Abstract
PrPSc is an infectious protein. The only experimentally verified difference between PrPSc and its normal cellular isoform (PrPC) is conformational. This work describes an approach to determining the presence of surface exposed or sequestered amino acids present in the PrPSc isoform. The N-hydroxysuccinimide esters of acetic acid and 4-trimethylammoniumbutyric acid were synthesized and reacted with detergent-solubilized brain extracts from Me7-infected mice, uninfected mice, 263K-infected hamsters or uninfected hamsters. These reaction mixtures were analyzed by western blots probed with the antibodies 3F4, 6D11, 7D9, AG4, AH6, GE8 or MAB5424. The 3F4, 6D11, AH6, and GE8 antibodies recognize an epitope that is encrypted in the PrPSc isoform, but exposed in the PrPC isoform. These reagents permit the detection of prion infected brain extracts without the need for proteinase K digestion. In addition they can be used, with an appropriate antibody, to determine which amino acids of PrPSc are exposed on the surface and which are encrypted, thus providing useful structural information. This approach was used to distinguish between the 263K and drowsy strains of hamster-adapted scrapie without the use of proteinase K.
Introduction
Prions (PrPSc) are infectious proteins that cause transmissible spongiform encephalopathies (TSEs).Citation1 They have the capacity to convert a normal cellular protein (PrPC) into a prion and thereby propagate an infection. PrPSc and PrPC possess identical covalent structures.Citation2 In addition, there are strains of PrPSc that have distinct phenotypes and physicochemical properties and yet possess identical covalent structures.Citation3 The experimentally determined structural difference between the two isoforms is entirely conformational.Citation1,Citation4
In a protein, the chemical environment of an amino acid is dependent upon the conformation of that protein. This means that the same amino acid can react differently with the same chemical reagent, depending upon the conformation of that protein.Citation5,Citation6 This phenomenon has been referred to as epitope protection.Citation7 Some antibodies selectively bind to one PrP isoform but not the other even though both isoforms possess identical covalent structures.Citation3,Citation8 This may result from the epitope being exposed in one conformation but not in the other. Alternately, since antibodies are large, the steric bulk of the antibody may prevent the antibody from binding to an exposed epitope on a PrP isoform in its native state. Thus the binding of an antibody may be dependent upon the native state conformation of the isoform as well as the surface availability of the epitope.
The structures of PrPC and PrPSc have been successfully chemically modified using activated esters (N-hydroxysuccinimide esters (NHS). These activated NHS ester reagents react primarily with the N-terminus of a protein, ε-amino group of lysine and hydroxyl groups of tyrosine, serine, and threonine.Citation9,Citation10 NHS ester based cross-linking reagents have been used to determine distance constraints in hamster PrPSc and to map an anti-bovine PrP epitope.Citation11,Citation12 NHS esters have been used to attach nanoparticles to human recombinant PrP.Citation13
These efforts indicate that there are a number of amino acids on the surface of PrP that will react with a small molecule NHS ester. If the reagent reacts with an antibody-recognized surface epitope, then it will be covalently modified. If the epitope is not on the surface, then it will not be covalently modified, i.e., the epitope will remain unchanged. In principle, western blotting with an antibody can be used to distinguish between these epitopes. Those epitopes that have reacted with the reagent will not be recognized by the antibody, while those that have not reacted will be recognized. In this way an antibody can be used in a novel way to determine which epitope is on the surface of PrPC or PrPSc and which is not. This approach relies on antibody recognition to identify accessible amino acids and is not dependent on mass spectrometry.
In order to test this approach, seven commercially available antibodies and two small molecule reagents were selected. The NHS esters of acetic acid (Ac) and 4-trimethylamoniumbutyric acid [(3-carboxypropyl)trimethylammonium chloride] (tMAB) were selected as the two small molecule reagents. The epitopes of these seven antibodies have been determined by other researchers and are shown in .Citation14-Citation19 Four of these antibodies, 3F4, 6D11, AH6, and GE8, were empirically determined to have an epitope that was no longer recognized by the antibody after reacting with either reagent. Amidation of ε-amino group of lysine is the most likely reason for the loss of recognition by the antibodies 3F4, 6D11, and GE8. Esterification of the hydroxyl of serine or tyrosine is the reason for the loss of recognition by the antibody AH6. The three other antibodies, MAB5424, 7D9, and AG4, were selected as controls, because their epitopes were not modified by the reagents.
