Abstract
The central role of tumor-specific TH1 cells in anticancer immune responses is becoming increasingly appreciated. However, little is known about how these cells are generated in vivo. Here, we used flow cytometry and gene expression microarrays to characterize the primary activation and TH1 differentiation of naïve tumor-specific CD4+ T cells in a mouse model of cancer immunosurveillance. We took advantage of T-cell receptor-transgenic mice in which CD4+ T cells recognize a tumor-specific antigen secreted by MHC class II-negative MOPC315 myeloma cells. Cancer cells were injected subcutaneously and T-cell activation was analyzed in draining lymph nodes and at the incipient tumor site 8 d later. Upon activation and migration to incipient tumor sites, tumor-specific CD4+ T cells exhibited the upregulation of 29 cell-surface molecules (CD2, CD5, CD11a, CD18, CD25, CD28, CD44, CD45, CD49d, CD51, CD54, CD69, CD71, CD83, CD86, CD90, CD95, CD102, CD122, CD153, CD166, CD200, CD249, CD254, CD274, CD279, Ly6C, MHC class I and CCR7) and the downregulation of five (CD27, CD31, CD45RB, CD62L and CD126). Activated CD4+ T cells produced interferon γ, a cytokine consistent with a TH1-polarized response, tumor necrosis factor α as well as interleukin (IL)-2, IL-3 and IL-10. The activation of naïve tumor-specific CD4+ T cells in draining lymph nodes resulted in the upregulation of 609 genes and the downregulation of 284 genes. The bioinformatic analysis of differentially expressed genes identified functional pathways related to tumor-specific TH1 cell activation. This study may represent a useful resource to guide the development of TH1-based immunotherapies against cancer.
Introduction
Tumor-specific CD4+ T cells have repeatedly been shown to play a central role in antitumor immune responses.Citation1-Citation6CD4+ T cells provide help to tumor-specific CD8+ T cells and B cells, and can promote the cytotoxic function of innate immune cells including macrophages. Tumor infiltration by CD4+ T cells has been reported to correlate with increased survival in patients affected by esophageal and lung cancer.Citation7,Citation8 The phenotype of tumor-infiltrating CD4+ T cells has also been shown to be important for antitumor immunity, with TH1 cells standing out as the most beneficial cell subset for patient survival.Citation9 Elucidating the molecular characteristics of primary activated tumor-specific CD4+ T cells may provide novel insights into the mechanisms underlying antitumor immune responses. Blocking cytotoxic T lymphocyte-associated antigen 4 (CTLA4), programmed death 1 (PD1) and PD-1 ligand 1 (PDL1), three immunosuppressive receptors that inhibit T-cell activation, is currently being tested in cancer patients as a strategy to boost T-cell responses.Citation10-Citation12 A comprehensive molecular characterization of activated tumor-specific CD4+ T cells may reveal additional targets for optimizing antitumor immune responses.
In order to study tumor-specific CD4+ T cells in a model of successful anticancer immunosurveillance, we used tumor-specific T-cell receptor (TCR)-transgenic mice.Citation13,Citation14 A single TCR specificity was ensured by introducing the severe combined immunodeficiency (SCID) mutation.Citation15 In these mice, all CD4+ T cells recognize a tumor-specific idiotypic peptide derived from the variable region of the immunoglobulin light chain expressed by MOPC315 myeloma cells, presented on MHC class II I-Ed molecules.Citation13,Citation14 Like most cancer cells, MOPC315 myeloma cells lack MHC class II molecules and cannot present antigens to the tumor-specific CD4+ T cells in a direct fashion.Citation16,Citation17 Instead, antigen presentation is mediated by professional antigen-presenting cells (APCs) such as dendritic cells (DCs) and macrophages.Citation6,Citation18 Tumor-specific TCR-transgenic SCID mice are protected against the subcutaneous (s.c.) inoculation of syngeneic MOPC315 myeloma cells, whereas non-transgenic mice develop fatal tumors.Citation15 Such protection is antigen-specific, CD4+ T cell-mediated and independent of both B and CD8+ T cells.Citation14,Citation15 In addition, the successful elimination of cancer cells requires antigens to be secreted by MOPC315 myeloma cells, as well as the migration of effector CD4+ T cells from the tumor-draining lymph node (LN) to the incipient tumor site.Citation6,Citation17,Citation19
The mechanism of cancer rejection by TCR-transgenic SCID mice has previously been studied by placing tumor cells in a collagen-containing gel (Matrigel) prior to administration. This strategy has allowed us to analyze the immune cells that infiltrate incipient neoplastic lesions, and to quantify the cytokines locally secreted during primary antitumor immune responses.Citation6,Citation20 Using this technique, we have shown that naïve tumor-specific CD4+ T cells first become activated in tumor-draining LN. During activation, tumor-specific CD4+ T cells acquire an effector phenotype with the upregulation of CD11a, CD44 and CD69, and the downregulation of CD62L, on their surface. These cells clonally expand, differentiate toward a TH1 phenotype and migrate to incipient tumor sites.Citation6,Citation20 Myeloma eradication is eventually achieved by the collaboration between tumor-specific TH1 cells and tumor-infiltrating, antigen-presenting M1 macrophages. Through these findings, we proposed a dual antitumor role of TH1-derived interferon γ (IFNγ).Citation20 First, IFNγ triggers the cytotoxic activity of tumor-infiltrating macrophages. Second, IFNγ induces macrophages to secrete the angiostatic chemokines C-X-C motif ligand 9 and 10 (CXCL9 and CXCL10), which may halt tumor progression by inhibiting angiogenesis and recruiting T cells. Collectively, our previous data suggest an antitumor role for the inflammatory response driven by tumor-specific TH1 cells.Citation6,Citation20,Citation21 Here, we performed a comprehensive molecular characterization of tumor-specific CD4+ T cells activated in vivo, both in tumor-draining LNs and at incipient tumor sites.
Results
Molecular changes on the surface of tumor-specific CD4+ T cells in draining lymph nodes upon activation in vivo
In order to characterize the activation of naïve tumor-specific CD4+ T cells in vivo, tumor-specific TCR-transgenic SCID mice were injected s.c. with MOPC315 myeloma cells suspended in Matrigel (). Eight days later, tumor-specific CD4+ T cells were collected from tumor-draining LNs and at incipient tumor sites (i.e., within Matrigel plugs) and analyzed by flow cytometry. In draining LNs, the expression of 89 molecules (listed in Table S1) on the cell surface was investigated. As a comparison, naïve tumor-specific CD4+ T cells from pooled LNs of non-injected TCR-transgenic SCID mice were analyzed in an identical manner (). Upon activation, 16 molecules were upregulated on the surface of tumor-specific CD4+ T cells in draining LNs, i.e., CD2, CD5 CD11a, CD18, CD27, CD44, CD45, CD54, CD69, CD71, CD86, CD153, CD200, CD249, CD278 and MHC class I (), while four molecules were downregulated, i.e., CD49d, CD62L, CD90 and CD126 (). Twelve additional molecules were equally expressed on activated and naïve tumor-specific CD4+ T cells: CD1d, CD4, CD28, CD31, CD45RB, CD51, CD95, CD102, CD122, CD274, Ly6A/E and Ly6C (). The remaining 57 molecules tested were not detected on the surface of either naïve or activated tumor-specific CD4+ T cells in LNs (data not shown).
