Abstract
Prions (infectious proteins) analogous to the scrapie agent have been identified in Saccharomyces cerevisiae and Podospora anserina based on their special genetic characteristics. Each is a protein acting as a gene, much like nucleic acids have been shown to act as enzymes. The [URE3], [PSI+], [PIN+] and [Het-s] prions are self-propagating amyloids of Ure2p, Sup35p, Rnq1p and the HET-s protein, respectively. The [b] and [C] prions are enzymes whose precursor activation requires their own active form. [URE3] and [PSI+] are clearly diseases, while [Het-s] and [b] carry out normal cell functions. Surprisingly, the prion domains of Ure2p and Sup35p can be randomized without loss of ability to become a prion. Thus amino acid content and not sequence determine these prions. Shuffleability also suggests amyloids with a parallel in-register b-sheet structure.
The Genesis of the Prion Concept from Studies in Mammals
The transmissible spongiform encephalopathies (TSEs) of mammals are inexorably fatal degenerative brain diseases whose etiology has long been debated,Citation1,Citation2 but are widely believed to be caused by an infectious protein. The unusual radiation-resistance of the scrapie agentCitation3 generated a flurry of speculation on its nature, including a surprisingly accurate early version of the protein-only hypothesis.Citation4 It was proposed that an altered form of a cellular protein binds a monomer of the normal form, and in this complex, changes the normal to the abnormal form. This is, in essence, the modern view. The key protein was identified genetically as the Sinc gene of mice controlling scrapie incubation period.Citation5 However, it was only 18 years later that Sinc was shown to be the gene encoding PrP,Citation6 the major component of the scrapie agent.Citation7
PrP is a nonessential proteinCitation8 located on the cell surface where it is bound by a GPI anchor.Citation9 Animals lacking the Prnp gene encoding PrP are immune to infection by the TSE agent,Citation10 showing neither pathology, nor substantial replication of infectivity. PrP from brains of TSE-infected animals is quite protease resistant, compared to the protease-sensistive normal protein. It accumulates significantly in diseased tissues because of reduced turnover. The precise structure of the TSE-form of PrP (called PrP-res or PrPSc) is not known, but it is clearly higher in β-sheet content than the normal protein. Amyloid deposits composed largely of PrP-res are observed in many but not all TSEs. The smallest infectious material is estimated to be a 14 to 28-mer, but most of the infectivity is much larger.Citation11 The protease-resistance of infectious material also suggests that it is amyloid in form, even if frank plaques are not always seen.
While extensive circumstantial evidence points to the TSEs being prion diseases, with the infectious agent nothing more than an altered PrP, definitive experiments are still not available, and there continues to be some debate on this point. The best evidence to date comes from studies in which amyloid formed in vitro from recombinant mouse PrP89–230 was injected into mice transgenic for PrP89–231. The mice developed a scrapie-like disease, albeit after an inordinately long incubation period, and their brains were infectious for normal mice.Citation12 The fact that this oft-attempted experiment has so far only worked with amyloid of PrP again indicates that amyloid is indeed the infectious material. Recently, Supattapone's group has demonstrated spontaneous in vitro generation of infectivity using Soto's PCR-like adaptation of Caughey's in vitro PrP-res propagation method. This may be the final proof.
The TSEs are infectious, hereditary and spontaneous. Brain extracts of one animal will readily infect another animal on injection or ingestion. Human hereditary Creutzfeldt-Jakob disease is caused by mutations in the gene for PrP, presumably making it more likely to spontaneously assume the prion form. Spontaneous cases are presumed due to spontaneous formation of infectious amyloid by the normal PrP protein.
Injection of infected brain extract into brains of uninfected animals produces disease with a long, but very characteristic incubation period. The incubation period is much longer for infections across species lines (the ‘species barrier’). Distinct TSE strains (or variants) have been defined, with different incubation periods, distinguishable signs and symptoms and biochemical characteristics of the altered PrP. These strain (or variant) characteristics are not due to different PrP sequences, but are thought to reflect different amyloid structures. The TSE strain (variant) also affects the species barrier: while one TSE strain may be unable to cross between a particular pair of species, another may readily do so (reviewed in ref. Citation13).
