Abstract
Adipocyte differentiation is a complex developmental process that involves the coordinated interplay of numerous transcription factors. PPARγ has emerged as a master regulator of adipogenesis and recent global target gene analysis demonstrated that PPARγ targets many genes encoding chromatin modification enzymes as well as genes of lipid metabolism and storage. Among such modification enzymes are histone lysine methyltransferases, which play important roles in transcriptional regulation. Histone methyltransferases are involved in PPARγ gene expression and subsequent adipogenesis. In addition, recent studies revealed that demethylation of histone H3 at lys9 is associated with resistance to obesity. We here review the role of histone methylation and demethylation in adipogenesis, metabolism, and obesity.
Introduction
In recent years, much attention has focused on modifications of chromatin because of their critical role in regulating gene expression and their active involvement in a number of cellular processes such as mitosis and cellular differentiation.Citation1 The accessibility of DNA in eukaryotic cells is determined by its organization in a DNA-protein complex known as chromatin. Chromatin structure is regulated in part through dynamic modifications of the constituent proteins, primarily histones. The fundamental unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around a histone octamer that is made up of two copies each of the four core histones, H2A, H2B, H3 and H4.Citation2 The N-terminal histone tails are subject to a variety of posttranslational modifications that include acetylation, methylation, phosphorylation and ubiquitination. Accumulating evidence has established a clear link between the pattern of histone modification found at particular loci and the transcription of those genes, thus leading to the statement of the histone code hypothesis.Citation3 Gene activation correlates with the hyperacetylation of histones H3 and H4, whereas hypoacetylation correlates with inactive chromatin.Citation4–Citation6
Modification of histones by methylation, which occurs at lysine and arginine residues, plays a role in many biological processes including transcriptional regulation, heterochromatin formation, X inactivation and genomic imprinting.Citation6,Citation7 Unlike acetylation, histone methylation can have both positive and negative effects on transcription, depending on the site of methylation. Methylation of Lys4 of histone H3 (H3K4), for instance, correlates with gene activation in most systems, whereas H3K9 and H3K27 are considered as hallmarks of a condensed chromatin state.Citation6,Citation8 Furthermore, H3K9 methylation by histone methyltransferase (HKMTs) has been shown to trigger heterochromatin formation and transcriptionally silence euchromatic regions by recruiting heterochromatin protein 1.Citation9 Reflecting the critical roles of methylated lysines at specific sites, multiple HKMTs have been identified that recognize the same lysine residue for mono-, di- and/or tri-methylations, although the biological role of each HKMT still remains elusive. Importantly, recent studies have demonstrated that histone methylation can also be enzymatically reversed by histone demethylases that include PADI4 (peptidyl arginine deiminase, type 4), LSD1 (lysine specific demethylase 1), and the JmjC (Jumonji C)-domain containing proteins.Citation10
Adipose tissue is an important metabolic organ that is crucial for whole body insulin sensitivity and energy homeostasis.Citation11 White adipose tissue (WAT), the predominant type of fat in adult humans, serves as a storage depot for excess energy, whereas brown adipose tissue (BAT) generates heat in newborns (and in animals such as rodents) through mitochondrial uncoupling of lipid oxidation.Citation12 Beyond the classical notion of the adipocyte as a storage depot for excess energy, the adipocyte also secretes a wide variety of bioactive molecules (referred to as adipokines) that regulate physiologic processes throughout the body. These include glucose metabolism, regulation of blood pressure, angiogenesis, immunity and reproductive function.Citation13 The main dysfunctions of adipose tissue, obesity and lipodystrophy, correlate with the development of diabetes, hypertension and dyslipidemia.Citation14 Obesity can be defined as an excess accumulation of white adipose mass, resulting from both an increase in adipocyte cell size and the development of new mature cells from undifferentiated precursors.
Adipocyte differentiation is orchestrated by an elaborate cascade of sequentially acting transcription factors and chromatin modifying co-regulators. These shape differentiation through the actions of hormones and other signaling pathways. The physiological program converting preadipocytes into adipocytes, called adipogenesis, has been well characterized—predominantly in cultured mouse cell lines.Citation11 A wide array of transcription factors participate in adipogenesis, although most attention has focused on several members of the CCAAT enhancer-binding protein (C/EBP) family and the nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ).
