Abstract
Labeling and tracing of specific sequences in living cells has been a major challenge in studying the spatiotemporal dynamics of native chromatin. Here we repurposed the prokaryotic CRISPR/Cas adaptive immunity system to specifically detect endogenous genomic loci in mouse embryonic stem cells. We constructed a catalytically inactive version of the Cas9 endonuclease, fused it with eGFP (dCas9-eGFP) and co-expressed small guide RNAs (gRNAs) to target pericentric, centric, and telomeric repeats, which are enriched in distinct nuclear structures. With major satellite specific gRNAs we obtained a characteristic chromocenter (CC) pattern, while gRNAs targeting minor satellites and telomeres highlighted smaller foci coinciding with centromere protein B (CENP-B) and telomeric repeat-binding factor 2 (TRF2), respectively. DNA sequence specific labeling by gRNA/dCas9-eGFP complexes was directly shown with 3D-fluorescent in situ hybridization (3D-FISH). Structured illumination microscopy (3D-SIM) of gRNA/dCas9-eGFP expressing cells revealed chromatin ultrastructures and demonstrated the potential of this approach for chromatin conformation studies by super resolution microscopy. This programmable dCas9 labeling system opens new perspectives to study functional nuclear architecture.
Introduction
Almost one hundred and fifty years since the original description of chromosomes, many genomes have been fully sequenced. While our knowledge of DNA sequences has reached base pair (bp) resolution, detailed information on the positioning, nuclear arrangement and interactions of specific gene loci in living cells is still limited.
Despite the absence of internal membranes, the nucleus is a highly organized organelle. With fluorescent in situ hybridization (FISH) all chromosomes in an interphase nucleus were mapped and shown to occupy distinct territories.Citation1 Additionally, gene rich chromosomes were found to be preferably located in the center of the nucleus, whereas gene poor chromosomes reside mostly in proximity to the nuclear periphery reflecting a functional nuclear organization.Citation2,Citation3 Clearly, FISH represents an important tool to label specific DNA sequences and to study nuclear architecture, but it is restricted to fixed specimens.
By now, several methods have been employed to label DNA in vivoCitation4 and to investigate chromatin spatiotemporal dynamics, for instance by the incorporation of fluorescently tagged chromatin proteins, like H2B-GFP.Citation5 However, these methods do not distinguish specific genomic sequences. To overcome these limitations, the lac operator and/or repressor recognition system has been developed.Citation6 This system, however, relies on artificially introduced sequences and does not provide information on endogenous genomic loci.
To date, targeting of specific endogenous genomic loci has been based on the sequence specific binding of Cys2His2 zinc finger modules (ZF),Citation7-Citation9 where individual ZFs bind to a distinct trinucleotide sequence and are combined into polydactyl zinc finger proteins (PZF). It has been shown, however, that the target specificity of ZFs can be affected by their neighboring modules, which requires evaluation of every newly designed PZF.Citation10,Citation11 PZFs have been widely replaced by designer transcription activator-like effectors (dTALEs), which have proven to be a powerful tool for genome engineering,Citation12,Citation13 influencing gene transcriptionCitation14-Citation16 and were recently applied for labeling genomic sequences in vivo.Citation17-Citation19 Yet, the fact that DNA binding of dTALEs is mediated by tandemly arranged repeats, whereby every repeat only differs in two residues, necessitates the use of elaborate cloning techniques.Citation20,Citation21
New experimental options became available with the discovery of the type II CRISPR/Cas system that is composed of clustered regularly interspaced short palindromic repeats (CRISPR) as well as CRISPR-associated (Cas) proteins and plays a vital role in prokaryotic adaptive immunity. Upon viral infection or plasmid uptake, short stretches (~30 bp) of foreign DNA (termed spacers) are incorporated between identical direct repeats into CRISPR arrays. Transcription of these arrays results in pre-CRISPR RNA (pre-crRNA), which subsequently interacts with a trans-activating crRNA (tracrRNA). This pre-crRNA/tracrRNA duplex forms a complex with the endonuclease Cas9, followed by further processing of pre-crRNA into crRNA. Endonuclease target-specificity is determined by the complementarity between spacer (crRNA) and protospacer (viral or plasmid) sequences.Citation22 As an important step toward the applicability of this system, a crRNA:tracrRNA chimera, named guide RNA (gRNA), has been shown to be able to replace the two RNA components and to specifically target Cas9 to user-defined DNA sequences.Citation23,Citation24 The CRISPR/Cas system has recently been applied as a versatile tool for genome editing in a wide range of organisms.Citation25-Citation29
Here, we present an approach for labeling specific endogenous genomic loci in living murine embryonic stem cells based on a modified CRISPR/Cas system.
Results
Adapting the CRISPR/Cas system for tracing specific DNA sequences in living cells
The CRISPR/Cas system features easily programmable sequence recognition, but combines it with an endonuclease activity. We, therefore, introduced mutations to inactivate the endonuclease activity of Cas9Citation30-Citation32 and fused it to the enhanced variant of GFP (eGFP) (). By co-transfecting a plasmid encoding this eGFP-tagged, nuclease deficient Cas9 (dCas9-eGFP) together with a gRNA expression vector, we aimed to target specific genomic loci in mouse embryonic stem cells. In this way, we expected to achieve specific targeting of dCas9-eGFP without cleavage of the underlying sequences ().