Discovery of Infectious Proteins (Prions) in S. Cerevisiae
When yeast is supplied with a good nitrogen source, such as ammonia, it turns off transcription of the genes encoding the enzymes and transporters (e.g., DAL5, encoding the allantoate transporter) needed to use poor nitrogen sources, like proline or allantoate (reviewed in refs. Citation14 and Citation15). This control mechanism is called nitrogen catabolite repression or nitrogen control and is mediated by Ure2p. [URE3] is a nonchromosomal gene whose dominant effect is to derepress these enzymes and transporters.Citation16 [PSI+] is a nonchromosomal gene discovered as a translational suppressor of nonsense mutations,Citation17 and Sup35p is a subunit of the translational termination factor.Citation18,Citation19 The molecular basis of [URE3] and [PSI+] was long a puzzle.
We proposed three genetic criteria to distinguish nucleic acid replicons such as viruses and plasmids from prionsCitation20 ().
Reversible curability.
While a virus, plasmid or prion may be curable (efficiently eliminated) by some treatment, a virus or plasmid is not likely to be regenerated de novo in less than geologic time. However, the protein capable of becoming a prion is still present in the cured strain and could spontaneously convert to the self-propagating altered prion form.
Overexpression of the protein increases frequency of prion generation.
Overproducing a chromosomally encoded protein will not increase the frequency with which a plasmid or virus arises de novo, but increasing the cellular content of a protein able to become a prion should increase the frequency of prion generation. The change must be self-propagating, and so should take over most of the population of molecules of that protein, converting them to the prion form.
Phenotype relation and gene dependence.
For viruses, plasmids and prions, the propagation of the nonchromosomal element always requires the activity of some chromosomal proteins. Prion propagation requires at least the gene encoding the protein. If the prion form of a protein were simply an inactive form of the normal protein, then the phenotype of the prion-carrying strains should resemble that of mutants in the gene encoding the protein. This contrasts with viruses or plasmids conferring a cellular phenotype (such as the mitochondrial DNA or killer virus). In these cases, the phenotype of mutation of a chromosomal gene needed for propagation of the nucleic acid replicon is that of absence of the replicon (e.g., killer-negative, glycerol minus).
[URE3] and [PSI+] are Prions
[URE3] has all of these properties if viewed as a prion of Ure2p, and [PSI+] qualifies as a prion of Sup35p.Citation20 [URE3] can be cured by guanidine but arises again at a low frequency.Citation20 The overproduction of Ure2p elevates the frequency of [URE3] by 20- to 200-fold.Citation20 Finally, the phenotype of [URE3] strains is very similar to that of ure2 mutants.Citation21 [PSI+] may be cured by high osmotic strength,Citation22 but [PSI+] derivatives of the cured strains are easily isolated.Citation23 Overproduction of Sup35p elevates the frequency of [PSI+] arising de novo,Citation24 and the [PSI+] phenotype resembles that of sup35 mutants, namely, nonsense suppression.Citation17
What Does it Take to be a Prion?