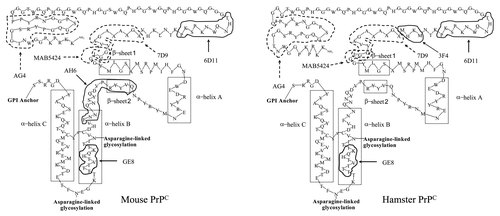
Figure 1. Cartoon of mouse and hamster PrPC. The epitopes of the antibodies 3F4 (MKHM), 6D11 (THNQWNKPSKP), AH6 (QVYYRPVDQYSNQNNF) and GE8 (IKQHTVTTT) are indicated with solid lines. The epitopes of AG4 (WNTGGSRYPGQGSPGGNRYPP), 7D9 (AGAAAAGAVVGGL), and MAB5424 (GGLG) are indicated by dashed lines. The positions of the two β-sheets (β-sheet 1 and β-sheet 2) and the three α-helicies (α-helix A, α-helix B, and α-helix C) are indicated with dotted lines. (Adapted from Dumpitak, Prusiner.Citation20,Citation21)
The reagents were prepared and reacted with detergent-solubilized extracts of brain homogenates from 263K-infected hamsters, drowsy (Dy)-infected hamsters, Me7-infected mice, uninfected mice, and uninfected hamsters and analyzed by western blotting. The results of these experiments are described.
Results
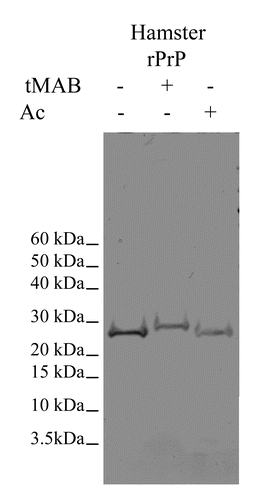
The NHS esters of acetic acid (Ac-NHS) and 4-trimentylammoniumbutyric acid (tMAB-NHS) were prepared by standard methods and reacted with recombinant Syrian hamster PrP (rSHaPrP). The reaction mixtures were run on SDS-PAGE and stained with Coomassie brilliant blue (). There is a distinct gel shift for the protein reacted with the tMAB-NHS reagent and not for the same reaction using the Ac-NHS reagent. Upon formation of a covalent bond with the protein, the tMAB-NHS reagent will add a positive charge to it; the Ac-NHS reagent will not. The extra positive charges on the protein would neutralize an equal number of negative charges from the SDS coating the protein, and result in an even larger apparent molecular weight when analyzed by SDS-PAGE. This is why there is a much larger gel shift for the protein reacted with the tMAB-NHS reagent.
Figure 2. SDS-PAGE gel of hamster recombinant PrP (rPrP), hamster rPrP reacted with tMAB-NHS (tMAB) or hamster rPrP reacted with Ac-NHS (Ac). This is a black and white image of a Coomassie stained SDS-PAGE gel.
The reaction of rSHaPrP with Ac-NHS was analyzed by mass spectrometry. This analysis showed that the reaction mixture is complex (). At least 13–20 amino acid residues reacted with the reagent (). rSHaPrP contains 12 primary amines, 11 lysine residues and the N-terminus of the PrP molecule. Since NHS esters react preferentially with primary amines,Citation10 this indicates that more than just primary amines are chemically reacting with the NHS reagents under these conditions. Trypsin digestion of the Ac-NHS treated rSHaPrP yielded mostly large fragments due to the amidation of ε-amino group of lysine. The reaction with the primary amines accounts for 12 of the observed modifications. The few fragments suitable for mass spectrometric analysis revealed the location of three more modifications of tyrosine and serine residues (PKPGGWNTGGSR, PKPGGWNTGGSR, YPGQGSPGGNR). The remaining five other covalent modifications remain unclear. This analysis also revealed that identifying a particular covalent modification is difficult to do by mass spectrometry even when the protein (rSHaPrP) possesses only a single post-translational modification (disulfide bond), let alone molecules as complex as PrPC and PrPSc.
