Abstract
Bidirectional transcription is an interesting feature of eukaryotic genomes; yet not all aspects of its mechanism are understood. Silkmoth choriogenesis is a model system for studying transcriptional regulation at the initiation level. As chorion genes comprise a large group of divergently transcribed gene pairs, we are presented with the possibility of investigating the intricacies of bidirectional transcription. Τheir well characterized 5’ regulatory regions and expression profiles lay the foundation for investigating protein:protein and protein:DNA interactions, and RNA polymerase function during oocyte development. In this article we summarize current knowledge on chorion gene regulation and propose an approach to modeling bidirectional transcription using chorion promoters.
Introduction
Bidirectional promoters control approximately 11% of genes identified in humansCitation1 and over 60% of the yeast genome.Citation2 This is an interesting feature of all eukaryotes,Citation3–Citation7 including the domesticated silkmoth, Bombyx mori.Citation8,Citation9 It must be noted, however, that currently the term bidirectional transcription does not refer exclusively to a pair of divergently transcribed genes as discussed in this review (), but is also used to describe production of both sense and antisense transcripts by unidirectional promoters (). In the latter, the antisense (or cryptic) transcript often assumes some regulatory role (for a review see Berretta and Morillon, 2009).Citation10 However, studying short bidirectional promoters such as those of chorion genes can provide some insight on the mechanistic aspect of the latter as well (plausible models shown in ). Moreover, generation of nongenic antisense transcripts from late chorion gene promoters has been reported,Citation11 but not been followed up to date.
Silkmoth choriogenesis, which is preceeded by vitellogenesis, is the final stage of oogenesis. Chorion genes are only active in nuclei of the follicular epithelium, strictly during oocyte development. They are turned on and off according to a program initiating at the onset of choriogenesis, and concluding as follicles mature.Citation12 The majority of chorion genes, which span a few hundred kbp on chromosome 2 where the chorion locus lies, is under the control of ∼250 bp-long bidirectional promoters.Citation13 Their transcriptional activity is temporal-specific, and the pairwise organization of an α- and a β-gene of the same developmental specificity (as in ) calls for their divergent transcription.Citation14–Citation16 Silkmoth chorion development provides a good model for addressing questions on bidirectional transcription since: (1) a single ovariole contains all developmental stages due to sequential follicular maturation; (2) α- and β-genes are well characterized;Citation16 and (3) a growing set of tools and resources for probing chorion gene function is available.Citation17–Citation19
Here we present a summary of current knowledge of the regulatory cascade governing chorion gene expression (for a review on choriogenesis see Papantonis A and Lecanidou R. Insect Mol Biol 2009; In press) by focusing on the A/B.L9 chorion gene pair which is expressed during early-middle choriogenesis. We go on to discuss some new questions and perspectives that arise from the synthesis of data available in this model system.
The Chorion Gene Regulatory Cascade
Silkmoth oogenesis is correlated with changes in levels of the hormone ecdysone; during early follicular development these are low and increase after mid-vitellogenesis.Citation20,Citation21 Choriogenesis initiates upon conclusion of vitellogenesis. The transition is instigated by a reduction in ecdysone levels, and by the regulation of various genes encoding for nuclear receptors and/or transcription factors (e.g., BmFTZ-F1).Citation20–Citation22 Several aspects of the molecular cascade governing choriogenesis are still elusive. However, recent data assist us in addressing some of its intricacies ().
It has been proposed that autocrine/paracrine signaling via prostaglandin triggers the onset of choriogenesis.Citation23 Other data suggests that C/EBP (CCAAT enhancer binding protein) acts as the critical activator and repressor.Citation24,Citation25 These functions could be complementary, since prostaglandin signals through cAMP and C/EBPs are cAMP-responsive genes, participating in co-regulative cascades alongside CREB (cAMP response element binding protein).Citation26,Citation27 In fact, the silkmoth CREB orthologue has recently been isolated; it produces three isoforms with expression peaks after pupation.Citation28 Additionally, typical cAMP response elements (5′-TGACGTCA-3′) have been identified in the C/EBP proximal promoter region (Papantonis A and Lecanidou R; unpublished data).