Figure 1. Labeling genomic loci with specific dCas9-eGFP/gRNA complexes. (A) Schematic representation of the dCas9-eGFP expression construct. A chicken β-actin promoter with a CMV enhancer (CAG, blue triangle) drives the expression of dCas9-eGFP. Inactivation of RuvC1 and HNH (red crosses) by amino acid substitutions (D10A and H840A within RuvC1 and HNH, respectively) is indicated. A second nuclear localization signal (NLS) (beige) is introduced upstream of the eGFP coding sequence (green). (B) Outline of the experimental design. dCas9-eGFP interacts with a co-expressed gRNA and is thereby guided to the genomic target sequence. Note that the presence of a protospacer adjacent motif (PAM) is a prerequisite for dCas9 binding. (C) Schematic representation of a mouse acrocentric chromosome. gRNAs were designed to target 20 bp protospacer sequences of telomeres (Tel, green), major satellites (MaS, yellow) and minor satellites (MiS, red) as indicated. (D, E, F) Co-expression of dCas9-eGFP and gRNAs complementary to MaS repeats (MaSgRNA, D), MiS (MiSgRNA, E) and Tel gRNAs (TelgRNA, F) in J1 mouse embryonic stem cells. MaSgRNA recruits dCas9-eGFP to chromocenters (CCs), MiSgRNA/dCas9-eGFP signals are observed in the periphery of CCs, while targeting of TelgRNA/dCas9-eGFP to telomeres results in distinct dCas9-eGFP foci, which can be detected throughout the nucleoplasm. Bar: 5 µm.
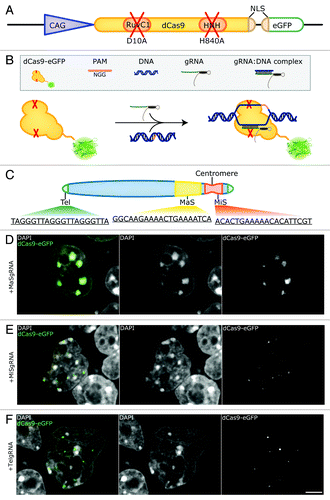
To test the feasibility of our method, we chose tandemly arranged repetitive DNA sequences, which enabled us to target dCas9-eGFP to extended genomic loci with a single gRNA construct. To this end, gRNAs directed to major (MaSgRNA) and minor satellite (MiSgRNA) repeats, as well as telomeres (TelgRNA) were designed. In mice, major satellite repeats consist of 234 bp repeat units that span within a 6 Mb region, whereas minor satellite repeats range between 0.6 to 1.2 Mb and consist of 120 bp repeat units.Citation33-Citation36 Telomeric repeats vary in length reflecting the cell’s replicative potentialCitation37 and in mouse amount to approximately 20–30 kb with the 6 bp repeat sequence TTAGGG ().
Effective tracing of repetitive DNA sequences using the CRISPR/Cas system
Mouse chromosomes are acrocentric as depicted in . Fluorescent in situ hybridization (FISH) experiments on metaphase chromosomes have shown that minor satellites are centromeric, whereas major satellite repeats occupy the subcentromeric part of the chromosome and are implicated in heterochromatin formation and sister chromatid cohesion.Citation38-Citation41 In interphase nuclei, centromeres cluster and form distinct chromocenters (CCs),Citation41,Citation42 which can be readily distinguished by enhanced DAPI-staining intensity due to their AT-richness.
J1 mouse embryonic stem cells were co-transfected with dCas9-eGFP and MaSgRNA encoding plasmids and imaged 48 h post-transfection. As depicted in , the CCs of MaSgRNA/dCas9-eGFP expressing cells show a bright eGFP signal, verifying the successful targeting of genomic DNA. The distribution exhibits remarkable specificity with very low background signals from freely diffusing dCas9-eGFP.
MiS repeats have been observed as individual focal entities at the periphery of CCs.Citation41 We were able to observe this characteristic distribution as eGFP fluorescent foci in the corresponding regions of MiSgRNA/dCas9-eGFP expressing cells ().
Telomeres are capping the ends of chromosomes and have been found localized throughout the nucleus.Citation43 Due to the acrocentric nature of mouse chromosomes, telomeres located at the acrocentric end can also be detected in the direct vicinity of CCs apart from remote nuclear sites. By expressing TelgRNA/dCas9-eGFP in J1 cells, small foci, often in the vicinity of CCs, were visible. The observed variable size of labeled telomeres is consistent with the fact that telomeres may form clustersCitation44 (). In some TelgRNA/dCas9-eGFP expressing cells, an elongated and fiber-like telomere-related eGFP signal was apparent, which was not observed in cells expressing MaS- or MiSgRNA/dCas9-eGFP. Taken together our results show that for all three targeted DNA sequences distinct labeling patterns were observed.