Peroxisome proliferator-activated receptor γ (PPARγ) is considered the master regulator of adipogenesis. It is a member of the nuclear receptor superfamily of ligand-activated transcription factors and is both necessary and sufficient for adipogenesis.Citation15,Citation16 The action of PPARγ is mediated through two protein isoforms: the ubiquitously expressed PPARγ1; and PPARγ2, which is restricted to adipose tissue. Expression of each isoform is driven by a specific promoter that confers distinct tissue-specific expression and regulation.Citation17 However, both isoforms are strongly induced during preadipocyte differentiation in vitro, and both are highly expressed in adipose tissues in animals. PPARγ1 is induced earlier than PPARγ2 and is maintained at high level during adipocyte differentiation.Citation18
CCAAT/enhancer-binding protein α is another principal adipogenic transcription factor and these two factors, PPARγ and C/EBPα mutually stimulate each other. They drive the transition of preadipocytes to mature adipocytes by activating numerous target genes required for maintaining the mature fat-laden adipocyte phenotype. Recently, several other transcription factors have been identified as regulators of adipogenesis. These include GATA2,Citation19 the Krüppel like factor (KLF) family,Citation20–Citation22 and Nr2f2.Citation23,Citation24 The regulation of adipogenesis by transcriptional cascade and the role of histone acetylation have been reviewed extensively elsewhere.Citation11,Citation16,Citation25 In this review, we focus on the role of methylation and demethylation of histones in adipogenesis and in the development of obesity.
Histone Methylations and Adipogenesis
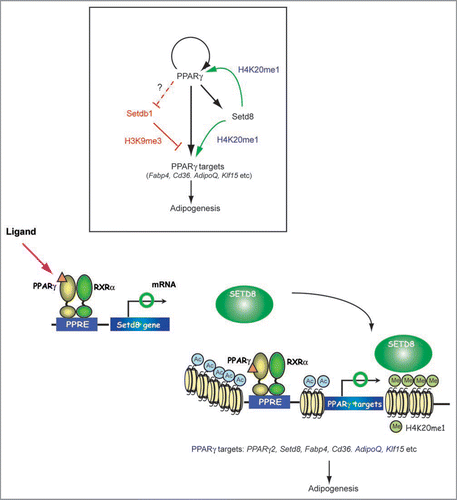
Regulation of adipogenesis by monomethylation of histone 4 at lysine 20 (H4K20) by PR-Set7/Setd8 and trimethylation of histone H3 at lysine 9 (H3K9) by Setdb1. Epigenetic determinants control the accessibility of promoter chromatin and establish lineage-specific heritable states of gene expression through the modulation of DNA methylation and posttranslational modification of core histones.Citation5,Citation26 Therefore, the expression and activities of histone-modifying enzymes should be distinctly regulated during adipocyte differentiation. It is tempting to speculate that epigenetic mechanisms also potentiate distinct functional states of PPARγ target genes and that PPARγ may regulate the expression of genes encoding histone modification enzymes. An epigenetic mechanism could, for example, act downstream of PPARγ and constitute a post-selection mechanism for potential PPARγ-responsive genes by allowing or preventing histone modification. By combining global gene expression analyses with a ChIP-chip approach, we have discovered that two well characterized HKMT, Setdb1 and Setd8 are coordinately regulated by PPARγ and that their expression leads to adipocyte differentiation through chromatin modificationCitation27 ().
Setdb1 and Setd8 are the histone lysine methyl transferases (HKMTs). Setdb1 tri-methylates histone H3K9 while Setd8 mono-methylates histone H4K20. Tri-methlylated H3K9 (H3K9me3) is considered the hallmark of the condensed chromatin state and transcriptionally silent euchromatin.Citation5 Monomethylated H4K20 (H4K20me1) has been implicated both in gene silencing and in transcriptional activation. Knockdown of these SET domain proteins confirmed that they are indeed involved in adipogenesis. Setdb1 mRNA levels decreased in concert with adipocyte differentiation. Knockdown of Setdb1 permitted the stimulation of adipogenesis even by an incomplete differentiation cocktail. By contrast, Setd8 mRNA was increased in abundance throughout adipocyte differentiation and knockdown of Setd8 impaired adipocyte differentiation. In this context, Setdb1 acts as an anti-adipogenic factor and Setd8 acts as pro-adipogenic factor and it is reasonable to think that Setdb1 is downregulated and Setd8 is upregulated during adipocyte differentiation. PPARγ may contribute to the transcriptional regulation of Setdb1 and thereby regulate H3K9me3, a silencing histone marker, to promote differentiation as described below.
While it remains to be determined whether Setdb1 is a directly downregulated by PPARγ, we demonstrated that Setd8 is a bona fide PPARγ target. PPARγ upregulates Setd8 and thereby regulates H4K20me1 to induce PPARγ and its targets to acquire the adipocyte phenotype. Intriguingly, Setdb1 and Setd8 are expressed in adipose tissue and reciprocally expressed in rodent models of obesity: downregulation of Setdb1 and upregulation of Setd8 suggests that these proteins play a role in regulating adiposity in the excess energy state.