From there on, chorion development relies on sequential activation and repression of genes encoding different protein constituents of the chorion layer;Citation12 these genes (i.e., chorion genes) are grouped as early, middle and late according to their time of expression. Molecular and biochemical analyses point to the sillkmoth orthologue of High Mobility Group protein A (HMGA) as the key factor orchestrating developmental progression.Citation29 HMGAs are known to shape chromatin in vivo by mediating protein:protein interaction and locally contorting DNA.Citation30–Citation32 During silkmoth choriogenesis, HMGA binding to chorion gene promoters mediates C/EBP, CHD1 and GATA interaction with cognate cis-elements.Citation29 Chromo-helicase-DNA binding domain protein 1 (CHD1) also plays a central role in the cascade.Citation9 CHD1 proteins are ATP-dependent chromatin remodelers which have been isolated from various eukaryotes.Citation33–Citation36 Silkmoth CHD1 repositions nucleosomes locally within the chorion locus, but also affects global chromatin architecture.Citation9 GATA, on the other hand, initially characterized as a dedicated activator of late genes,Citation37 probably has a repressive role during early and mid-choriogenesis, by antagonizing C/EBP to allow for interaction with HMGA and promoter elements.Citation25,Citation29,Citation38
Studies of the early-middle gene pair A/B.L9 (L12-type)Citation9,Citation15,Citation16,Citation29 provide a paradigm for the transcriptional circuit underlying chorion gene regulation (summarized in ). Prostaglandin pulses trigger second messages via cAMP, which (presumably) drive CREB binding to the C/EBP gene promoter; C/EBP protein molecules are produced and this creates a feedback loop enhancing C/EBP expression. GATA binding in later stages correlates with C/EBP downregulation. (C/EBP availability during choriogenesis follows a gradient: highest levels in early stages, lowest in late onesCitation24,Citation25). In turn, C/EBP homodimers bind to the chorion gene promoter to activate it; this association is mediated by HMGA, which also controls the association of GATA with the C/EBP promoter. Moreover, accessibility to the appropriate C/EBP element on the A/B.L9 promoter is dependent on CHD1 activity. Though there are four C/EBP elements present, only one seems to be activation-related () and a nucleosome masking the α-proximal half of the promoter is translocated further upstream. CHD1 recruitment is also mediated by HMGA and occurs long before activation of the respective gene.Citation9 Gene repression occurs as a consequence of GATA evicting C/EBP from the promoter. Again, the functional equilibrium between C/EBP and GATA seems to be regulated via HMGA. The cascade concludes as C/EBP factors contribute to the irreversible transcriptional inhibition of A/B.L9 by re-binding to more than one sites.Citation25 Therefore a closed transcriptional circuit controls both the C/EBP gene and the transcriptional fate of A/B.L9 during follicular development. Similar circuits control most chorion genes, although late genes (i.e., Hc.A/B) are an exception, as they are activated rather than repressed by GATA factors, in synergy with C/EBP.Citation24,Citation25,Citation37,Citation38
One could argue that the molecular events governing chorion gene regulation are fairly well described. On this basis, we propose that choriogenesis can be used for: (1) understanding exactly how the cellular machinery detects and transcribes discrete gene sets during different developmental periods (i.e., early—Er.A/B; early-middle—L12-type; middle-late—L11-type; late—Hc.A/B) although these are shuffled in the chorion locus, and (2) deciphering the mechanism of bidirectional (divergent) transcription.