Figure 1. Experimental set up. At day 0, T-cell receptor (TCR)-transgenic SCID mice were injected s.c. with MOPC315 myeloma cells (green cells) suspended in liquid Matrigel. When the Matrigel solution reached body temperature, it gelified and formed a plug embedding myeloma cells. Tumor-specific CD4+ T cells (red cells) became activated in the tumor-draining lymph node (LN), differentiated into TH1 cells, and subsequently migrated to incipient tumor sites (Matrigel plug). Eight d after the injection of tumor cells, mice were euthanized, and tumor-draining LNs and Matrigel plug were dissected out. The in vivo activation of tumor-specific CD4+ T cells was characterized by flow cytometry (in draining LNs and incipient tumor sites) and gene expression profiling (in draining LNs only).
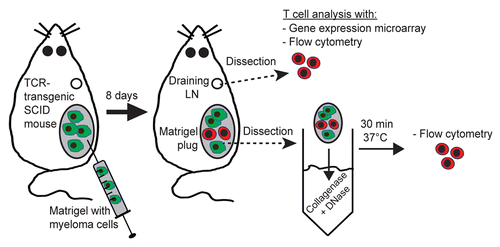
Figure 2. Expression pattern of molecules on the surface of tumor-specific CD4+ T cells in draining LN after in vivo activation. (A–C) T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells. Eight d later, the activation of tumor-specific (GB113+) CD4+ T cells from pooled tumor-draining lymph nodes (LNs) was analyzed by flow cytometry (blue curves). Filled gray areas indicate isotype-matched control stainings of activated T cells. For comparison, naïve tumor-specific CD4+ T cells from pooled LNs from non-injected TCR-transgenic SCID mice are shown (black curves). (A) Surface molecules that were upregulated after activation. (B) Surface molecules that were downregulated after activation. (C) Surface molecules that were expressed at similar levels on naïve and activated tumor-specific CD4+ T cells. Data are representative of 2–4 experiments.
The phenotype of tumor-specific CD4+ T cells at incipient tumor sites
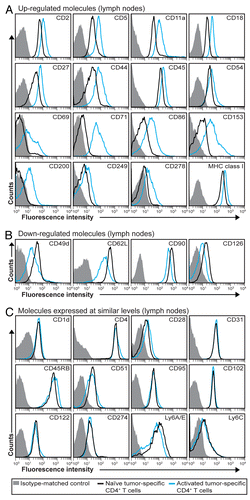
Matrigel-infiltrating tumor-specific CD4+ T cells were analyzed 8 d upon the injection of myeloma cells by flow cytometry and compared with activated and naïve T cells isolated from LNs. In this setting, we observed the upregulation of 29 cell-surface molecules, i.e., CD2, CD5, CD11a, CD18, CD25, CD28, CD44, CD45, CD49d, CD51, CD54, CD69, CD71, CD83, CD86, CD90, CD95, CD102, CD122, CD153, CD166, CD200, CD249, CD254, CD274, CD279, Ly6C, MHC class I and chemokine C-C motif receptor 7 (CCR7) and the downregulation of 5, i.e., CD27, CD31, CD45RB, CD62L and CD126 (). Many of these proteins were expressed at higher levels on the surface of tumor-infiltrating CD4+ T cells than on that of activated CD4+ T cells obtained from draining LNs (). Notably, several proteins not detected on T cells in LNs were observed on the surface of tumor-specific CD4+ T cells at incipient tumor sites, including CD25, CD83, CD166, CD254, CD279 and CCR7 (). Taken together, these observations indicate that tumor-specific CD4+ T cells that have migrated to incipient neoplastic lesions exhibit a higher activation or differentiation profile than activated tumor-specific CD4+ T cells found in LNs.
Figure 3. Molecular changes on the surface of activated tumor-specific CD4+ T cells that have migrated to incipient tumor sites. (A and B) T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells in Matrigel. Eight d later, the activation of Matrigel-infiltrating, tumor-specific (GB113+) T cells was analyzed by flow cytometry (red curves). Filled gray areas indicate isotype-matched control stainings of Matrigel-infiltrating T cells. For comparison, activated (blue curves) and naïve (black curves) tumor-specific CD4+ T cells from pooled lymph nodes (LNs) are shown. (A) Surface molecules that were upregulated after activation. (B) Surface molecules that were downregulated after activation. Data are representative of 2–4 experiments.
Activated tumor-specific CD4+ T cells differentiate toward a TH1 phenotype
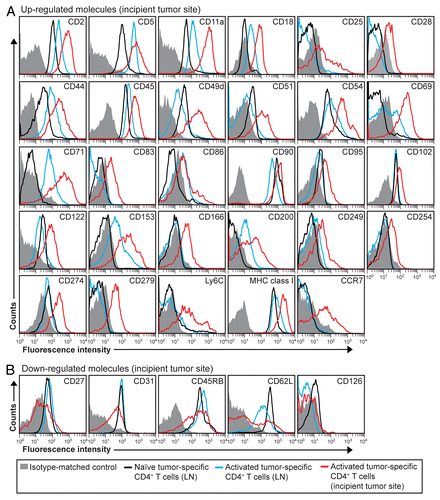
Effector CD4+ T cells mediate their functions mainly through the secretion of cytokines.Citation22 We have previously reported that Matrigel-infiltrating tumor-specific CD4+ T cells express T-bet, IFNγ and TNFα, consistent with a TH1 differentiation profile.Citation6,Citation20 To expand these findings, we analyzed the secretion of three additional cytokines (IL-2, IL-3 and IL-10) in both tumor-draining LNs and incipient tumor sites. In tumor-draining LNs, activated tumor-specific CD4+ T cells produced IFNγ, IL-2, IL-10 and TNFα (), while in incipient neoplastic lesions, tumor-specific CD4+ T cells secreted IFNγ, IL-3, IL-10 and TNFα but only low levels of IL-2 (). These data suggest that naïve tumor-specific CD4+ T cells first acquire a TH1 phenotype in draining LNs and then migrate as effector TH1 cells to incipient tumor sites. Notably, tumor-specific TH1 cells isolated 8 d after the inoculation of myeloma cells produced the pro-inflammatory cytokine TNFαCitation23 as well the (mainly) anti-inflammatory cytokine IL-10.Citation24
Figure 4. Intracellular cytokine expression in activated tumor-specific CD4+ T cells. (A and B) T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells in Matrigel. Eight d later, cytokine expression in tumor-specific (GB113+) CD4+ T cells was analyzed by intracellular flow cytometry, upon stimulation in vitro with ionomycin/phorbol myristate acetate (PMA) in the presence of monensin for 4 h. (A) Cytokine expression in activated tumor-specific CD4+ T cells in tumor-draining lymph nodes (LNs) (blue curves). For comparison, naïve tumor-specific CD4+ T cells from pooled LNs are shown (black curves). (B) Cytokine expression in tumor-specific (GB113+) T cells at incipient neoplastic lesions (Matrigel plugs) (red curves). Filled gray areas indicate isotype-matched control stainings. Data are representative of 2–3 experiments.
