Abstract
The specification of cell lineages and patterning in the embryo occurs sequentially as specific regions are increasingly restricted in their developmental fates. When and how this occurs is still not entirely clear. Nevertheless, the roles of epigenetic regulatory genes in partitioning the genome into active and inactive domains is evident in a variety of organisms and is highly conserved through evolution. The function of Pax2 in the kidney has been inferred by the phenotypic analysis of loss-of-function mutants in mice, fish, and humans. Although Pax2 and the related gene, Pax8, are essential for early intermediate mesoderm specification and are found in the epithelial lineage arising from that mesoderm, how these proteins regulate cell lineage restriction and gene expression patterns has remained obscure. Our recent data, suggests that Pax proteins help establish chromatin domains within cell lineages by providing the locus and tissue specificity for epigenetic imprinting complexes that modify histones. The novel protein PTIP is a key adaptor that links Pax proteins and possibly many other types of DNA binding proteins to a histone H3K4 methyltransferase complex. Given the prevalence of Pax2 expression in kidney development and in kidney disease, we now need to address the effects of epigenetics on renal disease states, on the stability of the terminal epithelial phenotype, and in the aging cell.
Introduction
How specialized cells and tissues differentiate from their more simple progenitors during embryonic development and become organized into a three-dimensional architectural framework remains a central problem in developmental biology and in the newly emerging field of regenerative medicine. Furthermore, the problem of maintaining the terminally differentiated state in adult tissues, cells of which may be renewing very slowly, if at all, is critical to proper physiological function and suppression of disease associated pathologies. Inherent to any differentiated state is the regulated expression of cell-type specific genes that define a particular cell phenotype. How are these patterns of gene expression established and remembered, in some cases through many cell divisions? How do cells receive and interpret the information that instructs them to assume specific fates and lineage pathways? Remarkable progress has been made in developmental biology that impacts our understanding of differentiation, cellular memory and gene regulation not only in normal systems but also in disease states. I will discuss how these emerging concepts impact our understanding of kidney development and renal disease and present some examples from my lab and others working in the field.
Patterning and Cell Lineages
Any complex metazoan has a body-plan, or pattern, that must be established during development. This body-plan consists of specialized, or differentiated, cells organized in a three-dimensional framework. This architectural plan can vary greatly amongst species and may be entirely independent of the cell types that make up tissues and organs. Indeed, when considering the variety of species even within a class of vertebrates, it is evident that many organisms have similar cell types and cell lineage pathways, yet their body patterns are very different. Are the genetic mechanisms that translate the genome into developmental programs similar among many species, or are they unique for each particular pattern generated?
Progress in understanding the molecular genetic basis of body patterning and cell lineage determination owes much to model organisms that are amenable to mutational screens and phenotypic analysis. In fact, many of the concepts and genes we now know to be essential for embryonic development in mammals trace their origins to a landmark paper by Christiane Nusslein and Eric WieschausCitation1 in which the fruit fly, Drosophila Melanogaster was used to identify mutations in genes controlling segmentation and polarity along the body axis. Before the individual genes were actually cloned, this study was able to postulate a hierarchy of segmentation genes that subdivided the embryonic body axis into increasingly smaller units. The concepts of segmentation did not directly touch upon the specification of different identities along the anterior-posterior axis. It was the pioneering work of Ed LewisCitation2,Citation3 that was critical to establishing the function of Drosophila homeotic or Hox gene complexes in determining the identities of individual segments. Thus, in simplest terms, there are genetic mechanisms that subdivide the developing embryo into regions, or segments, most likely by forming boundaries between adjacent groups of cells and there are mechanisms that assign a fate, or identity, to individual regions. Lastly, a group of epigenetic regulatory genes is essential for establishing the correct regionalization of homeotic gene expression in the fly embryo. These epigenetic regulators fall into two groups,Citation4 the Polycomb family, which is generally thought to repress homeotic gene expression at ectopic sites, and the Trithorax family, which is thought to activate or maintain homeotic gene expression at the correct site.
All metazoans require genetic mechanisms to specify body axes and cell types. Thus, it is no surprise that once nature has perfected the means to translate genetic information into a three-dimensional form, the basic regulatory machinery is highly conserved even if the biological outputs are vastly different. Mutational screens pointed to the existence of segmentation and homeotic genes in the fly. Once these were cloned, it quickly became apparent that mice and humans had many similar genes. My personal interest in development was sparked by this remarkable conservation of genetic mechanisms including the DNA binding proteins encoded by the mammalian Hox proteins and their Drosophila counterparts. But the Hox DNA binding domain was not the only region conserved over such vast evolutionary distance. As a post-doc in the lab of Peter Gruss, I was fortunate enough to be involved in the identification of the mammalian Pax gene family, which had a conserved DNA binding domain found in a subset of Drosophila segmentation genes.Citation5 These Pax genes were all expressed during embryonic development, some in obviously segmented structures, and others in tissue and regional specific patterns (). Remarkably, some of the Pax genes identified solely by DNA sequence conservation with Drosophila segmentation genes, mapped directly to genes mutated in congenital abnormalities including human aniridia (Pax6) and Wardenburg's syndrome (Pax3).Citation6 Thus, it became clear that the Pax genes were essential for forming specific structures very early in development.