We found that the N-terminal asparagine-rich part of Ure2p was necessary and sufficient to propagate and induce the [URE3] prion,Citation25,Citation26 and at the same time we reinterpreted the similar results of TerAvanesyan et al. on the Q/N rich N-terminal domain of Sup35p.Citation27 We call these the prion domains of the respective proteins. It is clear that mutations within the prion domain can affect prion propagation.Citation28–Citation31 Some of these changes do not prevent the protein from being a prion, but rather introduce a ‘species barrier’ between molecules.Citation32,Citation33
To examine whether there are sequence determinants of prion-formation ability, the prion domains of Ure2p and Sup35p were each shuffled, leaving amino acid content and codon usage unchanged.Citation34,Citation35 Five shuffled variants of each prion domain were generated and reintroduced into the chromosome in place of the normal prion domain (). It was found that each of the shuffled prion domains of Ure2p and of Sup35p were capable of being prions. The frequency of prion formation varied somewhat and in each case, one in five of the shuffled sequences produced only unstable prion variants. But all could be prions.Citation34,Citation35 In addition, each of the shuffled Ure2p species readily formed amyloid in vitro. In support of this picture, a detailed deletion analysis of the Ure2p prion domain showed that no single region of the prion domain is essential for prion-forming ability.Citation35
These results imply that the composition of the prion domain is the critical determinant of prion formation. It is very likely that the high Q/N content of the Ure2p and Sup35p prion domains is important. However, few of the many proteins with such Q/N-rich domains have been found capable of making prions. There are doubtless other compositional features of the prion domains that are important. Their relatively low content of charged residues and hydrophobic amino acids are probably important, but further work will be needed to define the critical features.
Because small deletions in the C-terminal domains of Ure2pCitation25 and larger deletions of Sup35pCitation36 C-terminus dramatically increase the frequency of prion formation, it was suggested that the prion domain and C-terminal domains interact, preventing the prion domains from interacting with eachother to form amyloid. However, no evidence for such an interaction could be detected,Citation37 and the Ure2p prion domain appears to be unstructured in its native (soluble) form. The fact that the prion domain can be shuffled and still support prion formation and propagation argues that if there is such an interaction, it is not important for this process.
Shuffleable Prion Domains Suggest Parallel In-Register Structure
Although amyloids have long been known to be rich in β-sheet, their more detailed architecture has been unclear. There are at least three mutually exclusive possibilities for the β-sheet architecture of amyloid. An antiparallel β-sheet has adjacent strands bonded to each other running in opposite orientations: N → C next to C → N, for example the amyloid of the Aβ (34–42) fragment.Citation38 In a parallel in-register β-sheet structure, adjacent bonded strands are in the same orientation: N → C next to N → C, and identical residues are bonded to each other, for example the amyloid of Aβ(1–40).Citation39–Citation42 Electron spin resonance indicates that amyloids of amylin and of a-synuclein also have parallel in-register β-sheet structure.Citation43,Citation44 A third possibility is some form of parallel out-of-register β-sheet, for example the β-helix structure of pectate lyases.Citation45 Here, like the antiparallel structure, nonidentical residues are paired.
Amyloid formation is much like a linear crystal, in that essentially a single species of protein is singled out to join the growing filaments. This specificity demands that there be some specificity in the bonding between chains. For anti-parallel β-sheets or β-helices, this could be large with small, positive with negative, hydrophobic with hydrophobic, hydrogen bonding (donor) with hydrogen bonding (recipient). In these cases, shuffling the sequence would disturb the alignment of complementary residues, and presumably prevent prion formation ().
For parallel in-register β-sheets, hydrogen bonding between Q/N residuesCitation46 or S/T residues, or hydrophobic with hydrophobic residues could provide specificity. However, charged residues (which are few in the prion domains of Ure2p and Sup35p) should tend to interfere with formation of this structure. Shuffling the residues of a parallel in-register β-sheet does not change the pairing, since identical residues are always paired. Thus, we argue that if a prion domain can be shuffled and still be a prion, it should have a parallel in-register β-sheet structure.Citation47
This suggests the sort of model shown in . The core of the amyloid is made up of Ure2p1–65,Citation48,Citation49 which should be a parallel in-register β-sheet.Citation47 Indeed Ure2p10–39, a fragment of the prion domain, has been shown to have such an architecture.Citation50 The folding of the β-sheet is demanded by the diameter of the amyloid filaments of the prion domain.Citation48 Ure2p66–95 is unstructured in both native and amyloid forms of Ure2p, and we call this the ‘tether’ (green in ). The C-terminal part of Ure2p apparently does not change its conformation on formation of amyloid.Citation51,Citation52 A similar parallel in-register β-sheet model can be proposed for Sup35p, since its prion domain is shuffleable and the charged M domain is likely to serve as a tether.