Most intriguingly, Setd8 is a target of PPARγ (). H4K20me1 levels increase robustly toward the end of adipocyte differentiation, and this is accompanied by increased numbers of genes modified by H4K20me1. In addition, H4K20me1 modification levels at PPARγ target genes correlate with PPARγ transcriptional activity. A combination of H4K20me1 ChIP-seq and transcriptome analyses revealed that more than 85% of genes modified by H4K20me1 are expressed at high levels, demonstrating a role for activating histone chromatin modification.Citation27 This is also supported by the recent ChIP analyses showing a preferential association of H4K20me1 with selected transcriptionally active or competent genes.Citation28,Citation29
Although the PPARγ1 gene is not modified by H4K20me1 before differentiation, an appreciable amount of PPARγ1 mRNA is detected. Towards the end of differentiation, PPARγ1 gene expression levels increase by four- to five-fold in correlation with the modification by H4K20me1. H4K20me1 may contribute to the robust gene expression required to progress to the adipocyte phenotype. Although several studies suggest association of H4K20 methylation with repressive chromatin, recent studies showed that H4K20me1 is enriched in promoter or coding regions of active genes.Citation28–Citation30 In addition, recent ChIP sequencing assays also revealed strong evidence that H4K20me1 is strongly correlated with gene activation in the regions downstream of the TSS, consistent with it being a marker of activation.Citation31 Therefore, we postulate that H4K20me1 functions to enhance gene transcription in adipogenesis.
Our data demonstrate that PPARγ is required for Pparγ2 gene expression. PPARγ/RXRα heterodimers bind directly to the Pparγ2 promoter and result in activating histone modifications of Pparγ2 gene, thereby activating transcription. Our results support a model in which a PPARγ-mediated transcriptional feedback-loop, acting through chromatin modification, is essential for the transcriptional activation of PPARγ2 and the subsequent maturation of adipocytes.
Histone 3 Lysine 9 Methytraseferase SETDB1
While SETDB1 is a PPARγ target that is downregulated during adipocyte differentiation and acts as an anti-adipogenic factor, Takada et al. independently demonstrated that SETDB1 is also activated by noncanonical Wnt 5a, which determines the fate of mesenchymal stem cells.
Osteoblasts and adipocytes differentiate from common pluripotent mesenchymal stem cells. Canonical Wnt signaling stimulates osteoblastic differentiation at several steps of cytodifferentiation while inhibiting adipogenesis.Citation32–Citation34 Canonical and noncanonical Wnt signaling pathways are activated by multiple Wnt ligands through binding to frizzled plasma membrane receptors. During activation of the canonical pathway, stabilization and nuclear translocation of the intracellular transducer β-catenin is induced, enabling it to associate with members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors and thus activate the transcription of target genes.Citation35 By contrast to canonical Wnt signaling, the signaling events downstream of the noncanonical signal are understood only vaguely and their physiological impact in cell fate decision of mesenchymal stem cell remains obscure. In addition, the molecular link of histone modification to the transcriptional cascade and response to change in the extracellular environment remains to be uncovered.
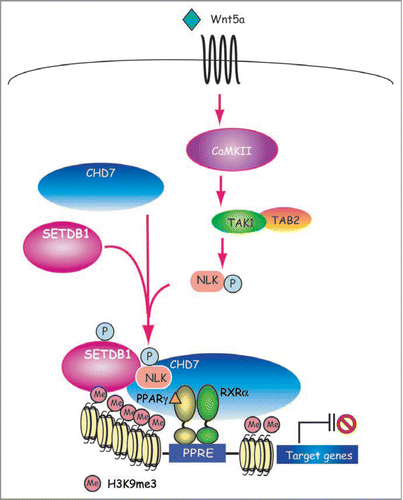
Since noncanonical Wnt ligand, Wnt5a, is expressed at significant levels and Wnt5a is capable of transrepressing PPARγ function induced by PPARγ agonists, Takada et al. explored the downstream signaling (). They demonstrated that PPARγ activation is repressed in trans by the Wnt5amediated activation of the CaMKII-TAK1/TAB2-NLK cascade and by activated NLK (Nemo like kinase). This thereby inhibits adipogenesis and stimulates osteogenesis through SETDB1.Citation36 An HDAC inhibitor, tricostatin A, was unable to reverse NLK-mediated suppression of PPARγ function in ST2 cells, a line of mesenchymal stem cells, indicating a possible role for other inactive histone-modifying enzymes.