Kidney Development
The developing kidney has been studied in detail ever since the pioneering work of Laurie SaxenCitation7 and his disciples. Remarkable progress has been made in refining the morphology and molecular underpinnings of early renal development. For the purposes of this article, I would like to briefly summarize the important early events and will refer the reader to some recent reviews for more in depth discussion.Citation8,Citation9 A summary of renal development must begin with gastrulation. In vertebrates, the process of gastrulation converts a single pluripotent sheet of embryonic tissue, the epiblast or embryonic ectoderm, into the three primary germ layers, the endoderm, the mesoderm and the ectoderm. In mammals, gastrulation is marked by a furrow called the primitive streak. At the most anterior end of the primitive streak is the node or organizer, called Hensen's node in the chick, and functionally equivalent to the blastopore lip or Speeman's organizer in the amphibian embryo. The node is a signaling center that expresses a potent combination of secreted factors for establishing the body axes and left-right asymmetry. The node moves anteriorly with the primitive streak to approximately the midpoint of the epiblast. The primitive streak then regresses and lays down the notochord along the ventral midline of the embryo. The notochord is a second critical signaling center for dorso-ventral and medio-lateral patterning of both neural plate and paraxial mesoderm. The paraxial mesoderm refers to the most medial mesodermal cells, which in response to regression of the streak become segmented into somites, blocks of cells surrounded by a simple epithelium (). At the level of the fourth somite and caudally, going medial to lateral, the notochord marks the midline, the somites abut the notochord on either side, and the unsegmented mesoderm is termed intermediate near the somite and lateral plate more distally. It is this region of intermediate mesoderm within which the kidney will form.
The earliest morphological indication of unique derivatives arising from the intermediate mesoderm is the formation of the pronephric duct, which is also called the primary nephric duct or Wolffian duct. This single cell thick epithelial tube runs bilaterally beginning at around the 12th somite in birds and mammals. The nephric duct extends caudally until it reaches the cloaca. As it grows, it induces a linear array of epithelial tubules, which are thought to derive from peri-ductal mesenchyme (). The tubules are referred to as pronephric or mesonephric, depending on their position and degree of development and represent an evolutionarily more primitive excretory system that forms transiently in mammals until it is replaced by the adult or metanephric kidney. Along the nephric duct, there is a graded evolution of renal tubule development with the most anterior, or pronephric tubules, being very rudimentary, and the mesonephric tubules becoming well developed with glomeruli and convoluted proximal tubule like structures.
The adult kidney, or metanephros, is formed at the caudal end of the nephric duct when an outgrowth, called the ureteric bud or metanephric diverticulum, extends into the surrounding metanephric mesenchyme (). Outgrowth or budding of the epithelia requires signals emanating from the mesenchyme. Once the ureteric bud has invaded the metanephric mesenchyme, inductive signals from the bud initiate the conversion of the metanephric mesenchyme to epithelium. The induced, condensing mesenchymal cells aggregate around the tips of the bud and will form a primitive polarized epithelium, the renal vesicle. Through a series of cleft formations, the renal vesicle forms first a comma then an S-shaped body, whose most distal end remains in contact with the ureteric bud epithelium and fuses to form a continuous epithelial tubule. This s-shaped tubule begins to express genes specific for glomerular podocyte cells at its most proximal end, markers for more distal tubules near the fusion with the ureteric bud epithelia and proximal tubules markers in between. Endothelial cells begin to infiltrate the most proximal cleft of the s-shaped body as the vasculature of the glomerular tuft takes shape. At this stage, the glomerular epithelium consists of a visceral and parietal component, with the visceral cells becoming podocytes and the parietal cells the epithelia surrounding the urinary space. The capillary tuft consists of capillary endothelial cells and a specialized type of smooth muscle cell, termed the mesangial cell, whose origin remains unclear.
While these renal vesicles are generating much of epithelia of the nephron, the ureteric bud epithelia continues to undergo branching morphogenesis in response to signals derived from the mesenchyme (for review, ref. Citation10). Branching follows a stereotypical pattern and results in new mesenchymal aggregates induced at the tips of the branches, as new nephrons are sequentially induced. This repeated branching and induction results in the formation of nephrons along the radial axis of the kidney, with the oldest nephrons being more medullary and the younger nephrons located towards the periphery. However, not all cells of the mesenchyme become induced and convert to epithelia, some cells remain mesenchymal and migrate to the interstitium. These interstitial mesenchymal cells, or stromal cells, are essential for providing signals that maintain branching morphogenesis of the ureteric bud and survival of the mesenchyme.
With the advent of molecular cloning and targeted mutagenesis in the mouse, some of the molecular events that drive the morphogenesis of the kidney are now at hand. Among the critical discoveries were the c-ret receptor tyrosine kinaseCitation11 and its ligand glial derived neurotrophic factor as mediators of ureteric bud outgrowth, the role of the Wilms' tumor suppressor gene WT1 in early kidney mesenchyme survival,Citation12 the function of Wnt signaling in the polarization of the mesenchymal aggregates to epithelial vesiclesCitation13 and the subsequent identification of Wnt9b as the inductive signal emanating from the ureteric bud.Citation14 Presently, the list of factors that are now known to regulate early kidney development is rather lengthy and includes both nuclear proteins with DNA binding capacity, cell signaling proteins, receptors, and cell matrix and matrix interacting proteins ().