A Possible Role for cis-Element Architecture
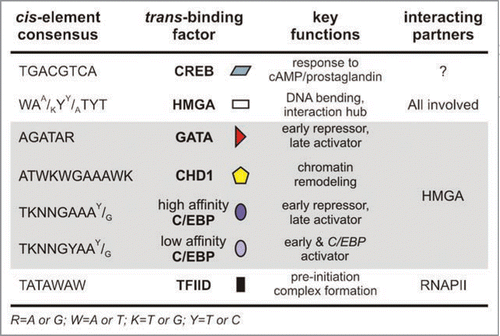
The recognition of discrete gene sets by the cellular machinery may be addressed by examining the relation between cis-element architecture (), association with trans-acting factors (), and transcriptional output. We infer that this is a C/EBP-driven cascade. HMGA and CHD1 binding sites are usually organized closer to the α-gene of a gene pair, whereas GATA sites lie closer to the β-gene. By contrast, C/EBP elements follow what appears to be a stochastic distribution and can be classified as early- or late-type sites, producing lower and higher affinity complexes with C/EBP homodimers, respectively.Citation24 By looking at number, relative positions, and the early to late-type ratio of C/EBP sites the following can be deduced: (1) Er.A/B promoters contain an average of three C/EBP binding sites; L12-type gene pair promoters consistently contain four sites (three early- and a late-type one); L11-type gene pair promoters have four late-type sites and Hc.A/B late gene promoters feature a single late-type binding element; (2) the ratio of C/EBP sites to promoter length is higher for early genes (Er.A/B), lower for middle ones and lowest for late Hc genes; (3) a C/EBP site positioned between the GATA site and the β-TATA box is “lost” in the transition from L12 to L11-type genes. C/EBP sites proximal to the α-TATA box also follow a specific pattern: a pair of an early-type and a latetype site in L12-type gene promoters; a pair of late-type sites in L11-type ones; a single late-type C/EBP site in Hc.A/B promoters in the same region as its counterpart in L11 genes (). From these observations we propose that promoter sequences are ‘marked’ by the arrangement of their modules, and thus recognized as of earlier or later developmental specificity and accordingly regulated.
Bidirectional Transcription: One or Two Polymerases?
In mammals, bidirectional promoters preferentially contain a specific subset of cis-elements (e.g., C/EBP binding sites) and histone post-translational modifications (e.g., H3 tri-methylated on lysine 4; H3K4me3).Citation1 Chorion genes comprise a suitable system for studying this aspect of bidirectional transcription since: (1) CHD1 binding to middle chorion promoters is correlated with temporal-specific appending of methyl-marks on H3K4,Citation9 (its tandem chromo-domains physically interact with modified histone tailsCitation39); (2) C/EBP plays a key role in gene regulation; and (3) all cis-elements involved in transcriptional initiation (and repression) of a given gene pair are constrained within just ∼250 bp (discussed above).
A key question in bidirectional promoter function is whether both genes in a chorion gene pair are transcribed simultaneously by a pair of RNA polymerase II holo-enzymes (RNAPII), or is each separately transcribed (). We shall hypothesize based on experimental data from the A/B.L9 gene pair. Initiation of transcription and nucleosome rearrangement are marked by TFIID recruitment, and H3K4 tri-methylation. In the A/B. L9 promoter two core octamers were detected: one masked the HMGA-C/EBP-CHD1 cis-element array (∼50 bp-long; red box in ); the second was centered on the β-TATA box, masking the GATA site (green box in ).Citation9 These positions describe the pre-choriogenic chromatin structure of the promoter, when the gene is inactive. Upon initiation of choriogenesis, the α-proximal nucleosome is translocated 20 bp closer to the α-gene, thus exposing the array. This is critical for C/EBP binding and formation of an enhanceosome.Citation40 Comparing pre- to post-choriogenic structure up to mid-choriogenesis, the β-half of the promoter is not significantly remodeled.Citation9 In vitro data suggest that a multimeric complex containing HMGA, CHD1 and C/EBP interaction may only form on the α-half of the promoter, but not on the β-half. Results from ChIP assays show that TFIID binding to the α-TATA sequence is correlated with gene expression, whereas detection of β-TATA:TFIID signal exhibits a lag (Papantonis A, Lecanidou R; unpublished data). It is, thus, most probable that the enhanceosome occupies the α-half (), and TFIID association with the α-TATA box is more prominent. The A/B.L9 promoter can be bent by HMGA in vitro.Citation29 If this applies in vivo, ‘fitting’ two RNAPII holocomplexes (15–40 nm)Citation41 into the space created by 200 bp (∼70 nm from α- to β-TATA) contorted by 90°, would produce a compact configuration (). It seems rather unlikely, due to both steric hindrance and TFIID recruitment patterns, that two RNAPII complexes can simultaneously be formed and initiate. Although RNA dot blot analyses have shown that both the α- and β-gene are equally activated during mid-choriogenesis,Citation12 their alternate transcription cannot be ruled out (). In fact, some data exist indicating the B.L11 gene is expressed slightly later than its A counterpart.Citation15 We propose that transcriptional activity relies on formation of a stable complex via interaction with the enhanceosome around the α-TATA box, and initiation on the β-TATA occurs separately, owing to close proximity with the enhanceosome and high local concentration of pre-initiation factors. In this manner, the α- and the β-gene of the pair would be alternatively transcribed, perhaps as stochastic events.