Early activation markers on tumor-specific CD4+ T cells activated in vivo
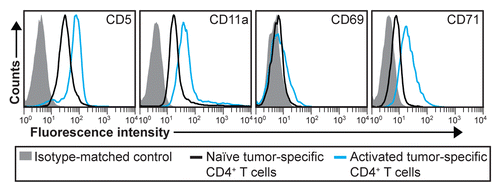
T-cell activation is often paralleled by the upregulation of CD25 and CD69 on the cell surface.Citation25,Citation26 Hence, these molecules are generally considered as early activation markers. However, CD25 and CD69 may be difficult to detect and alternative activation markers could prove useful. Driven by these considerations, we selected three markers (CD5, CD11a and CD71) that were strongly upregulated by CD4+ T cells 8 d after the inoculation of myeloma cells and analyzed their expression on activated tumor-specific CD4+ T cells in LNs at an earlier time point, namely on day 6 after tumor cell injection. We found that all three markers were clearly upregulated (). In contrast, CD69 was not as highly expressed on day +6 as on day +8. These data suggest that CD5, CD11a and CD71 may represent useful early activation markers for tumor-specific CD4+ T cells.
Figure 5. Early activation markers for tumor-specific CD4+ T cells. T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells. Six d later, the activation of tumor-specific (GB113+) CD4+ T cells from pooled tumor-draining lymph nodes (LNs) was analyzed by flow cytometry (blue curves). Filled gray areas indicate isotype-matched control stainings of activated T cells. For comparison, naïve tumor-specific CD4+ T cells from pooled LNs from non-injected TCR-transgenic SCID mice are shown (black curves). Data are representative of 2 experiments.
Gene expression in activated tumor-specific CD4+ TH1 cells
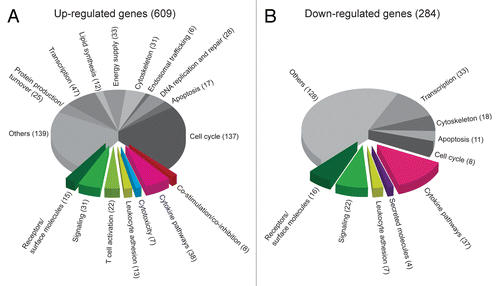
To obtain further insights into tumor-specific CD4+ T cells activated in vivo, we performed gene expression profiling. For this purpose, we compared the gene expression profile of activated tumor-specific CD4+ T cells obtained from tumor-draining LNs 8 d after the inoculation of myeloma cells with that of naïve tumor-specific CD4+ T cells. Using whole-genome microarrays, a total of 609 unique genes (856 probes) were found to be upregulated in activated tumor-specific CD4+ T cells (Table S2). The genes were manually annotated into functional pathways, and 134 of them were classified as related to the immune system, due to the involvement of their products in T-cell activation, co-stimulation, co-inhibition, leukocyte adhesion, cytokine pathways, cytotoxicity or immune signaling (). Among other functions, the remaining differentially expressed genes were linked to cell cycle, apoptosis, DNA replication and repair, endosomal trafficking, cytoskeleton, energy supply, lipid synthesis, transcription as well as protein production and turnover (). In addition, 284 unique genes (362 probes) were downregulated in activated tumor-specific CD4+ T cells (Table S3), of which 86 of the downregulated genes were linked to immune functions. Only a small fraction of downregulated genes were functionally associated with the cell cycle ().
Figure 6. Gene expression data arranged in functional pathways. (A and B) T-cell receptor (TCR)-transgenic SCID mice were injected s.c. with MOPC315 cells. Gene expression in activated tumor-specific CD4+ T cells isolated from pooled tumor-draining lymph nodes (LNs) 8 d later was compared with that of naïve tumor-specific CD4+ T cells. Genes were manually annotated into functional pathways. Genes important for or potentially associated with immune responses are raised from the pie chart. (A) Genes that are upregulated in activated tumor-specific CD4+ T cells as compared with naïve tumor-specific T cells. (B) Genes that are downregulated in activated tumor-specific CD4+ T cells as compared with naïve tumor-specific T cells.
Microarray data comparisons
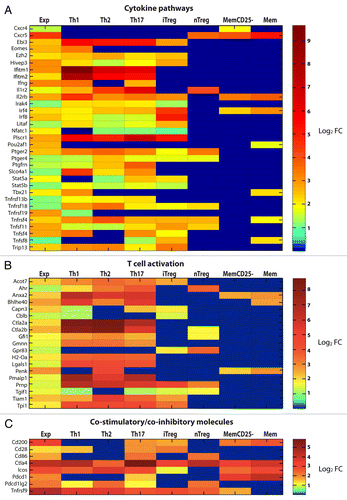
To obtain additional information from the mRNA profile of activated tumor-specific CD4+ T cells, up- and downregulated genes were compared with reference mRNA profiles of several subsets of polyclonal CD4+ T cells (obtained from the GEO database and the Immunological Genome Project).Citation27,Citation28 In particular, comparisons were made with TH1, TH2 and TH17 cells as well as with induced regulatory T cells (iTregs) that had been generated in vitro upon anti-CD3/CD28 stimulation of naïve CD4+ T cells in the presence of specific polarizing cytokines (; Figs. S1 and S2). Additional comparisons were made with CD4+CD25hi natural regulatory T cells (nTregs) and two populations of memory CD4+ T cells, namely CD25-CD44hiCD122lo (MemCD25-) and CD44hiCD62lo (Mem) cells, all of which had been isolated by cell sorting from wild-type C57BL/6J mice (; Figs. S1 and S2). The validity of this approach was supported by the observation that the Tbx21 gene, which encodes the TH1-specific transcription factor T-bet, was upregulated in both activated tumor-specific CD4+ T cells and reference TH1 cells but not in other T-cell subsets (except Mem) (). The gene expression profile of tumor-specific CD4+ T cells largely overlapped those of all in vitro polarized short-term T-cell cultures (i.e., TH1, TH2 and TH17 cells, and—to a lesser extent—iTregs), reflecting a core genetic program generally associated with early T-cell activation.