Pax Genes and Early Kidney Patterning
The essential nature of Pax genes in kidney development is evident from several key observations in genetically engineered mice. Among the earliest markers specific for the intermediate mesoderm are expression of the Pax2 and Pax8 genes, which are considered redundant at E9.5.Citation15 However at later stages, Pax2 but not Pax8 mutant mice exhibit complete agenesis of both kidneys and ureters ().Citation16 In humans, loss of one Pax2 allele can result in renal hypoplasia, vesicoureteral reflux and optic nerve colobomas.Citation17 These phenotypes are consistent with the Pax2 expression pattern in the epithelial components of the mesonephros, the nephric duct, the ureteric bud, and in the developing renal tubules.Citation18,Citation19 Both in vivo and in vitro, Pax2 is required for the conversion of renal mesenchymal cells to a primitive, proliferating epithelium.Citation20 Prior to induction, Pax2 protein expression demarcates the metanephric mesenchyme, activates GDNF expression such that ureteric bud epithelial outgrowth can occur, and controls the response to inductive signals.Citation21 Pax2 expression persists in the condensing mesenchyme and in the comma and s-shaped bodies, being down-regulated first in the most proximal loop of the s-shaped body and then in the epithelial cells of the proximal and distal tubules.
All the cumulative genetic and expression data point to an essential role for Pax2 in patterning the early intermediate mesoderm such that renal epithelial cells can be generated. In the chick embryo, ectopic Pax2, but not Pax8 could expand the region of intermediate mesoderm 15, whereas in the Xenopus embryo Pax2 and lim1 could achieve similar effects.Citation22 Thus, within the context of the developing mesoderm, Pax2 is sufficient to define the boundaries of the intermediate mesoderm along the medio-ventral axis. This early expression pattern suggests a fundamental role for Pax2/8 in mesodermal compartmentalization, perhaps similar to the boundary formation seen in Drosophila segmentation.
Despite all the information gleaned from genetic studies in mouse and humans, the biochemical nature of Pax proteins is less clear. The proteins contain a number of domains, as shown schematically in . Pax proteins bind DNA via the characteristic 128 amino acid paired-domain at the amino-terminus of the protein.Citation23–Citation25 The crystal structure of the Drosophila paired protein bound to DNACitation26 shows three amino-terminal α-helices that resemble a homeo-domain, followed by a carboxyl terminal region with three smaller α-helices. The amino terminal α-helices contact the 3′ part of the bipartite target sequence, whereas the carboxyl terminal tail recognizes the 5′ end of the DNA sequence. This results in a potentially large DNA recognition sequence that can span almost two full turns of the double helix. Furthermore, interactions between the paired-domain and the DNA recognition sequences can change the conformation of the Pax protein and the target DNAs.Citation27,Citation28 Genes thought to be up regulated by Pax2 include WT1Citation29, gdnfCitation21, wnt4Citation30 and sfrp2Citation31 in the developing kidney and engrailed-2Citation32 in the developing hindbrain. Yet, these are likely to represent only a small fraction of the potential Pax2 target genes.
In cell culture, Pax2 dependent transcription activation requires the serine-threonine rich carboxyl-terminal domain,Citation33 which is phosphorylated by the c-Jun N-terminal kinase (JNK) to enhance transactivation of Pax2 reporter genes (, C).Citation34 JNK translocates to the nucleus in response to a variety of external signals, including Wnt proteins and phosphorylates specific nuclear transcription factors. In the kidney, the most relevant upstream activators of JNK may be Wnt4 and Wnt9b, which are known regulators of kidney development.Citation13,Citation14 Compound mutants of JNK1, 2 and 3 show renal hypoplasia and optic nerve coloboma very similar to Pax2 hypomorphs.Citation35 Furthermore, kidneys cultured in the presence of JNK inhibitors show dramatic developmental arrest including inhibition of branching morphogenesis and failure of mesenchyme cells to condense at the ureteric bud tips (). These data suggest an important role for JNK in modifying the Pax2 transactivation domain and stimulating Pax2 dependent gene expression.
Pax proteins can also suppress gene expression by interactions with Groucho/TLE (Grg) proteins.Citation36 Grg4 suppresses Pax2 dependent gene activation in cell culture, can form a complex at the DNA binding site, and completely suppresses phosphorylation of the Pax2 transactivation domain, even in the presence of activated JNKs.Citation37 Long term repression by Groucho is thought to involve interactions with histone deacetylases and the chromatin silencing machinery.Citation38,Citation39 However, deacetylation of histones is not the only possible mechanism of Groucho mediated repression, as experiments with deletion mutants,Citation40 deacetylase inhibitorsCitation38 and anti-HDAC antibodiesCitation41 would suggest.
How we thought about Pax proteins as transcription factors was based solely on the presence of a DNA binding domain and on our own preconceptions of how transcription factors work. When the genes were first identified, the best characterized models for transcription factors were DNA binding proteins, such as NFKβ or AP1, which were identified based on their biochemical activities. Yet, Pax and Hox proteins were identified based purely on genetics and sequence homologies, not any known biochemical activity. Still, we tried to use existing models of transcription factors to understand their function. As new concepts are emerging from the chromatin and epigenetic field, it may be time to reassess our old models of developmental gene regulation taking into account these new discoveries.