This scenario requires further investigation and, of course, begs even more questions. By which mechanism do all these occur (provided they occur as described)? Can this be generalized for all chorion gene pairs or bidirectionally transcribed genes in other eukaryotes? Could this mechanism achieve the high resolution temporal profiling of gene pair transcription observed in vivo? We expect that silkmoth choriogenesis will serve as a model for addressing such questions in coming years, either by testing the in vivo function of cis-elements with transient expression assays, or by assessing, in greater detail, α- and β-gene transcription and transcription factor turnover rates on gene pair promoters.
Figures and Tables
Figure 1 Different kinds of bidirectonal transcription. (A) A pair of divergently transcribed pairs (α- and β-gene; as in chorion gene pairs) sharing a common 5′ regulatory region. Pre-initiation complex (PIC) formation and RNA polymerase II (RNAPII) recruitment precedes transcription, which occurs in both directions (arrows); the number of RNAPII molecules involved and mechanism remain unclear. Two alternative models for this mechanism are present in (C and D) (arrows). (B) A given gene X is under the control of a unidirectional promoter. PIC assembly and RNAPII recruitment close to the transcription start site (TSS) are followed by initiation, mainly producing functional transcripts of X (grey arrow), but occasionally transcribing the antisense strand (white arrows) to produce short non-coding RNA molecules. (C) One or two RNA polymerases per gene pair? Each gene is transcribed separately, in alternate pulses, and RNAPII holo-enzyme complex formation is based on the C/EBP-HMGA-CHD1 complex formed proximal to the α-TATA box. (D) Both genes of the pair are simultaneously transcribed by ‘dedicated’ polymerases, thus formation of two distinct RNAPII complexes—one on each TATA box—is required.
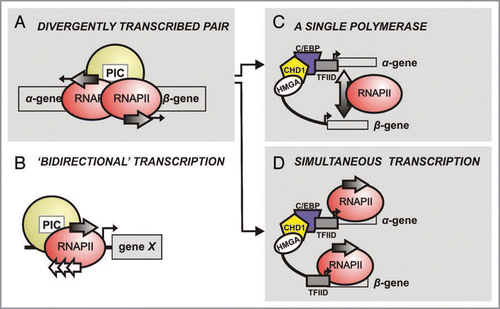
Figure 2 The chorion regulatory circuit. For a chorion gene pair of given developmental specificity, the cascade initiates upon prostaglandin pulses (extra-cellular) triggering second messaging via cAMP. The cAMP response factor CREB is then activated and binds the C/EBP gene promoter to initiate transcription (dotted arrow). C/EBP protein molecules (purple) produced contribute via a positive feedback loop; TFIID recruitment correlates with pre-initiation complex formation. C/EBP is finally switched off as GATA antagonises C/EBP for interaction with the HMGA-CHD1 complex (red line) and binds the promoter. In parallel, C/EBP switches the chorion gene pair on (both α- and β-gene), and proteins produced (grey arrows) integrate to the chorion layer. The target chorion gene is switched off by GATA (red line) and irreversibly ‘muted’ by multiple C/EBP binding events. In all cases, CHD1 acts as the chromatin remodeler to facilitate transcription factor binding.
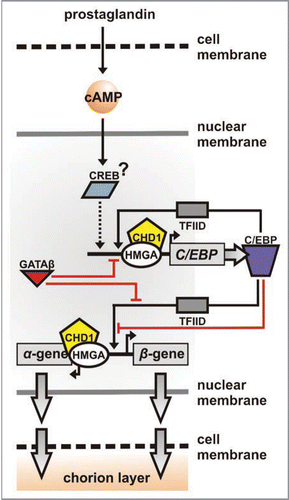
Figure 3 Chorion gene pair promoter regions. Transcription factor binding sites are presented for the early Er.A/B, the early-middle/middle L12-type, the middle/middle-late L11-type and the late Hc.A/B gene promoter regions. Orientation of C/EBP (ovals; light purple, lower affinity; dark purle, higher affinity), and GATA sites (triangles) is denoted. Theoretically predicted CHD1 (pentagons) and HMGA (white boxes) sites are indicated with a questionmark. Distances are drawn to scale (L12-type α- to β-TATA region corresponds to 211 bp); α-proximal arrays of CHD1-HMGA-C/EBP elements masked by a nucleosome appear boxed (red), as do β-proximal ones (green). The gradient bar represents choriogenic periods (E to L) over developmental time (vertical arrow).