Amyloid is the Prion Infectious Material, Not a Dead End (Side-) Product
In a ground-breaking study, infection of Podospora anserina with the [Het-s] prion by amyloid of recombinant HET-s protein was acheived.Citation53 Soluble protein was not infectious nor was heat- or acid-denatured aggregated protein. The transmissibility of [PSI+] by amyloid of recombinant Sup35p has also been demonstrated, and evidence was also obtained that the amyloid structure determines the prion variant.Citation54,Citation55 As mentioned above, amyloid of recombinant PrP has also shown some infectivity for mice.Citation12
We have now demonstrated the ability of amyloid formed in vitro from recombinant Ure2p to infect cells with the [URE3] prionCitation56 (). The low level infectivity of soluble Ure2p () is apparently due to filament formation while the experiment is in progress. Cells infected with amyloid of recombinant Ure2p show at least three prion variants, distinguishable by their mitotic stability and by the intensity of their phenotype (degree of DAL5 derepression). Extracts of [URE3] strains are also infectious, and transmit the [URE3] variant that was present in the strain from which the extract was prepared (). Remarkably, the amyloid made in vitro from recombinant Ure2p is as much as 1/3 as infectious as is an extract (on a per Ure2p molecule basis).Citation56 The extracts can be used to seed amyloid formation by soluble recombinant Ure2p, but the extent to which this amplification is variant-faithful is limited by the tendency of the ‘soluble’ Ure2p to spontaneously form amyloid filaments, the latter having a mixture of variant structures.Citation56
The Ure2p prion domain by itself, or fused to various other proteins can also form amyloid which is infectious.Citation56 Cells infected with these fusion proteins (or prion domain alone) show the same spectrum of prion variants as those infected with amyloid formed from the full length protein.
Preliminary size fractionation experiments indicate that infectious material is greater than 20 nm in diameter, indicating a filament length of >40 mer. Amyloid filaments must be sonicated to be infectious, apparently in order to get into yeast. However, while the largest size fraction of filaments has only low infectivity, resonication increases its infectivity many fold.Citation56 We suggest that this increase in infectivity is a combination of generation of new filament ends (which must be the growing point) and of allowing more facile entry into the cells.
The infectivity of amyloid (and not soluble or other aggregated forms) in all of the prion systems indicates that amyloid is not a dead-end or side product of the prion process. The structure of amyloid formed in vitro has long been recognized to be morphologically heterogeneous. Recently evidence for structural heterogeneity of Aβ amyloid has been obtained.Citation57 It is clear from the prion studies that prion variants are encoded by differences in amyloid structure. It will be particularly interesting to know what are these structural differences and how they propagate.
[PSI+] and [URE3] are Diseases of Yeast
It has been proposed, based on plate tests, that [PSI+] is an advantage to cells carrying it in surviving stressCitation58 and for evolvability.Citation59,Citation60 Some strains grow better under certain conditions if they are [PSI+] than if they are [psi-], although there are no conditions that uniformly favor [PSI+], and most conditions favor [psi-].Citation58–Citation60 The genetic basis for these phenotypes remains to be determined.
All of the conditions were measuring growth, but yeast may be spending most of its time in stationary phase. To what extent are the few conditions favoring [PSI+] represented in the wild? This question is almost impossible to answer directly, and it is further complicated by the fact that whether [PSI+] is favored or unfavored is very strain-dependent.
We examined the distribution of [URE3], [PSI+] and [PIN+] in 70 wild strains.Citation61 Prions arise de novo and spread by infection, so that even if they are a mild disadvantage to their host, they should be frequently found in the wild. As controls, we examined the distribution of parasitic DNA and RNA replicons of yeast: the 2 micron DNA plasmid, 20S RNA, 23S RNA and the L-BC virus (reviewed in ref. Citation62). We found that the mildly detrimental nucleic acid replicons were found in varying proportions of the wild yeast (). For example, 2 micron DNA has been shown to slow growth by 1.5–3.0%,Citation63 but is found in 38 of 70 wild strains.