Linking Pax Proteins to the Epigenetic Machinery
Chromatin biology and the study of epigenetic pathways have undergone a resurgence due to the remarkable progress made in defining the patterns of histone modifications and the identifying the enzymes responsible for methylation and acetylation of histone tails. While the Polycomb and Trithorax group of genes were defined genetically as repressors and activators respectively in flies, the true biochemical function of these proteins was not evident until the purification of histone methyltransferases and their associated proteins in yeast and tetrahymena (for review see Citation42). Thus, many of the Polycomb group proteins are part of complexes that methylate histone H3 at lysine 9 or lysine 27 or histone H4 at lysine 20; these modifications correlate with gene silencing or heterochromatin. The Trithorax group of proteins generally belong to complexes associated with histone H3 lysine 4 or lysine 36 methylation, modifications usually associated with active gene expression or euchromatin. These specific lysine residues can be mono-, di- or trimethylated, further increasing the combinatorial complexity of nucleosome modifications.
The modification of histones was first linked to cell lineage decisions in embryonic stem cells by large scale genomic chromatin immunoprecipitation analyses with antibodies specific for trimethylation at H3K4 and K3K27. In pluripotent ES cells, many key regulatory genes had low levels of both types of methylation marks, even though the genes were not expressed.Citation43,Citation44 These so called bivalent epigenetic marks were then resolved into high levels of trimethyl H3K4 or H3K27 depending on whether the genes were expressed or not in subsequent differentiated derivatives. These data are consistent with a model of cell lineage decision making that requires the compartmentalization of the genome into active and inactive domains. In mammalian development, the loss of pluripotency occurs as the epiblast undergoes gastrulation to form the primary germ layers. Strikingly, many of the epigenetic regulatory genes, including Polycomb and Trithorax homologues and de novo DNA methylases show embryonic defects at this time.
If an accumulation of positive and negative epigenetic marks is essential for differentiation along cell lineage pathways, then specific proteins must control the locus and tissue specificity for Polycomb and Trithorax complexes. In flies, the Polycomb response elements (PREs) are known cis-acting DNA sequences that bind directly to the complex.Citation45,Citation46 However, in mammals, PREs have not been described, nor is it clear how histone methyltransferase complexes recognize individual genes at the right time. I believe our studies with Pax2 have provided some insight into these questions.
The identification of the Pax2 interacting protein PTIP led us to reexamine the biochemical function of Pax proteins in development. PTIP is a ubiquitously expressed nuclear protein that is part of an H3K4 methylation complex and contains a carboxy-terminal Phospho-serine binding domain.Citation47–Citation50 In cell culture, Pax2 binding to DNA recruits PTIP and an MLL2/ALR complex that methylates H3K4 at that site.Citation50 The recruitment of ALR requires PTIP, suggesting that the Pax-PTIP interaction is a rate limiting step in nucleating the complex (). Thus, PTIP acts as an adaptor protein that links the H3K4 methylation machinery to a sequence specific binding protein. There are multiple Trithorax homologues in mammals. PTIP was not found in the MLL1/ALL complex that was first purified, rather it seems to co-purify with the Mll2/ALR complex, suggesting that there may be other adaptor molecules yet to be discovered.
Mice homozygous for a PTIP null allele are post-gastrulation lethal, disorganized and developmentally arrested.Citation51 This phenotype is more severe than any of the MLL mutations described to date. By E8.5 there is a global reduction in detectable levels of H3K4 di- and trimethylation. These data suggest that PTIP must interact with other DNA binding proteins beyond the Pax family. This is confirmed in the Xenopus gastrula where a PTIP homologue was shown to mediate activin signaling by binding to P-Smad2.Citation52 In the fly embryo, a PTIP homologue is also essential for early patterning, including the correct expression of segmentation genes prior to gastrulation and the activation of many Polycomb group target genes once repression is relieved.Citation53 We have also studied PTIP in embryonic stem cells, where it is necessary for retaining pluripotency during in vitro culture.Citation54
Given the discovery of PTIP and the link to histone methylation complexes, we have begun to model Pax2 biochemical activity along different lines (). Our hypothesis is that Pax2 marks specific regions of chromatin by modification of histones. These epigenetic marks can delineate both active and inactive regions of the genomes in a stable and heritable manner through potential interactions with nucleosome remodeling complexes and other chromatin effectors.Citation55 Thus, cell lineage restriction may become fixed at the level of chromatin structure. In the developing intermediate mesoderm and the kidney, this could establish chromatin domains in an active state such that transcription can occur in subsequent generations of daughter cells and it may also silence regions of chromatin to restrict transdifferentiation into other non-renal cell types. In order to examine this hypothesis, new tools and methods must be developed such that Pax2 target genes can be identified and analyzed at the level of chromatin structure.
Questions and Answers
Assistant Professor of Medicine, Washington University School of Medicine: Is phosphorylation of Pax2 required for the recruitment of the PTIP complex?
Collegiate Professor of Pathology, University of Michigan: We know that if we activate Wnt or the JNK pathway we see more recruitment of PTIP to the Pax2 DNA binding site. I can't say for certain what happens in the absence of signaling since there is always a level of phospho Pax2 present. We haven't mapped the serines that are responsible for that interaction, although that is something we are trying to do. Without deleting a specific serine, I can't say for certain that there is no interaction in the absence of phosphorylation. However, in the absence of signaling the interaction is less. Therefore, the data that we have suggest that activating the signaling pathways increases the amount of PTIP that is recruited to the DNA binding site.
Do you know where the phosphorylation takes place? Is it nuclear?