Figure 7. Comparison of upregulated genes in activated tumor-specific TH1 cells to reference data sets relative to several CD4+ T-cell subsets. (A–C) T-cell receptor (TCR)-transgenic SCID mice were injected s.c. with MOPC315 cells. Gene expression of activated tumor-specific CD4+ T cells isolated from pooled tumor-draining lymph nodes (LNs) 8 d later was analyzed. Upregulated genes were determined by comparing the gene expression in (1) activated tumor-specific CD4+ T cells to naïve tumor-specific CD4+ T cells (first column, annotated “Exp”) and (2) various activated reference CD4+ T-cell subsets to naïve reference CD4+ T cells. These reference sets were mouse CD4+ T cells from a previous study (annotated as TH1, TH2, TH17, iTreg and Treg in the heat-map) and memory T cells from the Immunological Genome Project (annotated as MemCD25-, and Mem in the heat-map). Genes that were upregulated (fold change, FC ≥ 2) in tumor-specific TH1 cells were selected for comparisons to reference data sets. Heat-map comparisons were categorized as the manual functional annotation categories shown in pie charts (). Each column represents expression comparisons for each CD4+ T-cell subset, and the scale bar indicates the level of differential gene expression (log2 transformed). Comparisons associated with a FC < 2 are in blue. (A) Genes involved in cytokine pathways. (B) Genes involved in T-cell activation. (C) Genes involved in co-stimulation/co-inhibition. Additional heat-maps can be found in Figures S1 and S2.
Correlation between gene and protein expression
Finally, we compared protein and mRNA expression levels for the molecules that had been successfully detected by flow cytometry on the surface of tumor-specific CD4+ T cells ( and ). For approximately half of these molecules, changes in protein levels were paralleled by similar shifts in mRNA expression levels (). In addition, several molecules that were not upregulated at the mRNA level in activated T cells, i.e., CD2, CD18, CD27, CD45, CD54 and CD69, were actually increased at the protein level, perhaps indicating the existence of post-transcriptional control mechanisms. Finally, for some molecules associated with a transcriptional upregulation in activated tumor-specific CD4+ T cells isolated from tumor-draining LNs, i.e., CD28, CD83, CD122 and CD279, protein levels were unchanged in the same cells but increased in activated tumor-specific CD4+ T cells infiltrating neoplastic lesions. This presumably reflects a delay between transcriptional upregulation and protein synthesis (or exposure on the cell surface).
Table 1. Changes in the expression of immune molecules in activated tumor-specific CD4+ T cells within lymph nodes
Discussion
Research on in vitro activated T cells has generated important findings regarding differentiation, intracellular signaling cascades and cell-to-cell interactions. However, T cells activated in vivo might display unique features due to the prominent influence of microenvironmental factors.Citation29 We have taken advantage of a model system in which tumor-specific CD4+ T cells activated in vivo can be studied while they eradicate tumor cells in the context of a primary anticancer immune response. This report is, to the best of our knowledge, the first comprehensive molecular characterization of tumor-specific CD4+ T cells activated in vivo in a setting of successful anticancer immunosurveillance.
A substantial part of the proteins that were either expressed or upregulated on activated tumor-specific CD4+ T cells in our study were adhesion molecules involved in the binding of T cells to APCs and endothelial cells, such as CD2, leukocyte function-associated antigen 1 (LFA-1, an integrin dimer composed of CD11a/CD18), CD44, the integrin α4 chain CD49d, the integrin αV chain CD51 and intercellular adhesion molecules 1 and 2 (ICAM-1 and ICAM-2; CD54 and CD102, respectively).Citation30,Citation31 CD44 has previously been shown to regulate the survival and memory development of TH1 cells.Citation32
The co-stimulatory molecules CD5, CD27, CD28 and CD278 are essential for T-cell activationCitation30,Citation33 and were upregulated on the surface of tumor-specific CD4+ T cells in our study. CD278 (inducible T-cell costimulator, ICOS) is upregulated upon TCR engagement and seems to be important for cell survival, polarization and memory.Citation34 We found the upregulation of CD278 particularly interesting as a tumor gene expression profile including high levels of CD278, presumably due to tumor-infiltrating T cells, has been associated with longer survival in metastatic melanoma patients.Citation35 CD27, a TNF receptor superfamily (TNFRSF) member, was upregulated in CD4+ T cells isolated from tumor-draining LNs but downregulated in cells infiltrating incipient tumor sites. CD27 has been suggested to modulate the generation of effector and memory T cells, with effector cells being CD27neg.Citation36 Hence, the downregulation of CD27 in incipient neoplastic lesions may indicate the presence of effector T cells. CD153 (ligand for the TNFRSF member CD30) was upregulated on activated tumor-specific CD4+ T cells. CD153 was involved in optimal TH1 responses against mycobacterial infections.Citation37 Moreover, CD153 expressing CD4+ T cells can interact with CD30-expressing T cells and hence boost IFNγ secretion.Citation37 The upregulation of CD153 in our study suggests that CD153 may play a role in tumor eradication, perhaps due to its impact on the maintenance of the TH1 effector phase through IFNγ production. Both TNFRSF members CD134 (OX-40) and CD137 (4–1BB)Citation30 were highly upregulated at the mRNA level. Surprisingly, we found CD86, a co-stimulatory molecule typically expressed by professional APCs,Citation38 to be upregulated on activated tumor-specific CD4+ T cells. The upregulation of CD86 at both the mRNA and protein levels strongly suggests that the CD86 molecules detected on the T-cell surface do not originate from trogocytosis (transfer of membrane molecules from APCs to T cells).Citation39 In fact, memory CD4+ T cells can express endogenously produced CD86 and provide co-stimulatory signals, indicating that CD86-expressing T cells may have a boosting effect on the activation of naïve T cells.Citation40,Citation41 Collectively, the data show that a successful antitumor immune response is accompanied by the expression of a high number of co-stimulatory and adhesion molecules on CD4+ T cells.
An appropriate stimulation of APCs is pivotal to initiate and maintain immune responses.Citation42 CD254, also known as TNF-related activation-induced cytokine (TRANCE), has been shown to enhance both DC survival and their capacity to secrete cytokines that are involved in inflammation, T-cell differentiation and proliferation.Citation43 The upregulation of CD254 on activated CD4+ T cells infiltrating incipient neoplastic lesions may indirectly ameliorate T-cell activation by stimulating DCs.