Combined gRNA/dCas9-eGFP labeling and 3D DNA-FISH
To validate the sequence specificity of the dCas9 labeling system, we verified the accurate targeting of all live-cell introduced fluorescent genomic tags by 3D DNA-FISH experiments. For this purpose, FISH probes specific for MaS, MiS and telomeric repeats were generated and immuno-FISH experiments were performed. We used a protocol optimized for eGFP epitope preservation and efficient probe hybridization,Citation45 yielding robust and homogeneous signals (). As it can readily be seen in , the probe for MaS repeats shows confined localization at the CCs, while MaSgRNA/dCas9-eGFP complexes exhibit strict co-localization with the probe. Accordingly, immuno-FISH experiments showed correct localization of MiSgRNA/dCas9-eGFP at MiS repeats (). Moreover, simultaneous hybridization with MaS and MiS probes showed that the gRNA/dCas9-eGFP complexes specifically labeled their respective target structure and did not affect the spatial integrity of neighboring structures (). In immuno-FISH of TelgRNA/dCas9-eGFP expressing cells, FISH probe and dCas9-eGFP were fully co-localized and exhibited the same variety of signal intensities with brighter foci possibly representing longer telomeres or telomere clusters ().
Figure 2. 3D-FISH shows precise targeting of dCas9-eGFP. (A, B, C, D) Immuno-FISH experiments in gRNA/dCas9-eGFP expressing cells. Hybridization of probes designed to target MaS (A, red and C, blue), MiS (B-C, red), Tel (D, red) demonstrate that dCas9-eGFP co-localizes with the respective sequences. Multicolor immuno-FISH in C (MiS, red; MaS, blue) highlights the restricted targeting of MiSgRNA/dCas9-eGFP and demonstrates that non-targeted neighboring nuclear structures maintain their integrity. Bars, 5 µm; insets, 1 µm.
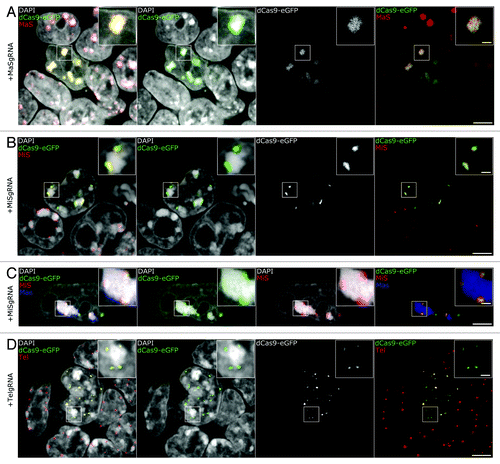
In each case, neighboring non-transfected cells in our specimens served as an internal control for the visualization of normal distribution of the FISH probes and comparison of signal strength. It must be noted here that on some occasions FISH probes did not fully penetrate to the core of well-associated stem cell colonies, while dCas9-eGFP signal was detected. Our observations confirm the specific and restricted localization of dCas9-eGFP to their targeted DNA sequences.
gRNAs/dCas9-eGFP complexes remain associated with mitotic chromosomes
During cell division, chromatin undergoes dramatic structural changes in order for chromosomes to compact and segregate properly. This chromatin rearrangement could possibly affect binding of recombinant macromolecular complexes. In addition, the targeted MiS repeats, forming the centromeres, are the sites of kinetochore formation and ensure normal chromosome segregation.Citation46 Likewise, pericentromeric regions, containing MaS repeats, play an important role in mitotic progression, as they have been linked to sister-chromatid cohesion.Citation47-Citation49 Furthermore, telomeric dysfunction has been shown to promote chromosome fusions, anaphase bridges and genome reorganization.Citation50-Citation52 Therefore, targeting of dCas9-eGFP at these essential regulatory sequences could potentially perturb chromosome arrangements and mitotic progression. Hence, we investigated, whether dCas9-eGFP stably maintained its association to targeted DNA sequences during mitosis. We were able to observe MaS, MiS, and TelgRNA/dCas9-eGFP expressing cells in different mitotic stages. A metaphase for the MaSgRNA/dCas9-eGFP targeting is depicted in , showing the robust labeling of the MaS domains and its restricted localization to the highly condensed CC regions. Similarly, two optical sections of a metaphase plate (z1, z2) showed that TelgRNA/dCas9-eGFP labeled both ends of mitotic chromosomes and multicolor FISH confirmed the pericentromeric and centromeric arrangement of MaS and MiS repeats, respectively ().
Figure 3. Association of gRNA/dCas9-eGFP to chromatin in mitotic cells. (A) Confocal optical section of a metaphase plate shows successful targeting of MaSgRNA/dCas9-eGFP. Note the robust eGFP signals at the CCs (DAPI bright regions, middle panel). Bar, 5 µm. (B) Two confocal optical sections (z1, z2) of a multicolor immuno-FISH stained metaphase show TelgRNA/dCas9-eGFP signals at the ends of chromosomes. The Integrity of (peri-) centromeric chromatin (MaS, MiS) is not compromised. Bar, 5 µm; insets, 1 µm. (C) Metaphase plate of a MiSgRNA/dCas9-eGFP expressing cell after immuno-FISH with MiS probe (red) acquired via 3D-SIM. Note the overlap between MiS probe and dCas9-eGFP. Bar, 5 μm; insets, 1 µm.