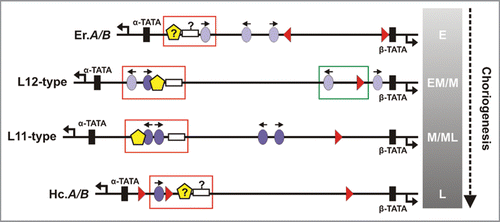
Figure 4 A summary of cis-elements of chorion genes and the protein factors that associate in trans with them. The cis-element consensus sequence, its cognate protein factor, the main function of the factor and proteins it interacts with during choriogenesis are presented. The symbols that accompany each factor are the ones used to depict the relevant binding sites in . Data for the CREB factor are based on binding site prediction and homologies to other insect systems. Symbols used to describe nucleotide composition are explained in the footnote.
Acknowledgements
We would like to thank Peter R. Cook for comments on the manuscript, and Luc Swevers for discussions on chorion regulation.
References
- Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, Weng Z. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res 2007; 17:818 - 827
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature 2009; 457:1033 - 1038
- Bayele HK. Trypanosoma brucei: A putative RNA polymerase II promoter. Exp Parasitol 2009; doi:10.1016/j.exppara.2009.08.007.
- Zanotto E, Häkkinen A, Teku G, Shen B, Ribeiro AS, Jacobs HT. NF-Y influences directionality of transcription from the bidirectional Mrps12/Sarsm promoter in both mouse and human cells. Biochim Biophys Acta 2 2009; 178:432 - 442
- Piontkivska H, Yang MQ, Larkin DM, Lewin HA, Reecy J, Elnitski L. Cross-species mapping of bidirectional promoters enables prediction of unannotated 5′ UTRs and identification of species-specific transcripts. BMC Genomics 2009; 10:189
- Wang Q, Wan L, Li D, Zhu L, Qian M, Deng M. Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinformatics 2009; 10:29
- Respuela P, Ferella M, Rada-Iglesias A, Aslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J Biol Chem 2008; 283:15884 - 15892
- Jones CW, Kafatos FC. Structure, organization and evolution of developmentally regulated chorion genes in a silkmoth. Cell 1980; 22:855 - 867
- Papantonis A, Tsatsarounos S, vanden Broeck J, Lecanidou R. CHD1 assumes a central role during chorion development. J Mol Biol 2008; 383:957 - 969
- Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 2009; 10:973 - 982
- Skeiky YA, Iatrou K. Silkmoth chorion antisense RNA: structural characterization, developmental regulation and evolutionary conservation. J Mol Biol 1990; 213:53 - 66
- Kafatos FC, Tzertzinis G, Spoerel NA, Nguyen HT. Goldsmith MR, Wilkins AS. Chorion genes: an overview of their structure, function and transcriptional regulation. Molecular Model System in the Lepidoptera 1995; 181 - 216 Cambridge University Press
- International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol 2008; 38:1036 - 1045
- Iatrou K, Tsitilou SG. Coordinately expressed chorion genes of B. mori: is developmental specificity determined by secondary structure recognition?. EMBO J 1983; 2:1431 - 1440
- Spoerel NA, Nguyen HT, Kafatos FC. Gene regulation and evolution in the chorion locus of Bombyx mori: structural and developmental characterization of four eggshell genes and their flanking DNA regions. J Mol Biol 1986; 190:23 - 35
- Spoerel NA, Nguyen HT, Towne S, Kafatos FC. Negative and positive regulators modulate activity of a silkmoth chorion gene during choriogenesis. J Mol Biol 1993; 230:151 - 160
- Goldsmith MR, Shimada T, Abe H. The genetics and genomics of the silkworm Bombyx mori. Ann Rev Entomol 2005; 50:71 - 100
- Cao J, Tong C, Wu X, Lv J, Yang Z, Jin Y. Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. Insect Biochem Mol Biol 2008; 38:1066 - 1071