CD31 (PECAM-1) is a widely used marker for endothelial cells. However, as we found CD31 to be expressed on both naïve and activated tumor-specific CD4+ T cells, our data support previous findings that CD31 is not endothelial cell-specific. CD31 contributes to cellular adhesion but is also involved in T-cell activation and immune tolerance, as Cd31−/− mice have been shown to reject cancer more efficiently than their wild-type counterparts.Citation44,Citation45
Transcription factors are key elements for the polarization of T cells and cytokine secretion. Our gene expression data clearly demonstrates that activated tumor-specific CD4+ T cells polarize toward a TH1 phenotype, hence manifesting the upregulation of the TH1-associated transcription factor T-bet (in line with previous data on T-bet protein levels).Citation20 The transcription factor eomesodermin, which has been shown to regulate IFNγ production by CD4+ T cells,Citation46 was also upregulated. Eomesodermin appears to play an essential role in preventing TH17 development.Citation47 In our study, the inhibition of TH2 development could be supported by the upregulation of the transcriptional coactivator POU domain class 2 associating factor 1 (Pou2af1).Citation48 CD25 and CD122 are the IL-2 receptor chains α and β, respectively. CD25 and CD122 generally associate with CD132 (IL-2 receptor common γ chain) to form the high affinity trimeric IL-2 receptor,Citation49 which (upon IL-2 binding) promotes T-cell proliferation and differentiation.Citation50 The expression pattern of transcription factors, cytokines and receptors in activated tumor-specific CD4+ T cells support the notion that myeloma cells are eradicated in this system mainly via a TH1-polarized immune response.
CD62L and CCR7 are essential molecules in lymphocyte migration and homing.Citation51 Not surprisingly, CD62L was downregulated on the surface of activated tumor-specific CD4+ T cells found in tumor-draining LNs. However, at the incipient tumor site, we observed two CD4+ T-cell populations with respect to CD62L expression, one CD62Lneg and one CD62Lpos. This indicates that some tumor-specific CD4+ T cells may have diverged toward a CD62LposCCR7+ central memory phenotype.Citation51
Re-establishing immune homeostasis after activation is essential to avoid chronic inflammation. This involves the reprogramming of immune cells via immunomodulatory molecules. CD152 (CTLA4), CD95 (FAS), CD200, CD274 and CD279 (PD1), and IL-10 all exert immunomodulatory functions,Citation24,Citation52-Citation56 and were all found to be upregulated at the protein or mRNA levels in our study. Blocking these molecules might constitute a strategy to avoid the downregulation of immune responses in the context of anticancer immunotherapy.
Several additional molecules were expressed on activated tumor-specific CD4+ T cells. A substantial number of these proteins are involved in lymphocyte activation, such as the tyrosine phosphatase CD45 and its isoform CD45RB.Citation57,Citation58 Although CD83 is generally considered as a DC-specific marker, CD83 transgenic T cells have been shown to exert enhanced effector functions as they produce high levels of IFNγ and IL-2 upon antigen stimulation.Citation59 We observed that activated tumor-specific CD4+ T cells that infiltrate incipient neoplastic lesions expressed CD83. CD90 (Thy1), which was downregulated in CD4+ T cells in tumor-draining LNs but upregulated at the tumor site, has also been shown to take part in T-cell activation.Citation60 Along similar lines, both CD166 (activated leukocyte cell adhesion molecule, ALCAM) and CD71 (transferrin receptor) appear to be upregulated in activated T cells.Citation61,Citation62 CD1d and MHC class I are molecules involved in antigen presentation. CD1d was expressed but not upregulated on activated CD4+ T cells, while MHC class I was upregulated.
By comparing the genes that were modulated in activated tumor-specific CD4+ T cells with reference gene expression data covering several CD4+ T cell subsets, we outlined similarities and differences between a TH1 antitumor immune response in vivo and TH1 cells from a reference set. The gene expression profile of tumor-specific TH1 cells was relatively similar to that of reference TH1, TH2 and TH17 cells and iTregs. However, all these cells were activated, proliferating cells, which may explain the concurrent expression of genes involved in T-cell activation, protein production and cell cycle. Tregs are known to express many T-cell activation markers such as CD25 and CTLA4, which are erroneously considered Treg-specific by some investigators.Citation63 There was an even greater resemblance between tumor-specific TH1 cells and reference TH1 cells, as both cell types were shown to upregulate several genes associated with cytotoxicity, in contrast to other all T-cell subset analyzed. We also observed some differences in the gene expression pattern of tumor-specific TH1 cells and reference TH1 cells, especially concerning genes involved in apoptosis, signaling, co-stimulation and co-inhibition. Activating T cells in vivo (as we did in our experiments) is obviously different from doing so in vitro (as it was the case for reference TH1 cells), which may explain the somewhat disparate gene expression patterns. However, the upregulation of Cd86, Cd200 and Pdcd1 (coding for PD1) in tumor-specific TH1 cells activated in vivo, but not by reference TH1 cells, is intriguing and could be related to the presence of malignant cells. It has been recently reported that PD1-expressing tumor-infiltrating T cells constitute a favorable prognostic factor in head and neck cancer patients, suggesting that PD1 may represent a marker for activated effector T cells in human tumors.Citation64
In conclusion, here we have provided a comprehensive molecular characterization of tumor-specific CD4+ T cells that are capable of orchestrating a successful attack against cancer in vivo. This broad characterization may have implications for the development of several immunotherapeutic strategies against cancer, including vaccines. The expression of co-stimulatory molecules and adhesion molecules, as well as the differentiation toward a TH1 profile, seem to be essential components of a successful antitumor immune response. Supporting a tumor microenvironment that enhances these features could therefore be advantageous for cancer immunotherapy.
Materials and Methods
Mice and cell lines
λ2315-specific TCR-transgenic SCID mice on a BALB/c backgroundCitation13 were bred in a heterozygous fashion. MOPC315 (IgA, λ2315) cells, constituting a transplantable BALB/c plasmacytoma,Citation65 were obtained from the American Type Culture Collection (ATCC) and propagated as in vitro growing cells. The study was approved by the National Committee for Animal Experiments (Oslo, Norway).
Analysis of T-cell activation by flow cytometry
Adult (7–12 weeks old) TCR-transgenic SCID mice were injected s.c. on the flank with 105 MOPC315 cells suspended in 250 μL ice-cold (4°C) growth factor-reduced Matrigel (BD Biosciences). Six or 8 d later, mice were euthanized, and Matrigel plugs and draining axillary LNs were excised. Control naïve T cells were obtained from pooled axillary, cervical, brachial, inguinal, lumbar and sacral LNs from non-injected TCR-transgenic SCID mice. Matrigel plugs were treated with 1 mg/mL collagenase and 0.3 mg/mL DNase (both from Sigma) at 37°C for 30 min. Single-cell suspensions were obtained by squeezing the Matrigel solutions and LNs through a stainless steel sieve (Sigma). Unspecific binding was blocked by incubation with 30% heat-inactivated (56°C, 30 min) rat serum and 100 μg/mL anti-FcγRII/III monoclonal antibody (mAb) (clone 2.4G2; ATCC). Cells were stained for 15 min on ice with specific mAb in phosphate-buffered saline (PBS, Gibco) supplemented with 0.5% bovine serum albumin (Biotest). Commercially available mAb used in this study are listed in Table S1. The following mAbs were affinity-purified and, if needed, biotinylated in our laboratory: anti-transgenic-TCR-clonotype (GB113), anti-FcγRII/III (2.4G2). mAbs were conjugated with either FITC, phycoerythrin, allophycocyanin or biotin. Biotinylated mAbs were detected with streptavidin conjugated to peridinin chlorophyll protein (BD Biosciences). For intracellular cytokine detection, cells were stimulated with phorbol myristate acetate and ionomycin (both from Sigma) in vitro for 4 h in cell culture medium supplemented with monensin, before staining with Cytofix/Cytoperm Plus reagents (BD Biosciences) and specific mAbs. Quadruple-stained cells were analyzed on a FACSCalibur instrument with CellquestPro (BD Biosciences) and FlowJo (Tree Star, Inc.) softwares.