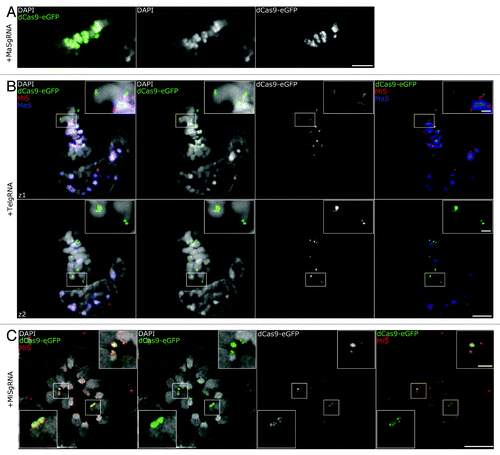
For a more detailed investigation of the MiS signals, we applied 3D-SIM microscopy and examined metaphases in MiSgRNA/dCas9-eGFP expressing cells (). We were able to distinguish distinct dCas9-eGFP foci in every chromosome and strict co-localization with the MiS probe. Using conventional microscopy, MiS clusters appear as diffraction-limited foci. With 3D-SIM, these clusters displayed ultrastructural organization, visible by FISH probe, as well as dCas9-eGFP labeling (, magnification insets).
Application of CRISPR/Cas labeling in super resolution studies
Next, we investigated whether the coverage of dCas9-eGFP labeling correlated with known proteins bound at these genomic loci, perturbed the canonical distribution of these proteins or prevented antibody binding. For this purpose, J1 cells expressing MiSgRNA/dCas9-eGFP were immunolabeled with anti-centromere protein-B (CENP-B) antibodies. CENP-B is a constitutive centromere protein located at the primary constriction by direct binding to MiS repeats at the CENP-B box.Citation53-Citation55 In mitosis it forms a link between the kinetochore and the underlying centromeric repeatsCitation56,Citation57 and regulates centromere formation.Citation58,Citation59 In interphase it has been visualized in association with MiS repeat clusters at the periphery of the CCs.Citation41
In MiSgRNA/dCas9-eGFP expressing cells, distinct CENP-B foci were consistently associated with dCas9-eGFP clusters (), suggesting that incorporation of the gRNA/dCas9-eGFP does neither interfere with the recruitment of CENP-B to MiS repeats, nor antibody binding upon fixation. Compared with wide-field deconvolution (wf), where MiS domains appeared as diffraction-limited foci, 3D-SIM revealed a sub-structural organization of the targeted repeats (, blowups 3D-SIM compared with wf).
Figure 4. 3D-SIM highlights the ultrastructure of MiS and Tel repeats. (A) Left panel depicts a mid z-section of a DAPI stained nucleus (gray) in MiSgRNA/dCas9-eGFP (green) expressing cells immunolabeled with anti-CENP-B antibodies (red). Four × magnifications of boxed areas (mid gallery) show the spatial association of dCas9-eGFP decorated domains to CENP-B assemblies. Wide-field (wf) deconvolved simulations of the corresponding 3D-SIM magnifications are shown for comparison (far right gallery). Bar, 5 µm; magnifications, 500 nm. (B) Left panel depicts a mid z-section of a DAPI stained nucleus (gray) in TelgRNA/dCas9-eGFP (green) expressing cells immunolabeled with anti-TRF2 antibodies (red). Four × magnifications of boxed areas (mid gallery) show overlapping pattern of dCas9-eGFP signals with TRF2. Wide-field (wf) deconvolved simulations of the corresponding 3D-SIM magnifications are shown for comparison (far right gallery). Note the elucidation of interconnected ovoid intensities within the telomere-cluster (mid-gallery, lower panel, and dCas9-eGFP). Bar, 5 µm; magnifications, 500 nm.
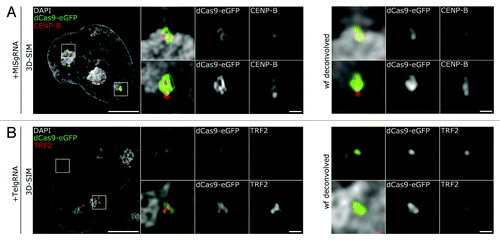
Consistent with the above findings, in TelgRNA/dCas9-eGFP expressing cells, the incorporation of the complexes to telomeric repeats allowed binding of anti-telomeric repeat-binding factor 2 (TRF2) antibodies (). Telomeric DNA sequences have been shown to form a T-loop structure, which is thought to protect the 3′-overhangs and regulate telomerase activity.Citation60,Citation61 By conventional fluorescence microscopy, telomeres are detected as diffraction-limited foci. In a recent study applying 3D-STORM, FISH detected telomeres appeared as ovoid clusters with an average diameter of 180 nm.Citation62 Although SIM does not offer the localization accuracy of STORM,Citation63 high-resolution microscopy of telomeres using the dCas9 labeling system revealed a consistent morphology to the above-mentioned study ().
With dCas9 labeling, we were able to visualize repetitive DNA sequences on fixed cells without the need of FISH probes and thus avoided flattening or destruction of chromatin due to sample denaturation. Furthermore, combination of 3D-SIM with immunostaining and dCas9 labeling highlighted ultrastructural properties of targeted MiS and Tel repeats.
Discussion
For modern cytogenetics and diagnostics, fluorescent in situ hybridization (FISH) has proven to be an indispensable method, but its application is still limited, due to its complexity and variability. Since its original introduction, numerous scientific publications have dealt with the optimization of probe and sample preparation. Although improvements have been made, harsh treatments, such as heat denaturation are required for probe hybridization that may compromise sample integrity.Citation64,Citation65 Furthermore, combining 3D-FISH with protein immunofluorescent detection remains a challenge and on occasion is not feasible due to detrimental effects on epitopes.Citation66 Therefore, even at the fixed cell level, live-cell genomic labeling systems offer superior sample preservation and their use is far less laborious compared with FISH probe creation and long hybridization and/or detection procedures. Noteworthy is the fact that at the dense stem cell colony level, we observed on occasion that FISH probes did not penetrate efficiently the core of the colony. In these cells, dCas9 labeling was uniform thereby rendering this method more efficient than FISH.