We found that none of the wild strains carried either [URE3] or [PSI+] (). Similarly, [PSI+] was absent from nine clinical isolates,Citation64 two industrial S. cerevisiae and eight other non-cerevisiae strains of Saccharomyces.Citation65 This indicates that these prions must be quite substantially detrimental to their host. As previously reported for two clinical isolates, [PIN+] is not rare in the wild (), but its frequency is similar to the parasitic DNA and RNA replicons, suggesting that it is a rather mild disease.
Our approach measures whether [URE3] or [PSI+] are advantageous or not without addressing specific conditions of growth. It remains possible that there is a natural situation in which [URE3] or [PSI+] are more of a help than a hindrance, just as the mild hemoglobin disease, Sickle Cell Trait, is an advantage in areas where malarial infection is prevalent. However, stress and the need to evolve are daily occurrences for yeast, and if [URE3] or [PSI+] helped in this regard, they would not be hard to find in the wild.
A Self-Activating Enzyme Acting As a Prion
The word ‘prion’ means ‘infectious protein’,Citation66 and although most prions are found to be self-propagating amyloids, this need not be the case.Citation4 If an enzyme were made as an inactive precursor, and the active form of the same enzyme were necessary for activation of the precursor, then this could appear as a prion system. The vacuolar protease B of S. cerevisiae is made as an inactive precursor, and is normally processed proteolytically to an active form by protease A (reviewed in ref. Citation67). However, in mutants deleted for protease A (pep4Δ), evidence for some transient self-activation was obtained.Citation68 We showed that this self-activation of protease B could be propagated indefinitely if cells were grown on nonfermentable carbon sources, under which conditions the gene encoding protease B is derepressed.Citation69
The inactive state of protease B is very stable, as is the active state. Spontaneous activation of the enzyme occurred only about once in 105 cells. Loss of the active state was more frequent, occuring in 1% or more of cells. The active state was transferable by cytoduction, and we called this nonchromosomal genetic element [β].Citation69
[β] has all the properties expected of a prion. Growth of cells on glucose media efficiently cures [β], but from cured cells it again arises de novo (reversible curability). Overproduction of the inactive protease B precursor increases the frequency of [β] generation de novo from about 10-5 to about 10-2 or higher.Citation69 The propagation of [β] depends on the PRB1 gene, but because the prion in this case is not an inactive form of the protein, the phenotype of [β] cells is the opposite of that of prb1 mutants.
Like the [Het-s] prion of Podospora anserina,Citation70 [β] is a prion with a function for the cells. Without [β], diploid cells fail to undergo meiosis and spore formation, and die more rapidly under starvation conditions.Citation69 Because [β] is only seen as a prion in the absence of protease A, one could view it as rather artificial. Alternatively, it could be seen as a prion so essential for the cell, that the protease B precursor has evolved to be protease A-cleavable, thus insuring that the prion (active protease B) is never lost. This amounts to duplication of function.
The importance of our findings is that there are many potentially self-modifying enzymes, including protein kinases, protein transacetylases, protein glycosyl transferases, protein methylases, and many others. We suggested that some of these enzymes might become prions under some circumstances. Indeed, we did not have to wait long.
A Possible Protein Kinase Prion
Crippled Growth is a nonchromosomal genetic element, called [C], of Podospora anserina, characterized by slow hyphal growth and dark pigmentation.Citation71 This trait has recently been shown to require for its propagation a gene encoding a MAP kinase kinase kinase.Citation72 Most strikingly, overproduction of the same enzyme increases the frequency with which the [C] nonchromosomal genetic element arises.Citation72 The Crippled Growth phenotype differs from that of mutation of the MAPKKK gene, as expected if it is due to activation of the MAPKKK enzyme, rather than inactivation. Interestingly, the MAPKKK protein has a 60 residue polyQ sequence near its N-terminus, but deletion of this sequence does not impair ability to propagate [C].Citation72 It is likely that [C] is a self-propagating self-activation of the MAPKKK,Citation73 but further work will be needed to confirm this conclusion.