Jun N-terminal kinase can be nuclear. Pax2 is never detected in the cytoplasm in any measureable way, so I think it is happening in the nucleus.
Assistant Professor of Medicine, St. Louis University School of Medicine: You mentioned that Grg4 is specifically upregulated in the kidney under certain circumstances and in certain cell types. Can you elaborate on that?
We and others have shown that podocyte precursor cells increase Grg4 expression at the s-shaped body stage. This is coincident with other podocyte markers and with the eventual downregulation of Pax2 expression.
I was really impressed by your data generated using the Pax2 reporter you put inside the cells. Have you been able to characterize the methylation pattern on any endogenous gene?
That is a good question. The short answer is no. The problem is we don't know much about endogenous genes in 293 cells in which these experiments are done. When we made these cells, we selected for cells that were responsive. We did that because there are a lot of cells that will integrate the reporter, but are unresponsive. It turns out they have repressed the integrated locus. Endogenous genes in the 293 cell are either going to be on or they are going to be off. If they are off, Pax2 is likely not to have access. If they are on, they are likely to already be modified. What we need is a cell that is undifferentiated and competent to respond to Pax2. We don't have that, but we do have ES cells which are presumably epigenetically naive. We have some potential targets that do show K4 methylation in response to Pax2, in the ES cell. I don't know whether that is PTIP dependent now, because we have not done a combination. It can be done, but the problem is you need to have a cell that is not already imprinted at that endogenous locus so that you can see the difference.
You can do whole genome gene chip experiments in Drosophila. Have you looked at the overall chip correlation between PTIP binding and K4 methylation?
No. This is because our fly antibody doesn't work very well. The chip experiments we preformed by introducing the mouse PTIP into the fly cells. It had an epitope tag that we could use. Other labs have done whole genome analysis of fly and of ES cells, but we don't have the capacity to do that right now.
Assistant Professor of Medicine, Washington University School of Medicine: Will you also clarify the signals or pathway to determine whether Grg4 or PTIP is going to regulate the activation of genes and if in PTIP knock outs you see Grg4 binding?
In PTIP knockouts we don't really know what happens to the Grg4. Most of the phenotype is manifests itself before Grg4 is activated, at least in the tissues with which we are familiar. The Grg4 complex includes a phosphatase that dephosphorylates Pax2, at least in vitro. It also includes a methyltransferase which methylates, arginine 3 of histone H4. That seems to promote methylation at lysine 20 of histone H4 and lysine 9 and 27 of histone H3. We don't know how it does that. However, the argentine methyltransferase is certainly part of the complex. The other polycomb methyltransferases are not part of the complex but they seem to be motivated by this arginine methylation event.
Professor of Medicine and Developmental Biology Washington University School of Medicine: You showed that the loss of PTIP progressed in the kidney at a very early stage. Probably this indicates that as is the case in ES cells, you lose the progenitor population in the metanephric mesenchyme. I have two questions. First, what happens to those cells in the PTIP knock outs? What choices do they make? Second, what do you think would happen if you were to use that inefficient Six2-cre and removed it just as the cells moved on to differentiate?
The PTIP germline mutants developmentally arrests in every tissue and it is very disorganized; there are some markers of differentiation, brachyury is detectable, early neural markers are detectable, occasionally you will see a somite or two, but you won't see much beyond that. Since the PTIP protein is everywhere it is not a transformation event in the sense that mesoderm was converted to ectoderm or something else. Everything is affected. Now if you were to knock it out in a mesenchyme stem cell population, my guess is those cells are going to be limited, they are not going to differentiate into any derivatives, they may stop growing, they may die, or they may actually be able to divide like in the neural tube knockouts I showed. Now with the Nestin-Cre driver, those cells actually are proliferating although PTIP is knocked out pretty early on or about e9, e10 and those sections were from E17. So that neural tube has grown and filled up with lots of progenitor cells. But if you do an expression analysis, you find all of the embryonic markers and very few of the terminal, neuronal and glial markers. Thus, the PTIP null cells appear stuck at a certain stage and are unable to differentiate further. I don't know if that will happen in the kidney but that is something that might be worth checking out.
Assistant Professor of Medicine, Washington University School of Medicine: Your elegant studies proposed to establish the key role of PTIP and Pax2 signaling pathway in epithelial cell lineage. A key marker for epithelial cell lineage is the forming of apical-basal polarity and asymmetrical cellular partition. A number of key ‘polarity’ genes have been identified, for example, crumbs, discs-large, scribble from Drosophila imaginal disc cells, the origin of the KC cell line used in your studies. Did you observe any change in polarity gene expression modulated by the PTIP-Pax2 signaling pathway? Is the PTIP-Pax2 pathway directly affecting epithelial cell polarity?
We haven't specifically looked at that. It speaks to the question of the identities of the Pax2 target genes where this is actually happening in vivo. It is not for lack of trying. We need to have a cell system where we can mimic this. We do have a couple of immortalized cell types from early kidney mesenchyme. We see things like intergrin alpha 6, or alpha 8, go up. We see various growth factors that are in early kidney development. If there is one message to take home, I would stress that I don't think Pax2 is transcriptionally activating all of these genes. Rather what it does is predispose the genome to allow superimposed secondary signals like WNT signals, BMP signals and hedgehogs to activate a different set of target genes in the intermediate mesoderm, than they would, say, in the neural plate. In other words, it predisposes the region to respond to these signals in a unique, kidney specific way. By definition you might expect that Pax2 would interact with some of the promoters or locus control regions or enhancers of the genes that you talked about. However, the actual activation of those genes could be many cell divisions later at which time they are responding to the polarity signal, or the WNT4, planar cell polarity signal. The question I am exploring is how these early linage decisions imprint regions of chromatin before gene expression actually takes place. I would like to be able to answer that question in an embryo, but I don't think we have the technology.