Here we report the successful labeling of endogenous centric, pericentric and telomeric chromatin loci in living mouse embryonic stem cells by repurposing the prokaryotic CRISPR/Cas system. During the preparation of this manuscript, a similar study has been published,Citation67 which showed that, besides repetitive sequences, also individual genes can be labeled by a catalytically inactive Cas9 endonuclease, confirming the potential of the dCas9 system to label genomic DNA in vivo. While their study focused on detection sensitivity, we performed a thorough comparison with 3D-FISH and immunolabeling methods. In addition we demonstrated potential applications in high resolution microscopy studies.
So far, dTALE based approaches are commonly applied as powerful tools for genome manipulation and live-cell tracing of chromatin. However, large-scale studies of nuclear organization require the design of dTALEs specific for many different target sequences including single copy genes. Due to laborious cloning procedures, large-scale production of these DNA binding proteins still remains a challenge. In contrast, the target specificity of the dCas9 labeling system relies on small and easily exchangeable gRNAs, which greatly expands the range of possible targets and is the basis for the convenience of dCas9 labeling. Moreover, combining dCas9 labeling with 3D-SIM microscopy enabled us to visualize the ultrastructure of diffraction limited chromatin clusters and their spatial relationship with known associated proteins.
Clearly, this programmable dCas9 DNA labeling system represents a powerful tool to monitor the spatiotemporal dynamics of endogenous genomic loci during cell cycle progression and differentiation and opens new perspectives to study functional nuclear architecture.
Materials and Methods
Cell culture and transfection
J1 embryonic stem cells (ESCs)Citation68 were cultivated at 37 °C and 5% CO2 on gelatin-coated petri dishes in Dulbecco’s modified Eagle’s medium supplemented with 16% fetal bovine serum (Biochrom), 0.1 mM β-mercaptoethanol (Invitrogen), 2 mM L-glutamine, 1x MEM non-essential amino acids, 100 U/ml penicillin, 100 µg/ml streptomycin (PAA Laboratories GmbH), 1000 U/ml recombinant mouse LIF (Millipore), 1 µM PD032501, and 3 µM CHIR99021 (Axon Medchem). Transfection of J1 cells was performed using Lipofectamin 2000 (Invitrogen) according to the manufacturer’s instructions. For FISH experiments 2.5 × 105, for all other experiments 5 × 105 cells were transfected. Cells were analyzed 48 h post-transfection.
Plasmid generation
For generating the dCas9-eGFP construct, plasmid hCas9_D10A was purchased from Addgene (addgene ID: 41816Citation24). Inactivation of the second nuclease domain (H840A) was performed by site-directed mutagenesis using primers dCas1-F, dCas2-R, dCas2-F, and dCas3-R. The resulting PCR-product (dCas9) was digested with BsrGI and XbaI and ligated into pCAG.Citation69,Citation70 The NLS-eGFP-sequence was amplified by PCR using primers eGFP1 and eGFP2, digested with AsiSI and NotI and ligated downstream of dCas9. pEX-A-U6-gRNA was synthesized at Eurofins MWG Operon according to Mali et al.Citation24 gRNA-expression vectors were generated by amplifying pEX-A-U6-gRNA with forward and reverse primers, which introduced the protospacer sequence for minor satellites repeats (MiS), major satellites repeats (MaS) and telomeres (Tel), respectively.
Nucleotide sequences:
dCas1-F: 5′-aaagcgatcg ctctagaatg gacaagaagt actccattgg g-3′
dCas2-R: 5′-ctggggcacg atagcatcca cgtcg-3′
dCas2-F: 5′-cgacgtggat gctatcgtgc cccag-3′
dCas3-R: 5′-tttgcggccg ctcattgtac aatcaccttc ctcttcttct tggggtc-3′
eGFP1: 5′-aaagcgatcg catccaaaga agaagagaaa ggtcatggtg agcaagggcg agg-3′
eGFP2: 5′-tttgcggccg cttacttgta cagctcgtcc atgcc-3′
MaSgRNA-F: 5′-ggcaagaaaa ctgaaaatca gttttagagc tagaaatagc aag-3′
MaSgRNA-R: 5′-tgattttcag ttttcttgcc cggtgtttcg tcctttccac -3′
TelgRNA-F: 5′-tagggttagg gttagggtta gttttagagc tagaaatagc aag-3′
TelgRNA-R: 5′-taaccctaac cctaacccta cggtgtttcg tcctttccac -3′
MiSgRNA-F: 5′-acactgaaaaa cacattcgtg ttttagagct agaaatagca ag-3′
MiSgRNA-R: 5′-acgaatgtgt ttttcagtgt cggtgtttcgt cctttccac- 3′
pEX-A-U6-gRNA: Sequence is available upon request.