Figures and Tables
Figure 1 Genetic criteria for prions. Reversible curing means that in a strain cured of a nonchromosomal genetic element, the same element can arise again. Overproducing a protein with the potential to become a prion increases the frequency with which the prion arises. If the prion form of the protein is an inactive form of the protein, then the phenotype of the presence of the prion is the same or similar to that of a mutant in the gene for the protein. Each of these three properties should be characteristic of prions but none of them are known (or expected) for nucleic acid replicons such as plasmids or viruses.
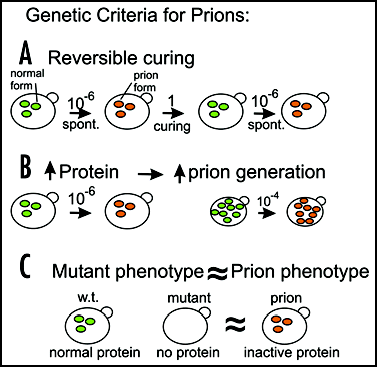
Figure 2 Scrambled prion domains can still be prions.Citation34,Citation35 In place of the normal Ure2 or Sup35 prion domains, shuffled prion domains (five of each) with the same amino acid content were constructed and integrated. Each of the shuffled prion domains could be a prion, although one of each was unstable.

Figure 3 A prion domain insensitive to scrambling should be a parallel in-register amyloid.Citation47 (A) Nonidentical residues are bonded in an anti-parallel β-sheet or β-helix. The specificity of amyloid propagation (similar to crystal growth) implies that there must be some complementarity of residues. Shuffling such a sequence would destroy any such complementarity and thus prion formation. Shuffling a parallel in-register β-sheet leaves identical residues paired with each other. If a prion domain can be shuffled and not lose prion-forming ability, it suggests a parallel in-register β-sheet structure. (B) Model of Ure2p amyloid structure (see text).
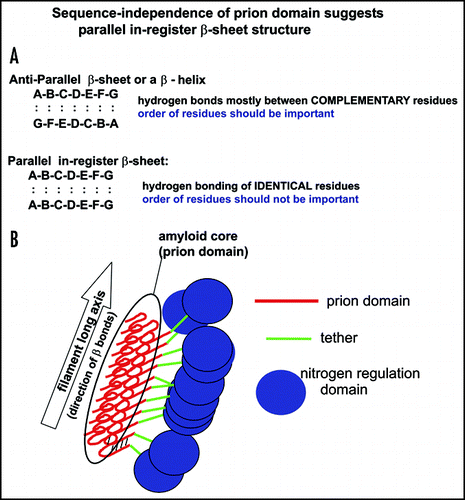
Figure 4 Amyloid of Ure2p is infectious.Citation56 Amyloid made in vitro from recombinant Ure2p (full length or the prion domain or fusions of the prion domain with other proteins) are infectious for yeast. (A) Filaments are sonicated (bar = 100 nm) and introduced into spheroplasts with a DNA plasmid and polyethylene glycol. (B) A large proportion of the clones transformed for the DNA plasmid were also infected with [URE3]. (C) The infected clones included several prion variants distinguished by stability and intensity of the phenotype, here indicated by activity of a DAL5-promoted ADE2 gene. Red clones have lost [URE3]. Extracts of each variant are infectious and transmit the variant of the strain from which they were made.Citation56
![Figure 4 Amyloid of Ure2p is infectious.Citation56 Amyloid made in vitro from recombinant Ure2p (full length or the prion domain or fusions of the prion domain with other proteins) are infectious for yeast. (A) Filaments are sonicated (bar = 100 nm) and introduced into spheroplasts with a DNA plasmid and polyethylene glycol. (B) A large proportion of the clones transformed for the DNA plasmid were also infected with [URE3]. (C) The infected clones included several prion variants distinguished by stability and intensity of the phenotype, here indicated by activity of a DAL5-promoted ADE2 gene. Red clones have lost [URE3]. Extracts of each variant are infectious and transmit the variant of the strain from which they were made.Citation56](/cms/asset/27c1d923-dd48-4447-84be-83999e492965/kprn_a_10904664_f0004.gif)
Figure 5 Enzymes needed for their own activation can be prions. “Prion” means “infectious protein”, not necessarily amyloid based. If an enzyme is essential for activation of its own precursor, then cells without the active form produce the same as progeny, and those with the active form produce offspring of the same kind. Transmission of just the active form (the protein only) from one cell to another lacking it, transmits the self-propagating activity, and so is an infectious protein. Two such systems have been described, the vacuolar protease B of S. cerevisiae,Citation69 and a protein kinase of Podospora anserina.Citation72