Gene expression analysis
Eight days after the inoculation of tumor cells, draining axillary LNs from TCR-transgenic SCID mice were dissected out and pooled, single-cell suspensions were made and staining with mAb was performed as described in the previous section. CD4+ GB113+ T cells were sorted (≥ 95% pure) by means of a FACSAria sorter in three independent experiments. Control T cells were obtained from pooled LNs excised from non-injected TCR-transgenic SCID mice and treated identically in two independent experiments. TRIzol (Invitrogen) was used to extract mRNA from sorted tumor-specific CD4+ T cells. Gene expression was quantified by gene microarrays (GeneChip Mouse Genome 430 2.0 Array, Affymetrix). The gene expression profiles were normalized and differentially expressed genes determined by comparing activated vs. naïve tumor-specific cells using the LIMMA package in R BioConductor.Citation66 Genes associated with a fold change (FC) ≥ 2 or ≤ 0.5 and with a p value < 0.05 were considered as differentially expressed. The microarray expression data sets were deposited in Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) under accession number GSE46453, and analyzed further with DAVID Bioinformatics Resources 6.7.Citation67,Citation68
Microarray data comparisons
Raw Affymetrix microarray data for CD4+ T cell subsets from a previous murine gene expression profiling study was downloaded from GEO (GSE14308). These CD4+ subsets comprised of naïve CD4+ cells, TH1, TH2, TH17, iTreg and nTreg arrays.Citation27 Reference TH1, TH2 and TH17 cells as well as iTregs were generated in vitro by anti-CD3/CD28 stimulation of naïve polyclonal CD4+ T cells in the presence of specific polarizing cytokines. CD4+CD25hi nTreg cells were isolated by cell sorting from the spleen and LNs of wild-type C57BL/6J mice. Each array consisted of 2 replicates, and data were normalized using the LIMMA software package in R BioConductor.Citation66 FC analysis was performed by comparing each of the T-cell subsets against naïve CD4+ T cells in the same study. Significance was defined using multiple testing at a false discovery rate cutoff of 0.1.
Additional comparisons were retrieved from the Immunological Genome Project (www.immgen.org). FC analysis was performed by comparing murine naïve CD4+ T cells (subcutanous LN naïve CD4+CD8−TCR+CD25−CD62LhiCD44low B6 6W) against two types of CD4+ memory T cells: (1) subcutanous LN memory-phenotype CD4+CD8-TCR+CD25−CD44hiCD122low B6 6W cells (annotated as MemCD25− in heat-maps) and (2) subcutanous LN memory-phenotype CD4+CD8−TCR+CD44hiCD62lowGFP-FOXP3− CD45.1-B6 cells (annotated as Mem in heat-maps).
| Abbreviations: | ||
| APC | = | antigen-presenting cell |
| CCR7 | = | chemokine C-C motif receptor 7 |
| CTLA4 | = | cytotoxic T lymphocyte-associated antigen 4 |
| DC | = | dendritic cell |
| FC | = | fold change |
| IFNγ | = | interferon γ |
| IL | = | interleukin |
| mAb | = | monoclonal antibody |
| LN | = | lymph node |
| PD1 | = | programmed death 1 |
| PDL1 | = | PD1 ligand 1 |
| s.c. | = | subcutaneous |
| SCID | = | severe combined immunodeficiency |
| TCR | = | T-cell receptor |
| TNFα | = | tumor necrosis factor alpha |
| TNFRSF | = | TNF receptor super family |
| Treg | = | regulatory T cell |
Additional material
Download Zip (724.8 KB)Acknowledgments
We thank Lise Aagaard, Hela Soltani and Siv Haugen Tunheim for technical help; Eivind Hovig for technical consultation on the microarray analysis; Anita Berge, Merete Høyem, Marianne Johannessen, Nina Kjølen, Maureen Lysaker, Thayaruby Narayanapillai, Mette Nielsen, Christine Olsen and Anneli Skogstad for helping type and for taking good care of the animals; Michael Bodd and Peter O. Hofgaard for critical reading of the manuscript. This work was supported by grants from South-Eastern Norway Regional Health Authority, The Research Council of Norway, the Norwegian Cancer Society, Kjøpmann Einar Unsgaard og hustru Kitty Unsgaards legat and Anders Jahres fund. K.B.L., O.A.W.H. and A.C. performed the experiments and collected the data. T.C. performed the bioinformatics analysis. K.B.L., O.A.W.H., T.C. and A.C. prepared the figures. All authors analyzed and discussed the data. B.B. and A.C. designed the study. K.B.L. wrote the manuscript. O.A.W.H., T.C., B.B. and A.C. contributed in writing the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here:
www.landesbioscience.com/journals/oncoimmunology/article/24383
Notes
† These authors contributed equally to this work.