Assistant Professor of Medicine, Washington University School of Medicine: You clearly showed that after Pax2 binding to DNA, the recruitment of PTIP and the methylation complex is associated with the activation of transcription. Once you have H3K4 methylation is Pax2 still required to contribute to activation? Specifically, is there any domain in Pax2 that you feel is required for continuing activation?
Another good question. In other words, at what point is the epigenetic pattern established? Do you need this protein at that point? We have not addressed that specifically. Looking at other systems, B cell development requires a very closely related protein called Pax5. Pax5 has been knocked out in a more terminally differentiated B cells by the Busslinger labCitation56 and the result is that you do need Pax5. B cells can revert to a more embryonic or non-B cell phenotype if you take it away. That would suggest either that there is a certain amount of plasticity along with lineage specification or that it also transcriptionally activates some genes more directly.
Is H3K4 methylation always associated with activation and K27 is always associated with repression? If so, I wonder how that happens.
This is a very active area of investigation, not by my lab but by many others in the chromatin field. I hesitate to say always. As with K4 there is an issue of the number of methyl groups. There is mono-, di- and trimethylation and I think the consensus is trimethylation almost always correlates with gene activity, dimethylation, not necessarily. However, there are proteins that interact with the methyl groups. In the case of trimethyl K4, there is a protein subunit of a nucleosome remodeling factor that recognizes it. A part of the protein actually recognizes the K4 methylation and another part of the same protein recognizes K9 acetylation. In the case of K9 there are the chromatin proteins, HP1, for example, that recognize the K9 methylation, at least the methylation of K9 within the context of that part of the histone tail. I think the current thinking is that these methyl markers are being recognized by protein modification or chromatin modifiers, either nucleoside remodeling complexes, in the case of K4, or by heterochromatic promoting proteins like HP1 in the case of the K9 and K27 methylation.
Abbreviations
| Pax | = | paired box containing gene |
| H3K4 | = | histone H3 lysine 4 |
| HMT | = | histone methyltransferase |
| PTIP | = | Pax transcription-activation domain interacting protein |
| JNK | = | c-Jun N-terminal kinase |
Figures and Tables
Figure 1 The mammalian Pax genes. A summary of the nine Pax genes found in mice and humans is outlined. All genes have the characteristic 128 amino acid paired DNA binding domain (PD) at the amino terminus. Based on other conserved sequences, the genes fall into several subfamilies, including the Pax3/7 group which also have a carboxy-terminal homeodomain (HD) and an octapeptide (OD), the Pax4/6 family, which have just the homeodomain, the Pax1/9 family which contains just the octapeptide, and the Pax2/5/8 group which has the octapeptdie and a partial homeodomain. The corresponding mouse and human mutations are indicated as are the key elements of the embryonic expression patterns.
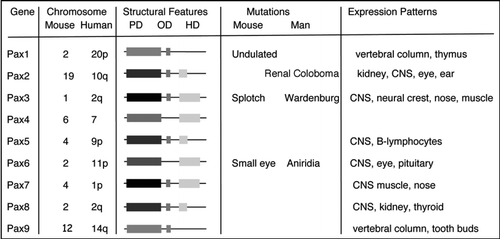
Figure 2 Early patterning of the kidney. (A) In a cross section through a post-gastrulation embryo, the position of the intermediate mesoderm is indicated with respect to the neural tube, notochord, somites and lateral plate. The schematic would correspond to a section posterior to the fourth somite. (B) A longitudinal rendition of the epithelial components from the intermediate mesoderm corresponding to approximately 10.5 days gestation (E10.5) in the mouse. (C) A schematic of the metanephric, or adult, kidney at approximately E12 once the ureteric bud has bifurcated. The cells condensed around the tips of the bud, known as the cap mesenchyme, are the epithelial progenitor cells for the nephrons and become polarized to form primitive renal vesicles.
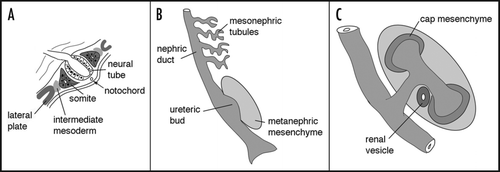
Figure 3 The intermediate mesoderm in Pax2 mutant mice. The region of intermediate mesoderm in wild-type (+/+) and Pax2 homozygous null (−/−) embryos was dissected out and stained with antibodies against cytokeratin, which marks the nephric duct (nd) and Pax2, which marks the metanephric mesenchyme (met) and the mesonephric tubules (mes) tubules. Note the lack of ureteric bud (ub) in Pax2 mutants, even though the nephric duct is initially formed.