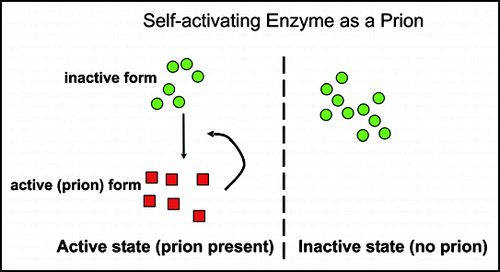
Table 1 Nonchromosomal genetic elements in wild Saccharomyces
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, NIDDK.
References
- Prusiner SB. Prion Biology and Diseases 2004; Cold Spring Harbor Cold Spring Harbor Laboratory Press
- Chesebro B. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br Med Bull 2003; 66:1 - 20
- Alper T, Haig DA, Clarke MC. The exceptionally small size of the scrapie agent. Biochem Biophys Res Commun 1966; 22:278 - 284
- Griffith JS. Self-replication and scrapie. Nature 1967; 215:1043 - 1044
- Dickinson AG, Meikle VMH, Fraser H. Identification of a gene which controls the incubation period of some strains of scrapie in mice. J Comp Path 1968; 78:293 - 299
- Carlson GA, Kingsbury DT, Goodman PA, Coleman S, Marshall ST, DeArmond S, Westaway D, Prusiner SB. Linkagae of prion protein and scrapie incubation time genes. Cell 1986; 46:503 - 511
- Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science 1982; 218:1309 - 1311
- Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature 1992; 356:577 - 582
- Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 1987; 51:229 - 240
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to Scrapie. Cell 1993; 73:1339 - 1347
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature 2005; 437:257 - 261
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science 2004; 305:673 - 676
- Collinge J. Variant Creutzfeldt-Jakob disease. Lancet 1999; 354:317 - 323
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: Connecting the dots. FEMS Microbiol Revs 2002; 26:223 - 238
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002; 290:1 - 18
- Lacroute F. NonMendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol 1971; 106:519 - 522
- Cox BS. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 1965; 20:505 - 521
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 1995; 14:4065 - 4072
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 1995; 14:4365 - 4373
- Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in S. cerevisiae. Science 1994; 264:566 - 569
- Drillien R, Lacroute F. Ureidosuccinic acid uptake in yeast and some aspects of its regulation. J Bacteriol 1972; 109:203 - 208
- Singh AC, Helms C, Sherman F. Mutation of the nonMendelian suppressor y+ in yeast by hypertonic media. Proc Natl Acad Sci USA 1979; 76:1952 - 2016
- Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res 1981; 37:173 - 182
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet 1993; 24:268 - 270
- Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 1995; 270:93 - 95
- Masison DC, Maddelein ML, Wickner RB. The prion model for [URE3] of yeast: Spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA 1997; 94:12503 - 12508
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the nonMendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 1994; 137:671 - 676
- Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2-mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 1994; 137:659 - 670
- DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998; 93:1241 - 1252
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 1999; 400:573 - 576
- Maddelein ML, Wickner RB. Two Prion-inducing regions of Ure2p are nonoverlapping. Mol Cell Biol 1999; 19:4516 - 4524
- Kochneva-Pervukhova NV, Paushkin SV, Kushnirov VV, Cox BS, Tuite MF, Ter-Avanesyan MD. Mechanism of inhibition of Y+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J 1998; 17:5805 - 5810
- Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J 2001; 20:6683 - 6691
- Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol 2004; 24:7206 - 7213
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA 2005; 102:12825 - 12830
- Kochneva-Pervukhova NV, Poznyakovski AI, Smirnov VN, Ter-Avanesyan MD. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharmyces cerevisiae. Curr Genet 1998; 34:146 - 151
- Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured?. Biochemistry 2005; 44:321 - 328
- Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, Kocisko DA, Hendsch ZS, Ashburn TT, Spencer RG, et al. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol 1995; 2:990 - 998
- Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer's β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc Natl Acad Sci USA 1998; 95:13407 - 13412
- Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer's β-amyloid fibrils. Proc Natl Acad Sci USA 2000; 97:13045 - 13050
- Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA 2002; 99:16742 - 16747
- Tycko R. Insights into the amyloid folding problem from solid-state NMR. Biochemistry 2003; 42:3151 - 3159
- Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem 2004; 279:48420 - 48425
- Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of a-synuclein fibrils studied by site-directed spin labeling. J Biol Chem 2003; 278:37530 - 37535
- Yoder MD, Jurnak F. Protein motifs: 3. The parallel β helix and other coiled folds. FASEB J 1995; 9:335 - 342
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA 1994; 91:5355 - 5358
- Ross ED, Minton AP, Wickner RB. Prion domains: Sequences, structures and interactions. Nat Cell Biol 2005; 7:1039 - 1044
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 1999; 283:1339 - 1343
- Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB, Steven AC. Architecture of Ure2p prion filaments: The N-terminal domain forms a central core fiber. J Biol Chem 2003; 278:43717 - 43727
- Chan JC, Oyler NA, Yau WM, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry 2005; 44:10669 - 10680
- Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA 2002; 99:5253 - 5260
- Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem 2004; 279:50025 - 50030
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA 2002; 99:7402 - 7407
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature 2004; 428:319 - 323
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature 2004; 428:323 - 328
- Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J 2005; 24:3082 - 3092
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science 2005; 307:262 - 265
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J 1999; 18:1974 - 1981
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 2004; 431:184 - 187
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 2000; 407:477 - 483
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA 2005; 102:10575 - 10580
- Wickner RB. Knipe DM, Howley PM. Fields Virology 2001; Philadelphia Lippincott, Williams and Wilkins 629 - 658
- Mead DJ, Gardner DCJ, Oliver SG. The yeast 2 m plasmid: Strategies for the survival of a selfish DNA. Mol Gen Genet 1986; 205:417 - 421
- Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol 2003; 49:1005 - 1017
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 2000; 35:865 - 876
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136 - 144
- Jones EW. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem 1991; 266:7963 - 7966
- Zubenko GS, Park FJ, Jones EW. Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics 1982; 102:679 - 690
- Roberts BT, Wickner RB. A class of prions that propagate via covalent auto-activation. Genes Dev 2003; 17:2083 - 2087
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA 1997; 94:9773 - 9778
- Silar P, Haedens V, Rossingnol M. Propagation of a novel cytoplasmic, infectious and deleterious determinant is controlled by translational accuracy in Podospora anserina. Genetics 1999; 151:87 - 95
- Kicka S, Silar P. PaASK1, a mitogen-activated protein kinase kinase kinase that controls cell degeneration and cell differentiation in Podospora anserina. Genetics 2004; 166:1241 - 1252
- Wickner RB, Edskes HK, Ross ED, Pierce MM, Shewmaker F, Baxa U, Brachmann A. Prions of yeast are genes made of protein: Amyloids and enzymes. Cold Spring Harb Symp Quant Biol 2004; 49:489 - 496