References
- Levitsky HI, Lazenby A, Hayashi RJ, Pardoll DM. In vivo priming of two distinct antitumor effector populations: the role of MHC class I expression. J Exp Med 1994; 179:1215 - 24; http://dx.doi.org/10.1084/jem.179.4.1215; PMID: 7908321
- Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4(+) T cells in the antitumor immune response. J Exp Med 1998; 188:2357 - 68; http://dx.doi.org/10.1084/jem.188.12.2357; PMID: 9858522
- Ossendorp F, Mengedé E, Camps M, Filius R, Melief CJ. Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J Exp Med 1998; 187:693 - 702; http://dx.doi.org/10.1084/jem.187.5.693; PMID: 9480979
- Pardoll DM, Topalian SL. The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol 1998; 10:588 - 94; http://dx.doi.org/10.1016/S0952-7915(98)80228-8; PMID: 9794842
- Marzo AL, Kinnear BF, Lake RA, Frelinger JJ, Collins EJ, Robinson BW, et al. Tumor-specific CD4+ T cells have a major “post-licensing” role in CTL mediated anti-tumor immunity. J Immunol 2000; 165:6047 - 55; PMID: 11086036
- Corthay A, Skovseth DK, Lundin KU, Røsjø E, Omholt H, Hofgaard PO, et al. Primary antitumor immune response mediated by CD4+ T cells. Immunity 2005; 22:371 - 83; http://dx.doi.org/10.1016/j.immuni.2005.02.003; PMID: 15780993
- Cho Y, Miyamoto M, Kato K, Fukunaga A, Shichinohe T, Kawarada Y, et al. CD4+ and CD8+ T cells cooperate to improve prognosis of patients with esophageal squamous cell carcinoma. Cancer Res 2003; 63:1555 - 9; PMID: 12670904
- Yoshida N, Abe H, Ohkuri T, Wakita D, Sato M, Noguchi D, et al. Expression of the MAGE-A4 and NY-ESO-1 cancer-testis antigens and T cell infiltration in non-small cell lung carcinoma and their prognostic significance. Int J Oncol 2006; 28:1089 - 98; PMID: 16596224
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012; 12:298 - 306; http://dx.doi.org/10.1038/nrc3245; PMID: 22419253
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711 - 23; http://dx.doi.org/10.1056/NEJMoa1003466; PMID: 20525992
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366:2443 - 54; http://dx.doi.org/10.1056/NEJMoa1200690; PMID: 22658127
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366:2455 - 65; http://dx.doi.org/10.1056/NEJMoa1200694; PMID: 22658128
- Bogen B, Gleditsch L, Weiss S, Dembic Z. Weak positive selection of transgenic T cell receptor-bearing thymocytes: importance of major histocompatibility complex class II, T cell receptor and CD4 surface molecule densities. Eur J Immunol 1992; 22:703 - 9; http://dx.doi.org/10.1002/eji.1830220313; PMID: 1547816
- Lauritzsen GF, Weiss S, Dembic Z, Bogen B. Naive idiotype-specific CD4+ T cells and immunosurveillance of B-cell tumors. Proc Natl Acad Sci U S A 1994; 91:5700 - 4; http://dx.doi.org/10.1073/pnas.91.12.5700; PMID: 7911244
- Bogen B, Munthe L, Sollien A, Hofgaard P, Omholt H, Dagnaes F, et al. Naive CD4+ T cells confer idiotype-specific tumor resistance in the absence of antibodies. Eur J Immunol 1995; 25:3079 - 86; http://dx.doi.org/10.1002/eji.1830251114; PMID: 7489746
- Lauritzsen GF, Bogen B. The role of idiotype-specific, CD4+ T cells in tumor resistance against major histocompatibility complex class II molecule negative plasmacytoma cells. Cell Immunol 1993; 148:177 - 88; http://dx.doi.org/10.1006/cimm.1993.1100; PMID: 8098665
- Corthay A, Lundin KU, Lorvik KB, Hofgaard PO, Bogen B. Secretion of tumor-specific antigen by myeloma cells is required for cancer immunosurveillance by CD4+ T cells. Cancer Res 2009; 69:5901 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-08-4816; PMID: 19567679
- Dembic Z, Schenck K, Bogen B. Dendritic cells purified from myeloma are primed with tumor-specific antigen (idiotype) and activate CD4+ T cells. Proc Natl Acad Sci U S A 2000; 97:2697 - 702; http://dx.doi.org/10.1073/pnas.050579897; PMID: 10706628
- Lorvik KB, Bogen B, Corthay A. Fingolimod blocks immunosurveillance of myeloma and B-cell lymphoma resulting in cancer development in mice. Blood 2012; 119:2176 - 7; http://dx.doi.org/10.1182/blood-2011-10-388892; PMID: 22383793
- Haabeth OA, Lorvik KB, Hammarström C, Donaldson IM, Haraldsen G, Bogen B, et al. Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat Commun 2011; 2:240; http://dx.doi.org/10.1038/ncomms1239; PMID: 21407206
- Haabeth OA, Bogen B, Corthay A. A model for cancer-suppressive inflammation. Oncoimmunology 2012; 1:1146 - 55; http://dx.doi.org/10.4161/onci.21542; PMID: 23170261
- Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood 2008; 112:1557 - 69; http://dx.doi.org/10.1182/blood-2008-05-078154; PMID: 18725574
- Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science 2002; 296:1634 - 5; http://dx.doi.org/10.1126/science.1071924; PMID: 12040173
- Li MO, Flavell RA. Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity 2008; 28:468 - 76; http://dx.doi.org/10.1016/j.immuni.2008.03.003; PMID: 18400189
- Robb RJ, Munck A, Smith KA. T cell growth factor receptors. Quantitation, specificity, and biological relevance. J Exp Med 1981; 154:1455 - 74; http://dx.doi.org/10.1084/jem.154.5.1455; PMID: 6975347
- López-Cabrera M, Santis AG, Fernández-Ruiz E, Blacher R, Esch F, Sánchez-Mateos P, et al. Molecular cloning, expression, and chromosomal localization of the human earliest lymphocyte activation antigen AIM/CD69, a new member of the C-type animal lectin superfamily of signal-transmitting receptors. J Exp Med 1993; 178:537 - 47; http://dx.doi.org/10.1084/jem.178.2.537; PMID: 8340758
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009; 30:155 - 67; http://dx.doi.org/10.1016/j.immuni.2008.12.009; PMID: 19144320
- Heng TS, Painter MW, Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008; 9:1091 - 4; http://dx.doi.org/10.1038/ni1008-1091; PMID: 18800157
- Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, et al. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol 2001; 19:23 - 45; http://dx.doi.org/10.1146/annurev.immunol.19.1.23; PMID: 11244029
- Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol 2009; 27:591 - 619; http://dx.doi.org/10.1146/annurev.immunol.021908.132706; PMID: 19132916
- Nandi A, Estess P, Siegelman M. Bimolecular complex between rolling and firm adhesion receptors required for cell arrest; CD44 association with VLA-4 in T cell extravasation. Immunity 2004; 20:455 - 65; http://dx.doi.org/10.1016/S1074-7613(04)00077-9; PMID: 15084274
- Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity 2010; 32:104 - 15; http://dx.doi.org/10.1016/j.immuni.2009.10.011; PMID: 20079666
- Ledbetter JA, Martin PJ, Spooner CE, Wofsy D, Tsu TT, Beatty PG, et al. Antibodies to Tp67 and Tp44 augment and sustain proliferative responses of activated T cells. J Immunol 1985; 135:2331 - 6; PMID: 3928743