Figure 3. Mass distribution of recombinant hamster PrP before and after reaction with the Ac-NHS reagent. Distributions are normalized and offset for clarity. Unreacted hamster recombinant PrP (b). Hamster recombinant PrP reacted with Ac-NHS reagent (20 mM concentration) (a). Ac is the mass difference corresponding to the addition of acetate (42 Da). [O] is the mass difference corresponding to the addition of oxygen (16 Da) to methionine.
![Figure 3. Mass distribution of recombinant hamster PrP before and after reaction with the Ac-NHS reagent. Distributions are normalized and offset for clarity. Unreacted hamster recombinant PrP (b). Hamster recombinant PrP reacted with Ac-NHS reagent (20 mM concentration) (a). Ac is the mass difference corresponding to the addition of acetate (42 Da). [O] is the mass difference corresponding to the addition of oxygen (16 Da) to methionine.](/cms/asset/b634c24c-b3db-4146-9324-7ad50658c6d3/kprn_a_10918795_f0003.gif)
The alternate approach of using antibodies with characterized epitopes to identify the modified amino acids was tested using western blot analysis. Uninfected and infected hamster (263K) and mouse (Me7) detergent-solubilized brain extracts were prepared and reacted with tMAB-NHS and Ac-NHS. The resulting reaction mixtures were analyzed by western blot. Each western blot was probed with one of seven antibodies. Four of these antibodies (3F4, AH6, 6D11 and GE8) recognized an epitope that was chemically modified by the tMAB-NHS and Ac-NHS reagents ( and ). The epitope recognized by the other three antibodies (MAB5424, AG4, and 7D9) was not chemically modified by either reagent (, , and ). The antibodies GE8, 6D11, AG4, 7D9, and MAB5424 were used to probe samples containing both hamster and mouse PrP. The antibodies 3F4 and AH6 were used to probe only hamster PrP or mouse PrP, respectively. In order to minimize secondary antibody artifacts, three goat-anti-mouse IgG secondary antibodies (Fc-specific, Fab-specific, and H+L) were used to probe the same blot. It was determined that the Fc-specific secondary antibody had no observable secondary antibody artifacts (data not shown).
Figure 4. western blots of infected (263K) or uninfected hamster brain extracts (HBH). Samples consist of brain extracts reacted with Ac-NHS, tMAB-NHS, proteinase K (PK) or brain extract controls with no added reagents. Identical amounts of the same reaction mixtures +/− tMAB-NHS, +/− Ac-NHS or PK with either uninfected HBH or 263K-infected HBH (263K) were probed with either the 3F4 (A), GE8 (B), 6D11 (C), AG4 (D), or 7D9 (E) antibodies. A comparison of samples of brain extract controls with no added reagents or samples treated with tMAB or PK (50 mg/mL; 37°C; 1hr) is also shown (F).
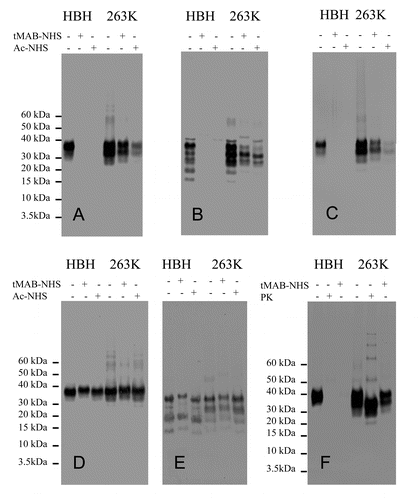
Figure 5. western blots of infected (Me7) or uninfected mouse brain extracts (MBH). Samples consist of brain extracts reacted with Ac-NHS, tMAB-NHS or brain extract controls with no added reagents. Identical amounts of the same reaction mixtures +/− tMAB-NHS or +/− Ac-NHS with either uninfected MBH or Me7-infected MBH (Me7) were probed with either the AH6 (A) GE8 (B), 6D11 (C), MAB5424 (D), AG4 (E), or 7D9 (F) antibodies.