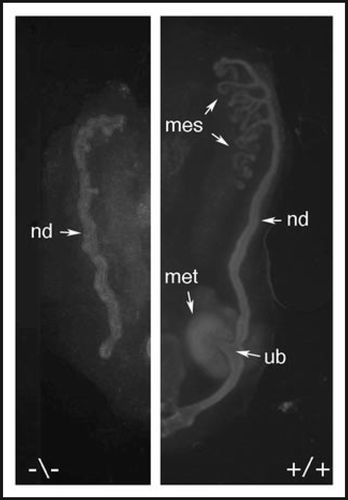
Figure 4 The Pax2 protein is phosphorylated by the c-Jun N-terminal kinase. (A) A schematic of the Pax2 protein showing the critical domains described. The activation domain is required for trans-activation potential in cell culture and contains many proline, serine and threonine residues. (B) The ERK, JNK and p38 kinases were activated in cell culture and used to phosphorylate recombinant GST-Pax2 proteins and control substrates with 32-P. Only the JNK kinase is able to significantly phosphorylate the Pax2 activation domain. (C) The Pax2 activation domain is as good a substrate for recombinant JNK enzyme as the c-Jun protein. (D) Organ cultures derived from E11.5 kidney rudiments and cultured for 48 hours in control media or with 20 µM of the JNK inhibitor SP600125. Cells were stained with antibodies against Pax2, to mark the mesenchyme, and cytokeratins, to mark the branching ureteric buds. Note the lack of condensation of Pax2 positive cells around the ureteric bud tips in the JNK inhibited culture.
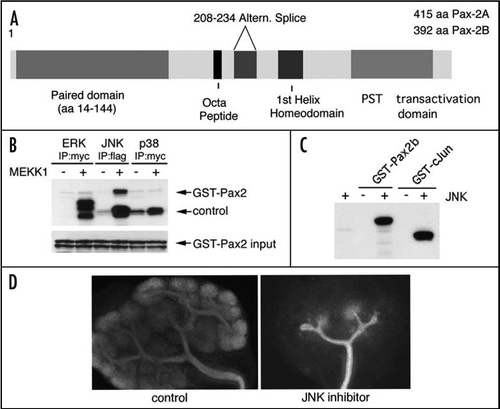
Figure 5 Pax2 and PTIP promote histone methylation. (A) Chromatin immunoprecipitation in cultured cells was done with the indicated antibodies, above the bar graphs and the precipitated DNA analyzed at an integrated Pax2 binding site. Cells were cultured with media alone (1, control), with Pax2 expression plasmids (2, Pax2), with a PTIP siRNA and Pax2 (3, SI Pax2) or rescued with a mouse PTIP that is resistant to the siRNA together with the siRNA and Pax2 (4, SI P + P). Note that Pax2 binds to its recognition sequence regardless of the presence of PTIP. PTIP localizes to the Pax2 recognition site only when Pax2 is expressed but not after knockdown with siPTIP. The H3K4 methyltransferase MLL2 localizes to the Pax2 recognition site in a Pax2 dependent manner and requires PTIP. PTIP knockdowns show no increase in H3K4 dimethylation, but the effect can be rescued with exogenous mouse PTIP. (B) A model of locus specificity for a histone methyltransferase (HMT) complex. Pax2 recognizes a specific sequence and recruits PTIP, potentially through the interaction with Phospho-serines. PTIP provides a link to the HMT complex, resulting in H3K4 methylation. These methylation marks provide docking sites for nucleosome remodeling factors that can promote sliding and assembly of RNA polymerase to activate or maintain gene expression.
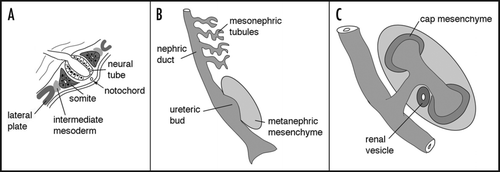
Table 1 Genes that regulate early kidney development and cell lineages
Acknowledgements
I thank Sanj Patel, Ming Fang, Yi Cai for their many contributions in the lab. This work has been supported by NIH grants DK054740 and DK073722 to G.R.D.
Note
Edited transcripts of research conferences sponsored by Organogenesis and the Washington University George M. O'Brien Center for Kidney Disease Research (P30 DK079333) are published in Organogenesis. These conferences cover organogenesis in all multicellular organisms including research into tissue engineering, artificial organs and organ substitutes and are participated in by faculty at Washington University School of Medicine, St. Louis, MO, USA.