NLK-containing complexes were purified from nuclear extracts of KCl treated HeLa cells that expressed FLAG-tagged NLK, using glycerol gradient centrifugation fractionation. These experiments lead to the identification of DEAH-box and CHD domain-containing ATPase protein, CHD7,Citation37 and SETDB1.Citation38,Citation39 The SETDB1 complex associated with PPARγ to methylate H3K9 in the promoters of PPARγ target genes, leading to chromatin inactivation through consequent histone-inactivating modification of H3K9me3. Complex formation of endogenous NLK, SETDB1 and CHD7 with PPARγ was seen only when ST2 cells were treated with Wnt5a.Citation36 Consistently, an increase in histone di- and tri-methylation at histone H3K9 was observed together with hypoacetylation of histone. In addition, noncanonical Wnt signaling activated by Wnt5a induces differentiation of adipocytes into osteoblasts in bone marrow. Thus, this complex is presumed to be a new type of HKMT corepressor complex for nuclear receptors active in signal transduction. These data also suggest that SETDB1 may be a nuclear target activated by signaling via cell membrane receptors to co-repress several classes of transcriptional factors.
Histone 3 Lysine 4 (H3K4) Trimethylation
As observed for histone acetylation, the methylation of H3K4 affects transcriptional activation. Several nuclear proteins, including transcription factors and chromatin modifying enzymes such as MLL3/4, PTIP, Wnt/β-catenin signal are reported that are associated with altered H3K4 methylation states in the promoters of PPARγ, CEBPα or other adipogenic genes.
H3K4-methyltransferases (H3K4MTs) include yeast and human Set1, MLL1, MLL2, MLL3/HALR, MLL4/ALR, Ash1 and Set7/9.Citation40 These proteins contain a SET domain, which is associated with an intrinsic histone lysine-specific methyltransferase activity.Citation41 Mammalian Set1 and MLL complexes belong to a highly conserved family of Set1-like complexes,Citation40 which also contain complex-specific subunits and a common core subcomplex consisting of RbBP5, ASH2L and WDR5.Citation42–Citation45 In particular, WDR5 mediates interactions of the H3K4MT unit with the histone substrate and also plays crucial roles in maintaining the integrity of the complex.Citation43–Citation45
Activating signal cointegrator-2 (ASC-2; also named NCOA6, AIB3, TRBP, TRAP250, NRC and PRIP) is a coactivator of numerous nuclear receptors and transcription factors.Citation46 Importantly, ASC-2 is an integral and unique component of a Set1-like complex named ASCOM (for ASC-2 complex). ASCOM contains MLL3 or MLL4,Citation42,Citation46 and indeed possesses H3K4MT activity.Citation42,Citation47–Citation49 More recent studies identified additional components of ASCOM, including UTX,Citation48,Citation49 a protein subsequently shown to be a H3K27-demethylase enzyme.Citation50–Citation53 Thus, ASCOM, unlike other Set1-like complexes, contains two distinct histonemodifying enzymes linked to transcriptional activation.
The importance of ASC-2 as a key coactivator of multiple nuclear receptors including PPARγ has been reported from studies with various ASC-2 mouse models. In further support for ASC-2 as a physiological coactivator of PPARγ is the observation that the transcriptional activity of PPARγ is impaired in ASC-2-null mouse embryonic fibroblasts (MEFs).Citation54–Citation56 In addition, ASC-2 plays essential roles for the adipogenic program directed by PPARγ as demonstrated by the finding that ASC-2-null MEFs are refractory to PPARγ-stimulated adipogenesis and fail to express the PPARγ-responsive, adipogenic marker gene aP2.Citation56 Interestingly, ASC-2 has been reported to play crucial roles in granulocyte differentiation as a coactivator of C/EBPα,Citation57 which also functions as a key adipogenic factor through its ability to trigger expression of PPARγ during adipogenesis.Citation58 Taken together, these results suggest that ASC-2 may exert its adipogenic function as a coactivator of at least 2 key adipogenic transcription factors, PPARγ and C/EBPα.
MLL3 and MLL4.