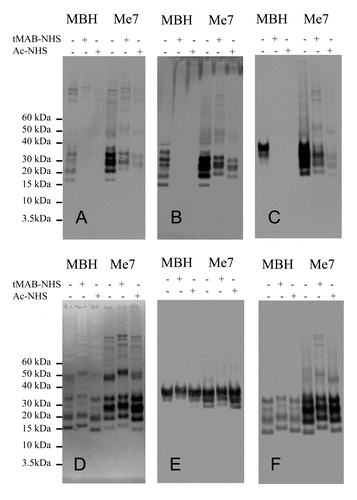
These results indicate that antibodies can be used to detect modified amino acids. The reaction mixtures of uninfected and infected (263K) detergent-solubilized extracts of hamster brain homogenates (HBH) were analyzed by western blot and probed with the antibodies 3F4, GE8, and 6D11 (Fig. Four A-C). In all cases the epitopes recognized by these antibodies are completely covalently modified by tMAB-NHS and Ac-NHS reagents when they are reacted with the uninfected HBH, but only partially modified when reacted with the 263K-infected HBH (Fig. Four A-C). It is also clear this result is not an artifact of the chemical reaction with PrP ( and ). If this were the case, then the western blots probed with antibodies that are not modified by the reagents ((MAB5424, AG4, and 7D9) would show substantial differences (vide infra) and this is not observed. Analogous results are observed with the uninfected and infected (Me7) detergent-solubilized extracts of mouse brain homogenates (MBH) (). Thus, antibodies can be used to detect amino acids that are covalently modified by the tMAB-NHS and Ac-NHS reagents.
This approach does not use proteinase K (PK), so a comparison between PK digestion and chemical modification was performed. Infected (263K) and uninfected HBH samples were reacted with tMAB or proteinase K (PK) (50 mg/mL; 37°C; 1hr). The samples were analyzed by western blot, probed with 3F4 (). The PrPC signal from the uninfected HBH was eliminated by either PK digestion or reaction with tMAB-NHS. The signal for PrPSc was present in samples treated with PK or reacted with tMAB-NHS. The PK treated sample showed the characteristic N-terminally truncated PK-resistant core referred to as PrP27–30. This indicates that these reagents can be used in place of PK to detect the presence of prions by using a western blot probed with an appropriate antibody.
The detergent-solubilized extract of brain homogenates from the 263K-infected hamster brain or the Me7-infected mouse brain is not identical to that of the respective uninfected controls. There may be molecules present in the infected extracts that are not present in the uninfected extracts. These molecules may prevent the reagents from completely reacting with the PrPC present in the sample and, thereby, generate artifactual results. In a separate experiment, the 263K-infected, Me7-infected and uninfected mouse brain extracts were spiked with rSHaPrP to determine if such molecules were present in the infected brain extracts. These results are shown in . The signal from the rSHaPrP is visible in the uninfected mouse brain extract, when probed with GE8. After reaction with either tMAB-NHS or Ac-NHS, the signal disappears (). The signals from the Me7-infected brain extract are more intense than that of the rSHaPrP, so it is not possible to observe a decline in signal intensity (). When the same samples are probed with the 3F4 antibody, there is a clear elimination of the signal from rSHaPrP in both the uninfected mouse brain extract and the Me7-infected mouse brain extract (). The signal from the rSHaPrP spiked into the 263K-infected brain extract is eliminated when the extract is reacted with tMAB-NHS (). This indicates that although the control brain extracts and the infected brain extracts are not identical, they are similar enough that the tMAB-NHS and Ac-NHS reagents react with all of the PrPC present in both the uninfected control and the infected samples of mouse and hamster brain extracts. Thus, the signals observed in the western blots are from epitopes that originated from PrPSc, since they did not react with the reagent.
Figure 6. western blots of infected (Me7 or 263K) or uninfected mouse brain extracts (MBH). Samples consist of brain extracts reacted with Ac-NHS, tMAB-NHS or brain extract controls with no added reagents. MBH and Me7 spiked with Hamster rPrP and then reacted with tMAB-NHS or Ac-NHS or control (no added reagent) and then probed with GE8 (A) or 3F4 (B). 263K-infected hamster brain extract (spiked with Hamster rPrP) +/− tMAB-NHS (C). Me7-infected mouse brain extract control or Me7-infected mouse brain extract reacted with tMAB-NHS or Ac-NHS and deglycosylated with PNGaseF or PNGaseF deglycosylated uninfected mouse brain extract (probed with GE8) (D). 263K-infected hamster brain extract control or 263K-infected hamster brain extract reacted with tMAB-NHS or Ac-NHS and deglycosylated with PNGaseF (probed with 3F4) (E).