Immunofluorescence staining (IF) and fluorescent in situ hybridization (FISH)
Immunostaining and FISH experiments were performed as described previously.Citation45,Citation64 Briefly, J1 ESCs grown on coverslips were washed 48 h post-transfection with phosphate buffered saline (PBS) and fixed with 4% formaldehyde for 10 min. After permeabilization with 0.5% Triton X-100 in PBS, cells were incubated with PBST (PBS. 0.02% Tween) supplemented with 2% BSA and 0.5% fish skin gelatin (blocking buffer) for 1 h. Both primary and secondary antibodies were diluted in blocking buffer and cells were incubated with the respective antibodies for 1 h in a dark humidified chamber at room temperature. Washings were performed with PBST. For immuno-FISH detection, cells were first incubated with primary (anti-GFP, Roche) and secondary antibodies, followed by postfixation and pre-treatment for hybridization. Hybridization was performed overnight at 37 °C. Following post-hybridization washings were performed with 2xSaline Sodium Citrate (SSC) at 37 °C and 0.1xSSC at 61 °C.Citation64 FISH probes for MiS and MaS were generated by PCR using mouse Cot1 DNA (Invitrogen) as a template (MaS-primers: 5′-GCG AGA AAA CTG AAA ATC AC-3′ and 5′-TCA AGT CGT CAA GTG GAT G-3′; MiS-primers: 5′-CAT GGA AAA TGA TAA AAA CC-3′ and 5′-CAT CTA ATA TGT TCT ACA GTG TGG-3′). The probe for telomeric repeats was produced by using self-annealing primers in the PCR (5′-TTA GGG TTA GGG TTA GGG TTA GGG TTA GGG-3′ and 5′-CCC TAA CCC TAA CCC TAA CCC TAA CCC TAA-3′). All probes were directly labeled by nick translation with Texas Red-dUTP or Cy3-dUTP and dissolved in hybridization mixture (50% formamide, 10% dextran sulfate, 1xSSC) at a concentration of 10–20 ng/ml. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), mounted with antifade medium (Vectashield, Vector Laboratories) and sealed with Covergrip sealant (Biotium). Primary antibodies used in this study were: anti-GFP (1:400; Roche 11814460001), anti-TRF2 (1:250, Abcam ab13579), anti-CENP-B (1:500, Abcam ab84489). The secondary antibodies were anti-rabbit conjugated to DyLight 594 (Jackson ImmunoResearch, 711-505-152), anti-mouse conjugated to Alexa 488 (Invitrogen, A21202) and anti-mouse conjugated to Alexa 594 (Invitrogen, A11032). Cells for 3D-SIM were grown on precision cover glass, thickness no. 1.5H (170 µm ± 5 µm; Marienfeld Superior) using immersion oil with a refractive index of 1.514 to minimize spherical aberration.
Microscopy and image acquisition
Optical sections for were acquired with an UltraVIEW VoX spinning disk confocal microscope (PerkinElmer), which was operated with the Volocity® software. For , , and , single optical sections or stacks of optical sections were collected with a Leica TCS SP5 confocal microscope using a Plan Apo 63×/1.4 NA oil immersion objective. High-resolution images ( and ) were obtained with a DeltaVision OMX V3 3D-SIM microscope (Applied Precision Imaging, GE Healthcare), equipped with a 60×/1.42 NA PlanApo oil objective and sCMOS cameras (Olympus). Images were acquired with a z-step size of 125 nm. Reconstruction and image deconvolution was applied to the SI raw data using the SoftWorX 4.0 software package (Applied Precision). Image processing and assembly was performed with FIJI and Photoshop CS5.1 (Adobe), respectively. Stacks of confocal optical sections were corrected for chromatic shifts with ImageJ plugins.Citation71
| Abbreviations: | ||
| CRISPR | = | Clustered Regulatory Interspaced Short Palindromic Repeats |
| Cas | = | CRISPR-associated |
| eGFP | = | enhanced-green fluorescent protein |
| 3D-FISH | = | 3D-fluorescent in situ hybridization |
| gRNA | = | guide RNA |
| bp | = | base pair |
| MaS | = | major satellite repeats |
| MiS | = | minor satellite repeats |
| CC | = | chromocenter |
| 3D-SIM | = | 3D-structured illumination microscopy |
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
The authors thank Katharina Thanisch (LMU Munich) for kindly providing the pEX-U6-gRNA cloning vector, George Church (Harvard Medical School) for the hCas9_D10A construct and En Li (China Novartis Institutes for BioMedical Research) for the J1 cell line. We also thank Irina Solovei (LMU Munich) for advice with FISH protocols. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, SFB 1064, Nanosystems Initiative Munich, NIM) and T.A. is a fellow of the Graduiertenkolleg GRK1721.
References
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005; 3:e157; http://dx.doi.org/10.1371/journal.pbio.0030157; PMID: 15839726
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 1999; 145:1119 - 31; http://dx.doi.org/10.1083/jcb.145.6.1119; PMID: 10366586
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res 2001; 9:541 - 67; http://dx.doi.org/10.1023/A:1012495201697; PMID: 11721953
- Martin RM, Leonhardt H, Cardoso MC. DNA labeling in living cells. Cytometry Part A: the journal of the International Society for Analytical Cytology 2005; 67:45-52.