Lee et al. demonstrated that MLL3 and MLL4 function as crucial, but redundant, H3K4MTs for adipogenesis, revealing an interesting connection between H3K4 trimethylation and adipogenesis.Citation59 They examined MLL3Δ/Δ miceCitation47 expressing an H3K4MT-inactivated mutant of MLL3 that bears an in-frame deletion of a 61-aa catalytic core region in the MLL3 SET domain. They found that these mice have a significantly decreased amount of WAT associated with a favorable overall metabolic profile, including improved insulin sensitivity and increased energy expenditure. These animals also develop ureter urothelium tumors that likely result from an ASCOM coactivator function for the tumor suppressor p53. They are also resistant to high-fat diet-induced fatty liver formation. The authors also showed that ASC-2, MLL3 and MLL4 are recruited to the PPARγ-activated aP2 gene during adipogenesis, and that PPARγ interacts directly with purified ASCOM. Their results raise the interesting possibility that novel antidiabetic and/or antisteatohepatitis therapeutics might be developed that modulate MLL3/4-H3K4MT activity in appropriate target tissues.
PTIP, a component of a histone lysine 4 methyltransferase (MLL4) complex.
Cho et al.Citation60 reported that the histone methylation regulator PTIP (Pax transactivation domain-interacting protein) is required for the expression of PPARγ and C/EBPα. PTIP is a component of a histone lysine methyltransferase (HKMT) complex that also contains the histone H3K4 methyltransferases MLL3 and MLL4 as well as the JmjC domain-containing protein UTX.Citation48,Citation49,Citation61 Recent studies demonstrated that most covalent histone lysine modifications are reversible and the jumonji C (JmjC)-domain-containing proteins possess demethylase activity. These authors reported that PTIP regulates PPARγ and C/EBPα expression in murine embryonic fibroblasts (MEFs) as well as during preadipocyte differentiation. In preadipocytes, PTIP is essential for the robust induction of PPARγ and C/EBPα (but not for C/EBPβ) during differentiation. On the other hand, before differentiation PTIP is dispensable for the basal level expression of PPARγ and C/EBPα. Increased trimethylation of Histone H3 by MLL4 in the promoters of the PPARγ and C/EBPα genes requires PTIP, as does occupancy of these promoters by RNA polymerase II (Pol II). These reports demonstrate the critical role of H3K4me3 on PPARγ.Citation60
Since knockout of PTIP is lethal in mice, adipose specific PTIP mice were generated by crossing PTIP conditional KO mice with aP2-Cre mice expressing Cre under the control of adipose specific aP2 (adipocyte-specific fatty acid-binding protein 4, also known as FABP4) promoter. Although KO mice had body weight similar to the control mice, they displayed over 50% decrease of BAT mass. Expression markers common to WAT and BAT (PRDM16, cidea, ntrk3),Citation62 brown fat thermogenic genes (UCP1 and PGC1α), and mitochondrial components (cox5b and cox8b) also decreased significantly in the residual BAT of KO mice.Citation63 By contrast, the mass of WAT did not differ significantly between the PTIP KO mice and wild type. These results were consistent with previous reports that deletion of PPARγ in BAT leads to markedly decreased tissue weight in mice.Citation64 The main function of BAT is to burn fatty acids to generate heat. When these mice were exposed to environmental cold, the KO mice were unable to maintain body temperatures and were thus cold intolerant. This result was consistent with previous reports that ablation of BAT led to cold intolerance in mice. A recent study showed that BAT shares precursors (Myf5 positive cells) with muscle cells but not with white adipocytes. Induction of PRDM16 expression in Myf5 positive cells directs them to develop into BAT. PRDM16 and PPARγ physically interact during BAT differentiation.Citation65
Wnt, β-catenin and COUP-TFII.
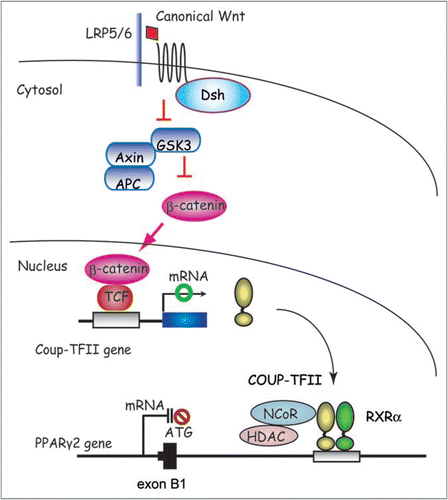
Canonical Wnt/β-catenin signaling prevents the induction of PPARγ and CEBPα gene expression and thereby inhibits adipogenesis.Citation66 This inhibition does not involve the rapid and transient upstream induction of C/EBPβ and C/EBPδ. The endogenous Wnt isoform involved has been proposed to be Wnt10b.Citation66,Citation67 It is secreted by preadipocytes, stabilizes β-catenin, prevents 3T3-L1 differentiation and shifts development of bipotential stromal progenitors away from adipogenesis and toward osteoblastogenesis. The molecular details by which canonical Wnt10b inhibits the downstream induction of C/EBPα and PPARγCitation67 including transcription cascade and epigenetics remain fully elucidated. Thus, it has been a big challenge in the field of Wnt signaling and adipogenesis to integrate known components of the Wnt signaling pathway with the downstream effectors controlling adipogenesis and PPARγ activity.