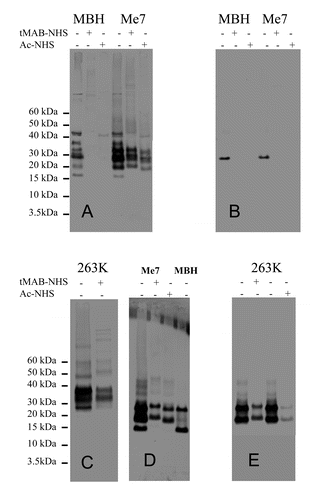
This is further confirmed by close examination of the apparent shift to higher molecular weights as a result of the reaction with tMAB-NHS ( and ). The covalent attachment of the tMAB residue would be expected to increase the molecular weight of the protein and add a positive charge to the protein. This combination of addition of mass and positive charge (cancelling out the negative charges of the some of the SDS molecules) results in this observed gel shift (). A comparison of the same material reacted with an equal amount of Ac-NHS shows no such shift for mouse and hamster samples ( and ). The mouse samples probed with antibodies, MAB5424, AG4, and 7D9, whose epitopes are unaffected by reaction with tMAB-NHS, show this shift in both uninfected and Me7-infected mouse brain extracts (). The hamster samples probed with the antibodies AG4 and 7D9 show analogous shifts ( and ).
These results indicate that the reagents Ac-NHS and tMAB-NHS have different reactivity toward PrPSc. The intensity of the western blot signals is dependent upon the reagent and the location of the epitope. When 263K-infected hamster brain extract is reacted with tMAB-NHS the signal is more intense for the same extract reacted with the Ac-NHS when the blot is probed with 3F4 or 6D11 ( and ). When the same material is probed with the antibody GE8, the signal intensity is similar for 263K-infected extracts reacted with either tMAB-NHS or Ac-NHS (). When the same material is probed with antibodies that do not recognize an epitope that is modified by tMAB-NHS or Ac-NHS (AG4 and 7D9), the signal intensities are comparable ( and ). Analogous results are observed in Me7-infected mouse brain extracts (). This suggests that structural information may be obtained by the use of reagents with different chemical structures and chemical reactivity.
The signals from the western blot indicate that prions have a number of surface exposed functionalities that react with the reagent, which account for the observed gel shifts when the tMAB-NHS reagent is used. Both PrPC and the PrPSc contain a GPI anchor and asparagine-linked sugar antennae.Citation2 In principle the sugar residues in both the sugar antennae and GPI anchor could react with the tMAB-NHS reagent, so PNGase F was used to cleave the asparagine-linked sugar antennae,Citation22,Citation23 in order to determine if the observed gel shifts were due to covalent attachment of the tMAB-NHS reagent to the protein or the sugars comprising the sugar antennae. A western blot of the PNGase F treatment of Me7-infected mouse brain extract control, reacted with tMAB-NHS, or reacted with Ac-NHS and uninfected mouse brain control is shown in . An analogous western blot of PNGase F treated 263K-infected hamster brain extract is shown in . PNGase F treatment removes the sugar antennae, but it does not alter the gel shift, which indicates that the gel shift is a result of the reaction with the protein and not the sugar residues. It is possible that the sugar components of the GPI anchor may react with the tMAB-NHS reagent, but since they contain the same sugars, this is unlikely. The observed gel shift from the PNGase F treated samples is similar to that observed when recombinant hamster PrP is reacted with the tMAB-NHS reagent ().
This approach was evaluated for its ability to distinguish between two strains of hamster adapted scrapie (263K and Dy). HBHs were prepared for both strains and each was reacted with either tMAB-NHS or Ac-NHS or no reagent was added. Western blot analysis was performed and probed with the antibodies 3F4, GE8, 6D11, or AG4 (). The intensity of the signals from both the Dy and the 263K strain reacted with tMAB-NHS are more intense than those reacted with Ac-NHS when the blots were probed with the mAb 6D11 or 3F4. There is a specific difference in signal intensity that is noticeable in the western blot probed with the antibody 6D11, where the signal for the diglycosylated band is present in the Dy sample reacted with Ac-NHS and significantly reduced in the analogous 263K sample ().
Figure 7. western blots of 263K or drowsy (Dy) strain of hamster-adapted scrapie infected hamster brain extracts (HBH). Samples consist of brain extracts reacted with Ac-NHS, tMAB-NHS, or brain extract controls with no added reagents. Identical amounts of the same reaction mixtures +/− tMAB-NHS, +/− Ac-NHS with either 263K or Dy-infected HBH were probed with either the 3F4 (A), GE8 (B), 6D11 (C), or AG4 (D) antibodies.