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol 2001; 153:1341 - 53; http://dx.doi.org/10.1083/jcb.153.7.1341; PMID: 11425866
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 1996; 135:1685 - 700; http://dx.doi.org/10.1083/jcb.135.6.1685; PMID: 8991083
- Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys 2010; 43:1 - 21; http://dx.doi.org/10.1017/S0033583510000089; PMID: 20478078
- Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 2001; 70:313 - 40; http://dx.doi.org/10.1146/annurev.biochem.70.1.313; PMID: 11395410
- Segal DJ, Barbas CF 3rd. Design of novel sequence-specific DNA-binding proteins. Curr Opin Chem Biol 2000; 4:34 - 9; http://dx.doi.org/10.1016/S1367-5931(99)00048-4; PMID: 10679372
- Segal DJ, Dreier B, Beerli RR, Barbas CF 3rd. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci U S A 1999; 96:2758 - 63; http://dx.doi.org/10.1073/pnas.96.6.2758; PMID: 10077584
- DeFrancesco L. Move over ZFNs. Nat Biotechnol 2011; 29:681 - 4; http://dx.doi.org/10.1038/nbt.1935; PMID: 21822235
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011; 29:143 - 8; http://dx.doi.org/10.1038/nbt.1755; PMID: 21179091
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 2011; 39:9283 - 93; http://dx.doi.org/10.1093/nar/gkr597; PMID: 21813459
- Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy DK, Zhu JK. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol 2012; 78:311 - 21; http://dx.doi.org/10.1007/s11103-011-9866-x; PMID: 22167390
- Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res 2012; 40:5368 - 77; http://dx.doi.org/10.1093/nar/gks199; PMID: 22387464
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 2011; 29:149 - 53; http://dx.doi.org/10.1038/nbt.1775; PMID: 21248753
- Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol 2013; 20:1321 - 4; http://dx.doi.org/10.1038/nsmb.2680; PMID: 24096363
- Ma H, Reyes-Gutierrez P, Pederson T. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci U S A 2013; 110:21048 - 53; http://dx.doi.org/10.1073/pnas.1319097110; PMID: 24324157
- Thanisch K, Schneider K, Morbitzer R, Solovei I, Lahaye T, Bultmann S, Leonhardt H. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res 2013; http://dx.doi.org/10.1093/nar/gkt1348; PMID: 24371265
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011; 39:e82; http://dx.doi.org/10.1093/nar/gkr218; PMID: 21493687
- Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res 2011; 39:5790 - 9; http://dx.doi.org/10.1093/nar/gkr151; PMID: 21421566
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet 2012; 46:311 - 39; http://dx.doi.org/10.1146/annurev-genet-110711-155447; PMID: 23145983
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819 - 23; http://dx.doi.org/10.1126/science.1231143; PMID: 23287718
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823 - 6; http://dx.doi.org/10.1126/science.1232033; PMID: 23287722
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227 - 9; http://dx.doi.org/10.1038/nbt.2501; PMID: 23360964
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013; 31:233 - 9; http://dx.doi.org/10.1038/nbt.2508; PMID: 23360965
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 2013; 23:720 - 3; http://dx.doi.org/10.1038/cr.2013.46; PMID: 23545779
- Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 2013; 10:741 - 3; http://dx.doi.org/10.1038/nmeth.2532; PMID: 23817069
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31:230 - 2; http://dx.doi.org/10.1038/nbt.2507; PMID: 23360966
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 2013; 23:1163 - 71; http://dx.doi.org/10.1038/cr.2013.122; PMID: 23979020
- Kearns NA, Genga RM, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 2014; 141:219 - 23; http://dx.doi.org/10.1242/dev.103341; PMID: 24346702
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152:1173 - 83; http://dx.doi.org/10.1016/j.cell.2013.02.022; PMID: 23452860
- Kipling D, Ackford HE, Taylor BA, Cooke HJ. Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics 1991; 11:235 - 41; http://dx.doi.org/10.1016/0888-7543(91)90128-2; PMID: 1685135
- Kipling D, Wilson HE, Mitchell AR, Taylor BA, Cooke HJ. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma 1994; 103:46 - 55; http://dx.doi.org/10.1007/BF00364725; PMID: 8013255
- Hörz W, Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res 1981; 9:683 - 96; http://dx.doi.org/10.1093/nar/9.3.683; PMID: 6261227
- Kuznetsova IS, Prusov AN, Enukashvily NI, Podgornaya OI. New types of mouse centromeric satellite DNAs. Chromosome Res 2005; 13:9 - 25; http://dx.doi.org/10.1007/s10577-005-2346-x; PMID: 15791408
- Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 2005; 26:867 - 74; http://dx.doi.org/10.1093/carcin/bgh296; PMID: 15471900
- Pidoux AL, Allshire RC. The role of heterochromatin in centromere function. Philos Trans R Soc Lond B Biol Sci 2005; 360:569 - 79; http://dx.doi.org/10.1098/rstb.2004.1611; PMID: 15905142
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science 2001; 294:2539 - 42; http://dx.doi.org/10.1126/science.1064027; PMID: 11598266
- Wong AK, Rattner JB. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res 1988; 16:11645 - 61; http://dx.doi.org/10.1093/nar/16.24.11645; PMID: 3211746
- Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 2004; 166:493 - 505; http://dx.doi.org/10.1083/jcb.200403109; PMID: 15302854
- Joseph A, Mitchell AR, Miller OJ. The organization of the mouse satellite DNA at centromeres. Exp Cell Res 1989; 183:494 - 500; http://dx.doi.org/10.1016/0014-4827(89)90408-4; PMID: 2767161
- Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res 2003; 11:485 - 502; http://dx.doi.org/10.1023/A:1025016828544; PMID: 12971724
- Nagele RG, Velasco AQ, Anderson WJ, McMahon DJ, Thomson Z, Fazekas J, Wind K, Lee H. Telomere associations in interphase nuclei: possible role in maintenance of interphase chromosome topology. J Cell Sci 2001; 114:377 - 88; PMID: 11148139
- Markaki Y, Smeets D, Cremer M, Schermelleh L. Fluorescence in situ hybridization applications for super-resolution 3D structured illumination microscopy. Methods Mol Biol 2013; 950:43 - 64; PMID: 23086869
- Pidoux AL, Allshire RC. Centromeres: getting a grip of chromosomes. Curr Opin Cell Biol 2000; 12:308 - 19; http://dx.doi.org/10.1016/S0955-0674(00)00094-6; PMID: 10801468
- Stephens AD, Snider CE, Haase J, Haggerty RA, Vasquez PA, Forest MG, Bloom K. Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J Cell Biol 2013; 203:407 - 16; http://dx.doi.org/10.1083/jcb.201307104; PMID: 24189271
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks?. Nat Rev Genet 2008; 9:923 - 37; http://dx.doi.org/10.1038/nrg2466; PMID: 19002142
- Gartenberg M. Heterochromatin and the cohesion of sister chromatids. Chromosome Res 2009; 17:229 - 38; http://dx.doi.org/10.1007/s10577-008-9012-z; PMID: 19308703
- De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol 2005; 70:197 - 204; http://dx.doi.org/10.1101/sqb.2005.70.032; PMID: 16869754
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al, ALT Starr Cancer Consortium. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 2012; 8:e1002772; http://dx.doi.org/10.1371/journal.pgen.1002772; PMID: 22829774
- Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett 2005; 579:859 - 62; http://dx.doi.org/10.1016/j.febslet.2004.11.036; PMID: 15680963
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 1989; 109:1963 - 73; http://dx.doi.org/10.1083/jcb.109.5.1963; PMID: 2808515
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 1992; 116:585 - 96; http://dx.doi.org/10.1083/jcb.116.3.585; PMID: 1730770
- Pluta AF, Saitoh N, Goldberg I, Earnshaw WC. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J Cell Biol 1992; 116:1081 - 93; http://dx.doi.org/10.1083/jcb.116.5.1081; PMID: 1740467
- Suzuki N, Nakano M, Nozaki N, Egashira S, Okazaki T, Masumoto H. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J Biol Chem 2004; 279:5934 - 46; http://dx.doi.org/10.1074/jbc.M306477200; PMID: 14612452
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 2006; 8:458 - 69; http://dx.doi.org/10.1038/ncb1397; PMID: 16622419
- Kitagawa K, Masumoto H, Ikeda M, Okazaki T. Analysis of protein-DNA and protein-protein interactions of centromere protein B (CENP-B) and properties of the DNA-CENP-B complex in the cell cycle. Mol Cell Biol 1995; 15:1602 - 12; PMID: 7862152
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell 2007; 131:1287 - 300; http://dx.doi.org/10.1016/j.cell.2007.10.045; PMID: 18160038
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell 1999; 97:503 - 14; http://dx.doi.org/10.1016/S0092-8674(00)80760-6; PMID: 10338214
- Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J Cell Biol 2004; 166:161 - 5; http://dx.doi.org/10.1083/jcb.200403118; PMID: 15249582
- Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 2013; 155:345 - 56; http://dx.doi.org/10.1016/j.cell.2013.09.048; PMID: 24120135
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol 2010; 190:165 - 75; http://dx.doi.org/10.1083/jcb.201002018; PMID: 20643879
- Solovei I, Cremer M. 3D-FISH on cultured cells combined with immunostaining. Methods Mol Biol 2010; 659:117 - 26; http://dx.doi.org/10.1007/978-1-60761-789-1_8; PMID: 20809307
- Chaumeil J, Micsinai M, Skok JA. Combined immunofluorescence and DNA FISH on 3D-preserved interphase nuclei to study changes in 3D nuclear organization. J Vis Exp 2013; •••:e50087; PMID: 23407477
- Molenaar C, Wiesmeijer K, Verwoerd NP, Khazen S, Eils R, Tanke HJ, Dirks RW. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J 2003; 22:6631 - 41; http://dx.doi.org/10.1093/emboj/cdg633; PMID: 14657034
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155:1479 - 91; http://dx.doi.org/10.1016/j.cell.2013.12.001; PMID: 24360272
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992; 69:915 - 26; http://dx.doi.org/10.1016/0092-8674(92)90611-F; PMID: 1606615
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991; 108:193 - 9; http://dx.doi.org/10.1016/0378-1119(91)90434-D; PMID: 1660837
- Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259 - 64; http://dx.doi.org/10.1038/embor.2009.201; PMID: 19798101
- Ronneberger O, Baddeley D, Scheipl F, Verveer PJ, Burkhardt H, Cremer C, Fahrmeir L, Cremer T, Joffe B. Spatial quantitative analysis of fluorescently labeled nuclear structures: problems, methods, pitfalls. Chromosome Res 2008; 16:523 - 62; http://dx.doi.org/10.1007/s10577-008-1236-4; PMID: 18461488