By combining global gene expression analyses with a ChIP-chip microarray approach, Okamura et al. demonstrated that Wnt/β-catenin signaling activates the expression of the nuclear receptor Coup-TFII which in turn recruits the SMRT co-repressor complex to the first introns located downstream from the first exons of both PPARγ1 and γ2 mRNAs (). This maintains the local chromatin in a hypoacetylated state accompanied by the repression of H3K4me3 and thereby represses expression of PPARγ.Citation24
COUP-TFII has an important role downstream of Hedgehog signaling to inhibit adipogenesis by acting at the C/EBPα promoter.Citation23 When combined with the study by Okamura et al. the results define COUP-TFII as a molecular hub that integrates the input of two key developmental signaling pathways, Wnt and Hedgehog, with PPARγ gene expression and adipocyte differentiation. The impact of Hedgehog on epigenetic regulation remains to be elucidated.
H3K4 Dimethylation
The promoters of adipogenic genes harbor the H3K4 dimethylation signal that is already present in preadipocytes, labeling these genes as silent but poised for transcription.Citation68 This mark is restricted to the promoters in preadipocytes. During the differentiation process, H3K4 dimethylation increases in the promoters and is found also in the coding region of these same genes, coinciding with the start of transcription.Citation68 On the other hand, H3K4 trimethylation in the promoters of apM1, lep and glut4 can be detected only after the start of their transcription, while in the coding regions this mark was delayed, being only detectable at low levels in fully differentiated adipocytes. Mursri et al. reported that treatment with a low dose of the methyltransferase inhibitor methylthoadenosine erased this epigenetic mark from the promoters studied and resulted in dramatically decreased adipogenesis.Citation68 This indicates the importance of this histone posttranslational modification in the regulation of adipogenesis. However, the transcription factor(s) involved in maintaining adequate levels of histone methylation at the adipogenic genes remain unknown.
H3K27 Methylation
Recent mapping of histone methylation in pluripotent and lineage-committed cells have revealed an unexpectedly high frequency of colocalizaion of “activating” H3K4me3 and the “repressive” H3K27me3 on promoters of developmental regulators.Citation69 Genomic regions containing both of these marks are termed “bivalent domains” and are of great interest due to their role in maintaining a poised transcriptional state.Citation69,Citation70 A characteristic of pluripotent cells is the presence of a bivalent histone mark in the chromatin of regulatory regions. Interestingly, H3K4me3 and H3K27me3 also colocalize on the PPARγ1 promoter in MEFs.Citation69 Because genes containing bivalent domains are not considered transcriptionally active, it is possible that NR response elements contain these marks in the absence of a ligand. In response to hormone stimulation, removal of the silencing H3K27 mark would allow rapid activation. PTIP associates with both histone H3K4 methyltransferases MLL3 and MLL4 and histone H3K27 demethylase UTX.Citation53,Citation71 However, the role of UTX and H3K27 methylation in adipogenesis is largely unknown.
Histone Demethylase JHDM2A and Obesity
Recent studies demonstrated that most covalent histone lysine modifications are reversible and the jumonji C (JmjC)-domaincontaining proteins have been shown to possess such demethylase activities.Citation10,Citation72 However, there is little information available on the biological roles of histone lysine demethylation in intact animal model systems. JHDM2A (JmjC-domain-containing histone demethylase 2A, also known as JMJD1A) catalyzes removal of H3K9 mono- and dimethylation through iron and α-ketoglutarate dependent oxidative reactionsCitation73 and plays essential roles for spermatogenesis in mice.Citation74
Two groups have recently reported the role of histone demethylase JHDM2a in the protection from obesity.Citation75,Citation76 Mice deficient in JHDM2a (JHDM2a-/-) develop adult onset obesity, hypertriglyceridemia, hypercholesterolemia, hyperinsulinemia, and hyperleptinemia, which are hallmarks of metabolic syndrome.Citation77 The phenotype of the JHDM2A-/- mice is essentially the same; however, the molecular mechanisms of development of obesity are a little differently understood. Tateishi et al. identified Pparα and Uncoupling protein 1 (Ucp1), two of the important genes involved in controlling energy balance, as direct targets of JHDM2a in skeletal muscle or BAT.Citation75 They also showed that Jhdm2a expression is regulated by the β-adrenergic signaling pathway. Because the expression of these two genes as well as Jhdm2a, is induced by β-adrenergic stimulation, they suggested that JHDM2a contributes to the cellular responses downstream of β-adrenergic signaling. Based on these findings, they proposed that the obese phenotype is due to the loss of JHDM2a which is critical in regulating metabolic control through Pparα and β-adrenergic signaling pathways.