- Simpson TR, Quezada SA, Allison JP. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS). Curr Opin Immunol 2010; 22:326 - 32; http://dx.doi.org/10.1016/j.coi.2010.01.001; PMID: 20116985
- Bogunovic D, O’Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A 2009; 106:20429 - 34; http://dx.doi.org/10.1073/pnas.0905139106; PMID: 19915147
- Arens R, Tesselaar K, Baars PA, van Schijndel GM, Hendriks J, Pals ST, et al. Constitutive CD27/CD70 interaction induces expansion of effector-type T cells and results in IFNgamma-mediated B cell depletion. Immunity 2001; 15:801 - 12; http://dx.doi.org/10.1016/S1074-7613(01)00236-9; PMID: 11728341
- Tang C, Yamada H, Shibata K, Muta H, Wajjwalku W, Podack ER, et al. A novel role of CD30L/CD30 signaling by T-T cell interaction in Th1 response against mycobacterial infection. J Immunol 2008; 181:6316 - 27; PMID: 18941223
- Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol 2005; 23:515 - 48; http://dx.doi.org/10.1146/annurev.immunol.23.021704.115611; PMID: 15771580
- Davis DM. Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat Rev Immunol 2007; 7:238 - 43; http://dx.doi.org/10.1038/nri2020; PMID: 17290299
- Hakamada-Taguchi R, Kato T, Ushijima H, Murakami M, Uede T, Nariuchi H. Expression and co-stimulatory function of B7-2 on murine CD4+ T cells. Eur J Immunol 1998; 28:865 - 73; http://dx.doi.org/10.1002/(SICI)1521-4141(199803)28:03<865::AID-IMMU865>3.0.CO;2-T; PMID: 9541581
- Jeannin P, Herbault N, Delneste Y, Magistrelli G, Lecoanet-Henchoz S, Caron G, et al. Human effector memory T cells express CD86: a functional role in naive T cell priming. J Immunol 1999; 162:2044 - 8; PMID: 9973476
- Gallucci S, Matzinger P. Danger signals: SOS to the immune system. Curr Opin Immunol 2001; 13:114 - 9; http://dx.doi.org/10.1016/S0952-7915(00)00191-6; PMID: 11154927
- Josien R, Wong BR, Li HL, Steinman RM, Choi Y. TRANCE, a TNF family member, is differentially expressed on T cell subsets and induces cytokine production in dendritic cells. J Immunol 1999; 162:2562 - 8; PMID: 10072496
- Ma L, Mauro C, Cornish GH, Chai JG, Coe D, Fu H, et al. Ig gene-like molecule CD31 plays a nonredundant role in the regulation of T-cell immunity and tolerance. Proc Natl Acad Sci U S A 2010; 107:19461 - 6; http://dx.doi.org/10.1073/pnas.1011748107; PMID: 20978210
- Newton-Nash DK, Newman PJ. A new role for platelet-endothelial cell adhesion molecule-1 (CD31): inhibition of TCR-mediated signal transduction. J Immunol 1999; 163:682 - 8; PMID: 10395658
- Suto A, Wurster AL, Reiner SL, Grusby MJ. IL-21 inhibits IFN-gamma production in developing Th1 cells through the repression of Eomesodermin expression. J Immunol 2006; 177:3721 - 7; PMID: 16951332
- Yang Y, Xu J, Niu Y, Bromberg JS, Ding Y. T-bet and eomesodermin play critical roles in directing T cell differentiation to Th1 versus Th17. J Immunol 2008; 181:8700 - 10; PMID: 19050290
- Brunner C, Sindrilaru A, Girkontaite I, Fischer KD, Sunderkötter C, Wirth T. BOB.1/OBF.1 controls the balance of TH1 and TH2 immune responses. EMBO J 2007; 26:3191 - 202; http://dx.doi.org/10.1038/sj.emboj.7601742; PMID: 17568779
- Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science 2005; 310:1159 - 63; http://dx.doi.org/10.1126/science.1117893; PMID: 16293754
- Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol 2011; 23:598 - 604; http://dx.doi.org/10.1016/j.coi.2011.08.003; PMID: 21889323
- Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999; 401:708 - 12; http://dx.doi.org/10.1038/44385; PMID: 10537110
- Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994; 1:405 - 13; http://dx.doi.org/10.1016/1074-7613(94)90071-X; PMID: 7882171
- Brenner D, Krammer PH, Arnold R. Concepts of activated T cell death. Crit Rev Oncol Hematol 2008; 66:52 - 64; http://dx.doi.org/10.1016/j.critrevonc.2008.01.002; PMID: 18289867
- Minas K, Liversidge J. Is the CD200/CD200 receptor interaction more than just a myeloid cell inhibitory signal?. Crit Rev Immunol 2006; 26:213 - 30; http://dx.doi.org/10.1615/CritRevImmunol.v26.i3.20; PMID: 16928187
- Keir ME, Francisco LM, Sharpe AH. PD-1 and its ligands in T-cell immunity. Curr Opin Immunol 2007; 19:309 - 14; http://dx.doi.org/10.1016/j.coi.2007.04.012; PMID: 17433872
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000; 192:1027 - 34; http://dx.doi.org/10.1084/jem.192.7.1027; PMID: 11015443
- James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature 2012; 487:64 - 9; PMID: 22763440
- Birkeland ML, Kraus T, Tardelli L, Puré E. Progressive changes in CD45RB phenotype and lymphokine production by murine CD4+ T cells after alloantigen exposure. Immunology 1992; 75:632 - 8; PMID: 1350569
- Wolenski M, Cramer SO, Ehrlich S, Steeg C, Fleischer B, von Bonin A. Enhanced activation of CD83-positive T cells. Scand J Immunol 2003; 58:306 - 11; http://dx.doi.org/10.1046/j.1365-3083.2003.01303.x; PMID: 12950676
- Haeryfar SM, Hoskin DW. Thy-1: more than a mouse pan-T cell marker. J Immunol 2004; 173:3581 - 8; PMID: 15356100
- Bowen MA, Bajorath J, D’Egidio M, Whitney GS, Palmer D, Kobarg J, et al. Characterization of mouse ALCAM (CD166): the CD6-binding domain is conserved in different homologs and mediates cross-species binding. Eur J Immunol 1997; 27:1469 - 78; http://dx.doi.org/10.1002/eji.1830270625; PMID: 9209500
- Caruso A, Licenziati S, Corulli M, Canaris AD, De Francesco MA, Fiorentini S, et al. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry 1997; 27:71 - 6; http://dx.doi.org/10.1002/(SICI)1097-0320(19970101)27:1<71::AID-CYTO9>3.0.CO;2-O; PMID: 9000587
- Corthay A. How do regulatory T cells work?. Scand J Immunol 2009; 70:326 - 36; http://dx.doi.org/10.1111/j.1365-3083.2009.02308.x; PMID: 19751267
- Badoual C, Hans S, Merillon N, Van Ryswick C, Ravel P, Benhamouda N, et al. PD-1-expressing tumor-infiltrating T cells are a favorable prognostic biomarker in HPV-associated head and neck cancer. Cancer Res 2013; 73:128 - 38; http://dx.doi.org/10.1158/0008-5472.CAN-12-2606; PMID: 23135914
- Eisen HN, Simms ES, Potter M. Mouse myeloma proteins with antihapten antibody acitivity. The protein produced by plasma cell tumor MOPC-315. Biochemistry 1968; 7:4126 - 34; http://dx.doi.org/10.1021/bi00851a048; PMID: 4177521
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3:e3; http://dx.doi.org/10.2202/1544-6115.1027; PMID: 16646809
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44 - 57; http://dx.doi.org/10.1038/nprot.2008.211; PMID: 19131956
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1 - 13; http://dx.doi.org/10.1093/nar/gkn923; PMID: 19033363