- Uchino K, Sezutsu H, Imamura M, Kobayashi I, Tatematsu K, Iizuka T, et al. Construction of a piggy- Bac-based enhancer trap system for the analysis of gene function in silkworm Bombyx mori. Insect Biochem Mol Biol 2008; 38:1165 - 1173
- Swevers L, Iatrou K. The ecdysone regulatory cascade and ovarian development in lepidopteran insects: insights from the silkmoth paradigm. Insect Biochem Mol Biol 2003; 33:1285 - 1297
- Swevers L, Iatrou K. The ecdysone agonist tebufenozide blocks progression of the ecdysteroid-regulated gene expression cascade during silkmoth oogenesis and causes developmental arrest at mid-vitellogenesis. Insect Biochem Mol Biol 1999; 29:955 - 963
- Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol Cell 1999; 3:143 - 149
- Machado E, Swevers L, Sdralia N, Medeiros MN, Mello FG, Iatrou K. Prostaglandin signaling and ovarian follicle development in the silkmoth, Bombyx mori. Insect Biochem Mol Biol 2007; 37:876 - 885
- Sourmeli S, Papantonis A, Lecanidou R. A novel role for the Bombyx Slbo homologue, BmC/EBP, in insect choriogenesis. Biochem Biophys Res Commun 2005; 337:713 - 719
- Papantonis A, Sourmeli S, Lecanidou R. Chorion gene activation and repression is dependent on BmC/EBP expression and binding to cognate cis-elements. Biochem Biophys Res Commun 2008; 369:905 - 909
- Wilson HL, Roesler WJ. CCAAT/enhancer binding proteins: do they possess intrinsic cAMP-inducible activity?. Mol Cell Endocrinol 2002; 188:15 - 20
- Manna PR, Dyson MT, Stocco DM. Role of basic leucine zipper proteins in transcriptional regulation of the steroidogenic acute regulatory protein gene. Mol Cell Endocrinol 2009; 302:1 - 11
- Song H, Sun Y, Zhang Y, Li M. Molecular cloning and characterization of Bombyx mori CREB gene. Arch Insect Biochem Physiol 2009; 71:31 - 44
- Papantonis A, vanden Broeck J, Lecanidou R. Architectural factor HMGA induces promoter bending and recruits C/EBP and GATA during silkmoth chorion gene regulation. Biochem J 2008; 416:85 - 97
- Foti D, Iuliano R, Chiefari E, Brunetti A. A nucleoprotein complex containing Sp1, C/EBPbeta and HMGI-Y controls human insulin receptor gene transcription. Mol Cell Biol 2003; 23:2720 - 2732
- Yie J, Liang S, Merika M, Thanos D. Intra- and intermolecular cooperative binding of HMGI(Y) to the beta-interferon promoter. Mol Cell Biol 1997; 17:3649 - 3662
- Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 2001; 277:63 - 81
- Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromo- and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci USA 1993; 90:2414 - 2418
- Stokes DG, Tartof KD, Perry RP. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci USA 1996; 93:7137 - 7142
- Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo-domain protein CHD1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J 2000; 19:2323 - 2331
- McDaniel IE, Lee JM, Berger MS, Hanagami CK, Armstrong JA. Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics 2008; 178:583 - 587
- Skeiky YA, Drevet JR, Swevers L, Iatrou K. Protein phosphorylation and control of chorion gene activation through mobilization of a promoter DNA binding factor from the cytoplasm into the nucleus. J Biol Chem 1994; 269:12196 - 12203
- Sourmeli S, Kravariti L, Lecanidou R. In vitro analysis of Bombyx mori early chorion gene regulation: stage specific expression involves interactions with C/EBP-like and GATA factors. Insect Biochem Mol Biol 2003; 33:525 - 540
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 2007; 317:1087 - 1090
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev 2001; 11:205 - 208
- Marenduzzo D, Micheletti C, Cook PR. Entropydriven genome organization. Biophys J 2006; 90:3712 - 3721