References
- Nuesslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature 1980; 287:795 - 801
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 1978; 276:565 - 570
- Lewis EB. The 1991 Albert Lasker Medical Awards. Clusters of master control genes regulate the development of higher organisms. JAMA 1992; 267:1524 - 1531
- Schuettengruber B, Chourrout D, Vervoort M, et al. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735 - 745
- Deutsch U, Dressler GR, Gruss P. Pax 1, a member of a paired box homologous murine gene family, is expressed in segmented structures during development. Cell 1988; 53:617 - 625
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet 2002; 18:41 - 47
- Saxen L. Barlow PW, Green PB, White CC. Organogenesis of the Kidney. Dev Cell Biol 1987; 19 ed Cambridge, UK Cambridge University Press Series 19
- Moritz KM, Wintour EM, Black MJ, et al. Factors influencing mammalian kidney development: implications for health in adult life. Adv Anat Embryol Cell Biol 2008; 196:1 - 78
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol 2006; 22:509 - 529
- Costantini F. Renal branching morphogenesis: concepts, questions and recent advances. Differentiation 2006; 74:402 - 421
- Schuchardt A, D'Agati V, Larsson-Blomberg L, et al. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 1994; 367:380 - 383
- Kreidberg JA, Sariola H, Loring JM, et al. WT1 is required for early kidney development. Cell 1993; 74:679 - 691
- Stark K, Vainio S, Vassileva G, et al. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 1994; 372:679 - 683
- Carroll TJ, Park JS, Hayashi S, et al. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell 2005; 9:283 - 292
- Bouchard M, Souabni A, Mandler M, et al. Nephric lineage specification by Pax2 and Pax8. Genes Dev 2002; 16:2958 - 2970
- Torres M, Gomez-Pardo E, Dressler GR, et al. Pax-2 controls multiple steps of urogenital development. Development 1995; 121:4057 - 4065
- Sanyanusin P, Schimmenti LA, McNoe LA, et al. Mutation of the Pax2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet 1995; 9:358 - 364
- Dressler GR, Deutsch U, Chowdhury K, et al. Pax2, a new murine paired-box containing gene and its expression in the developing excretory system. Development 1990; 109:787 - 795
- Dressler GR, Douglass EC. Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci USA 1992; 89:1179 - 1183
- Rothenpieler UW, Dressler GR. Pax-2 is required for mesenchyme-to-epithelium conversion during kidney development. Development 1993; 119:711 - 720
- Brophy PD, Ostrom L, Lang KM, et al. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 2001; 128:4747 - 4756
- Carroll TJ, Vize PD. Synergism between Pax-8 and lim-1 in embryonic kidney development. Dev Biol 1999; 214:46 - 59
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev 1993; 7:2048 - 2061
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol Cell Biol 1995; 15:2858 - 2871
- Epstein JA, Glaser T, Cai J, et al. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev 1994; 8:2022 - 2034
- Xu W, Rould MA, Jun S, et al. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell 1995; 80:639 - 650
- Epstein J, Cai J, Glaser T, et al. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem 1994; 269:8355 - 8361
- Chalepakis G, Wijnholds J, Gruss P. Pax-3-DNA interaction: flexibility in the DNA binding and induction of DNA conformational changes by paired domains. Nucleic Acids Res 1994; 22:3131 - 3137
- Dehbi M, Ghahremani M, Lechner M, et al. The paired-box transcription factor, PAX2, positively modulates expression of the Wilms' tumor suppressor gene (WT1). Oncogene 1996; 13:447 - 453
- Torban E, Dziarmaga A, Iglesias D, et al. PAX2 activates Wnt4 expression during mammalian kidney development. J Biol Chem 2005; 281:12705 - 12712
- Brophy PD, Lang KM, Dressler GR. The secreted frizzled related protein 2 (SFRP2) gene is a target of the Pax2 transcription factor. J Biol Chem 2003; 278:52401 - 52405
- Song DL, Chalepakis G, Gruss P, et al. Two Pax-binding sites are required for early embryonic brain expression of an Engrailed-2 transgene. Development 1996; 122:627 - 635
- Lechner MS, Dressler GR. Mapping of Pax-2 transcription activation domains. J Biol Chem 1996; 271:21088 - 21093
- Cai Y, Lechner MS, Nihalani D, et al. Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. J Biol Chem 2002; 277:1217 - 1222
- Weston CR, Wong A, Hall JP, et al. JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev 2003; 17:1271 - 1280
- Eberhard D, Jimenez G, Heavey B, et al. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J 2000; 19:2292 - 2303
- Cai Y, Brophy PD, Levitan I, et al. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J 2003; 22:5522 - 5529
- Courey AJ, Jia S. Transcriptional repression: the long and the short of it. Genes Dev 2001; 15:2786 - 2796
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene 2000; 249:1 - 16
- Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev 1998; 12:1931 - 1940
- Dasen JS, Barbera JP, Herman TS, et al. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev 2001; 15:3193 - 3207
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693 - 705
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 326
- Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8:532 - 538
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 2007; 134:223 - 232
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol 2008; 20:266 - 273
- Manke IA, Lowery DM, Nguyen A, et al. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 2003; 302:636 - 639
- Issaeva I, Zonis Y, Rozovskaia T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 2007; 27:1889 - 1903
- Cho YW, Hong T, Hong S, et al. PTIP associates with MLL3-and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 2007; 282:20395 - 20406
- Patel SR, Kim D, Levitan I, et al. The BRCT-domain containing protein ptip links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Dev Cell 2007; 13:580 - 592
- Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol 2003; 23:1666 - 1673
- Shimizu K, Bourillot PY, Nielsen SJ, et al. Swift is a novel BRCT domain coactivator of Smad2 in transforming growth factor beta signaling. Mol Cell Biol 2001; 21:3901 - 3912
- Fang M, Ren H, Liu J, et al. Drosophila PTIP is essential for anterior/posterior patterning in development and interacts with the PcG and TrxG pathways. Development 2009; 136:1929 - 1938
- Kim D, Patel SR, Xiao H, et al. An essential role for PTIP in maintaining embryonic stem cell pluripotency. Stem Cells 2009; in press
- Ruthenburg AJ, Li H, Patel DJ, et al. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev 2007; 8:983 - 994
- Mikkola I, Heavey B, Horcher M, et al. Reversion of B cell commitment upon loss of Pax5 expression. Science 2002; 297:110 - 113