marker of the nuclear matrix) localized to to nuclear speckles within euchromatic regions. Moreover, RNA polymerase II, N-WASP, and transcription/splicing factors p54nrb and PSF were validated as Pknox1 interactors by tandem affinity purification. PSF co-precipitated with THOC and nuclear actin, both of which co-localize in nuclear speckles. Although latrunculin A (an agent that disrupts microfilament organization in cultured cells) slightly decreased the general level of HoxB gene expression, inhibition of nuclear actin polymerization by cytochalasin D blocked the expression of HoxB transcripts in a collinear manner. The study supports the hypothesis that nuclear actin polymerization is involved in the activation of HoxB gene expression in nuclear speckles ().
DNA quadruplexes spread by retrotransposons may serve as genome regulators
Transposable elements (TEs) are ubiquitously present in eukaryotic genomes and increasing evidence suggests that they are involved in regulatory networks of eukaryotic cells and contribute to genome evolution. It was recently reported that many plant long-terminal-repeat (LTR) retrotransposons contain DNA quadruplex-forming sequences at precise positions and that quadruplexes are better preserved in evolutionary younger elements. Since quadruplexes can modulate molecular processes, it was proposed that quadruplexes found at specific distances upstream and downstream from the endogenous TE promoter can affect transcription of the element. Moreover, quadruplexes found in solo LTRs, as well as in 3′ ends of 5′-truncated copies of LINE-1 elements, were shown to affect expression of neighboring genes. Following up on these previous findings, Drs Kejnovsky and Lexa recently proposed a model of quadruplex spread by retrotransposons involving the possible role of quadruplexes in genome organization. The authors found that quadruplexes can fulfill a dual regulatory role, on the one hand affecting the retrotransposon life-cycle as well as the activity of neighboring genes, replication of DNA and chromatin status, and on the other hand playing a role in recombination-based genome rearrangements.
Genome chaos
Genome chaos describes a process of complex, rapid genome re-organization resulting in the formation of chaotic genomes, and is followed by the potential to establish stable genomes. Initially detected through cytogenetic analyses, the phenomenon of genome chaos has recently been confirmed by whole-genome sequencing efforts which identified multiple subtypes including “chromothripsis,” “chromoplexy,” “chromoanasynthesis,” and “chromoanagenesis.” Genome chaos occurs commonly in tumors, but both the mechanism and detailed aspects of the process are unknown due to the inability of observing its evolution over time in clinical samples. Dr Henry Heng and colleagues recently developed an experimental system to monitor the evolutionary process of genome chaos and elucidate its mechanism. They found that genome chaos occurs following exposure to chemotherapeutics with different mechanisms, which act collectively as stressors. Characterization of the karyotype and its dynamic changes prior to, during, and after induction of genome chaos demonstrated that chromosome fragmentation occurs just prior to chaotic genome formation. Chaotic genomes seemed to form by random rejoining of chromosomal fragments, in part through non-homologous end joining. Stress induced genome chaos resulted in increased karyotypic heterogeneity. Such increased evolutionary potential was demonstrated by the identification of increased transcriptome dynamics associated with high levels of karyotypic variance. The authors suggest that in contrast to impacting on a limited number of cancer genes, re-organized genomes may lead to new system dynamics essential for cancer evolution. Genome chaos acts as a mechanism of rapid, adaptive, genome-based evolution that plays an essential role in promoting rapid macroevolution of new genome-defined systems during crisis, which may explain some unwanted consequences of cancer treatment ().
References
- Li L-C. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 2014; 9:45 - 52; http://dx.doi.org/10.4161/epi.26830; PMID: 24149777
References
- Naum-Onganía G, Díaz-Cortez VM, Blasi F, Rivera-Pomar R. Nuclear actin polymerization from faster growing ends in the initial activation of Hox gene transcription: Are nuclear speckles involved?. Transcription 2014; 4:260 - 72; http://dx.doi.org/10.4161/trns.27672; PMID: 24406343
References
- Kejnovsky E, Lexa M. Quadruplex-forming DNA sequences spread by retrotransposons may serve as genome regulators. Mob Genet Elements 2014; 4:e28084; http://dx.doi.org/10.4161/mge.28084; PMID: 24616836
References
- Liu G, Stevens JB, Horne SD, Abdallah BY, Ye KJ, Bremer SW, Ye CJ, Chen DJ, Heng HH. Genome chaos: survival strategy during crisis. Cell Cycle 2014; 13:528 - 37; http://dx.doi.org/10.4161/cc.27378; PMID: 24299711