The other group, Inagaki et al.Citation76 demonstrated that JHDM2a also regulates metabolic genes related to energy homeostasis including anti-adipogenesis (Nr2f2 also known as CoupTFII,Citation23,Citation24 and GATA2,Citation19), regulation of fat storage (Apoc1,Citation78), glucose transport (Slc2a4,Citation79), and a gene associated with susceptibility to type 2 diabetes (ADAMTS9,Citation80) in WAT. Their JHDM2a-/- mice furthermore exhibit fasting-induced hypothermia, indicating reduced energy expenditure. These mice also have a higher respiratory quotient, indicating less fat utilization for energy production, which may also make them more prone to obesity.
Thus, these two reports demonstrated H3K9 demethylase JHDM2a is a crucial regulator of genes involved in energy expenditure and fat storage, which further suggests that it is a previously unrecognized key regulator of obesity and metabolic syndrome.
Conclusion Remarks
Histone methylation is catalyzed by histone methyltransferases and reversed by histone demethylases. Recent studies have uncovered that changes in histone modification are a key component of an epigenetic network controlling adipogenesis and energy homeostasis. Further work is required to unravel the causal relationship between diet-induced obesity and histone modification in genes associated with nutritional balance. It is interesting to speculate that reduced JHDM2a activity may contribute to the pathogenesis of common forms of obesity and insulin resistance. Together with the finding of the link between H3K4 methyltranferase MLL3 with the metabolic phenotypes,Citation59 the finding that H3K9 demethylase protects against obesity indicates that modulation of histone lysine methylation in chromatin may be a new target in the treatment of obesity and metabolic syndrome. Moreover, studies of the role of epigenetic changes in histone modification to human nutrition and obesity will be a fruitful area for further research.
Abbreviations
| BAT | = | brown adipose tissue |
| C/EBP | = | CCAAT/enhancer binding protein |
| HKMT | = | histone lysine methyltransferase |
| NR | = | nuclear receptor |
| PPAR | = | peroxisome proliferator-activated receptor |
| PPRE | = | PPAR response element |
| WAT | = | white adipose tissue |
Figures and Tables
Figure 1 Models for the coordinate regulation of transcription and histone modification by PPARγ for adipogenesis. Two well characterized HKMT, Setdb1 and Setd8 are coordinately regulated by PPARγ and their increased activity facilitates terminal adipocyte differentiation through chromatin modification. PPARγ also drives induction of PPARγ2 via a feedback loop and many other of the target genes via two pathways; one through transcription and the other, by way of an epigenetic pathway. PPARγ requires Setd8 to acquire H4K20me1 modification in order to enhance its transcription, while Setd8 requires PPARγ to be transcriptionally induced. These two are both required for the expression of PPARγ targets. Setdb1 is an anti-adipogenic factor whose expression is downregulated toward the end of differentiation. Setdb1 is also identified as a PPARγ target, however, it remains to be determined whether it is a direct downregulated target genes.
Acknowledgements
We thank Rob Rawson for critical reading of the manuscript.
References
- Wolffe AP. Transcriptional regulation in the context of chromatin structure. Essays Biochem 2001; 37:45 - 57
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999; 98:285 - 294
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 1080
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol 2003; 15:172 - 183
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code?. Curr Opin Genet Dev 2005; 15:163 - 176
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6:838 - 849
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell 2005; 18:263 - 272
- Bannister AJ, Kouzarides T. Reversing histone methylation. Nature 2005; 436:1103 - 1106
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410:116 - 120
- Klose RJ, Kallin EM, Zhang Y. JmjC-domaincontaining proteins and histone demethylation. Nat Rev 2006; 7:715 - 727
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006; 444:847 - 853
- Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes Dev 2008; 22:1269 - 1275
- Trayhurn P, Wood IS. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br J Nutr 2004; 92:347 - 355
- Friedman JM. Obesity in the new millennium. Nature 2000; 404:632 - 634
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 2002; 16:22 - 26
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab 2006; 4:263 - 273
- Zhu Y, Qi C, Korenberg JR, Chen XN, Noya D, Rao MS, Reddy JK. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR-gamma) gene: alternative promoter use and different splicing yield two mPPARgamma isoforms. Proc Natl Acad Sci USA 1995; 92:7921 - 7925
- Morrison RF, Farmer SR. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem 1999; 274:17088 - 17097
- Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 2000; 290:134 - 138
- Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab 2008; 7:339 - 347
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab 2005; 1:27 - 39
- Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem 2005; 280:12867 - 12875
- Xu Z, Yu S, Hsu C-H, Eguchi J, Rosen ED. The orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II is a critical regulator of adipogenesis. Proc Natl Acad Sci USA 2008; 0707082105
- Okamura M, Kudo H, Wakabayashi K, Tanaka T, Nonaka A, Uchida A, et al. COUP-TFII acts downstream of Wnt/β-catenin signal to silence PPARγ gene expression and repress adipogenesis. Proc Natl Acad Sci USA 2009; 106:5819 - 5824
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7:885 - 896
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33:245 - 254
- Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 2009; 29:3544 - 3555
- Talasz H, Lindner HH, Sarg B, Helliger W. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem 2005; 280:38814 - 38822
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 2006; 26:9185 - 9195
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18:1251 - 1262
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 837
- Gong Y, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001; 107:513 - 523
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 2002; 346:1513 - 1521
- Fujino T, et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA 2003; 100:229 - 234
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006; 127:469 - 480
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPARgamma transactivation. Nat Cell Biol 2007; 9:1273 - 1285
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 2004; 36:955 - 957
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002; 16:919 - 932
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 2003; 12:475 - 487
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25:15 - 30
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006; 75:243 - 269
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol 2003; 23:140 - 149
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 2006; 13:713 - 719
- Steward MM, Lee JS, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 2006; 13:852 - 854
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005; 121:859 - 872
- Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth and development. Endocr Rev 2005; 26:583 - 597
- Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci USA 2006; 103:15392 - 15397
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 2007; 282:20395 - 20406
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 2007; 27:1889 - 1903
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007; 449:689 - 694
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007; 318:447 - 450
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007; 449:731 - 734
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA 2007; 104:18439 - 18444
- Kuang SQ, Liao L, Zhang H, Pereira FA, Yuan Y, DeMayo FJ, et al. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem 2002; 277:45356 - 45360
- Antonson P, Schuster GU, Wang L, Rozell B, Holter E, Flodby P, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol 2003; 23:1260 - 1268
- Qi C, Surapureddi S, Zhu YJ, Yu S, Kashireddy P, Rao MS, et al. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem 2003; 278:25281 - 25284
- Hong S, Lee MY, Cheong J. Functional interaction of transcriptional coactivator ASC-2 and C/EBPalpha in granulocyte differentiation of HL-60 promyelocytic cell. Biochem Biophys Res Commun 2001; 282:1257 - 1262
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev 2000; 14:1293 - 1307
- Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA 2008; 105:19229 - 19234
- Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, et al. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab 2009; 10:27 - 39
- Kim D, Wang M, Cai Q, Brooks H, Dressler GR. Pax transactivation-domain interacting protein is required for urine concentration and osmotolerance in collecting duct epithelia. J Am Soc Nephrol 2007; 18:1458 - 1465
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab 2007; 6:38 - 54
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 2003; 100:15712 - 15717
- Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 1993; 366:740 - 742
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev 2008; 22:1397 - 1409
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science 2000; 289:950 - 953
- Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, et al. Regulation of Wnt signaling during adipogenesis. J Biol Chem 2002; 277:30998 - 31004
- Musri MM, Corominola H, Casamitjana R, Gomis R, Parrizas M. Histone H3 lysine 4 dimethylation signals the transcriptional competence of the adiponectin promoter in preadipocytes. J Biol Chem 2006; 281:17180 - 17188
- Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448:553 - 560
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 326
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 2004; 14:155 - 164
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007; 25:1 - 14
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006; 125:483 - 495
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450:119 - 123
- Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 2009; 458:757 - 761
- Inagaki T, Tachibana M, Magoori K, Kudo H, Tanaka T, Okamura M, et al. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 2009; 14:991 - 1001
- Koza RA, Nikonova L, Hogan J, Rim J-S, Mendoza T, Faulk C, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet 2006; 2:81
- Jong MC, Voshol PJ, Muurling M, Dahlmans VE, Romijn JA, Pijl H, et al. Protection from obesity and insulin resistance in mice overexpressing human apolipoprotein C1. Diabetes 2001; 50:2779 - 2785
- Rossetti L, Stenbit AE, Chen W, Hu M, Barzilai N, Katz EB, et al. Peripheral but not hepatic insulin resistance in mice with one disrupted allele of the glucose transporter type 4 (GLUT4) gene. J Clin Invest 1997; 100:1831 - 1839
- Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40:638 - 645