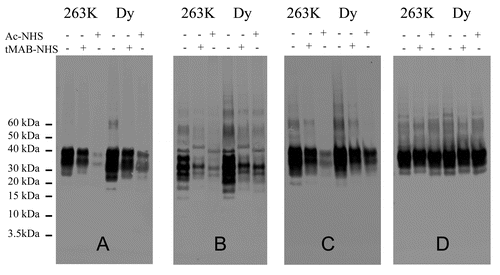
In order to verify this observation, four western blots, analogous to that shown in were prepared and probed with the 6D11 antibody. The intensity of the signals was measured by densitometry. The ratio of the intensity of the diglycosylated band to that of the monoglycosylated band was determined for the Dy and 263K strains treated with nothing, tMAB-NHS or Ac-NHS. The values of this ratio for the 263K strain treated with nothing, tMAB-NHS, or Ac-NHS were determined to be 1.3 ± 0.1, 1.3 ± 0.1, and 0.9 ± 0.1, respectively. The analogous values for the Dy strain were determined to be 1.3 ± 0.1, 1.3 ± 0.1, and 1.3 ± 0.1. A t-test confirmed that there is a statistically significant difference (p > 0.99) between the ratio intensity of diglycosylated band to that of the monoglycosylated band when Dy and 263K are treated with Ac-NHS. Thus, this approach can be used to distinguish between strains without the use of PK.
The intensities of the western blot signals are reduced after the reaction with tMAB-NHS or the Ac-NHS as show in ; . The intensity of the western blot signal remains fairly constant when hamster samples are probed with the antibodies AG4 or 7D9. A similar result is observed when the mouse samples are probed with the antibodies MAB5424, AG4, or 7D9. Since these three antibodies bind to epitopes that are unaffected by tMAB-NHS or the Ac-NHS reagents, the loss of signal is not an artifact of the chemical reaction ( and and ). Since PrPC is present in infected brain extracts, part of this reduction in signal intensity is from the loss of the signal from the PrPC. Another possible contribution to this loss of signal intensity may be due to the reaction of smaller PrPSc multimers, the so-called proteinase K sensitive portion of the mixture.Citation24,Citation25 The differences in signal intensity for those samples reacted with tMAB-NHS or Ac-NHS are due to the structural differences of the reagents’ non-reactive side chains (methyl vs. propyltrimethylammonium), since both possess identical activated NHS ester groups.
Discussion
The results of this small study indicate that PrPC and PrPSc are covalently modified by tMAB-NHS and Ac-NHS reagents to different extents. These modifications can be employed to distinguish PrPSc from PrPC using western blots probed with monoclonal antibodies. This differentiation is accomplished without the use of PK (Figs. Four and 5). When rSHaPrP was spiked into infected mouse (Me7) and hamster (263K) samples and the samples were reacted with tMAB-NHS or Ac-NHS, the signal from rSHaPrP was no longer observed (), indicating that the infected homogenates do not possess molecules that would prevent their complete reaction with the PrPC present in those samples. Furthermore, this approach can be used to distinguish two strains of hamster-adapted scrapie, 263K and Dy, without the use of PK (). Based on the observed gel shifts, a number of additional amino acid residues in both the PrPC and the PrPSc isoforms react with these reagents ( and ), which suggests that if this set of antibodies was enlarged more information on the structural differences between the isoforms could be obtained. This approach can be used to distinguish between PrP isoforms, including prion strains, without the use of PK.
In addition to distinguishing between isoforms this approach can be used to identify the specific amino acid residues that react in a conformation-dependent manner with these reagents (). The epitope for the 3F4 antibody is found in hamster but not mouse PrP.Citation26-Citation28 The lysine present in this epitope (MKHM) is the only amino acid that forms a stable covalent bond with these reagents and its covalent modification would account for the observed signal loss when probed with 3F4 (). The epitope for the AH6 antibody is (QVYYRPVDQYSNQNNF) is found in mouse, but not hamster PrP.Citation14,Citation18 The sole difference between the two regions is the replacement of the serine (S) with an asparagine (N) in hamster. This suggests that either the serine, the adjacent tyrosine or both is/are covalently modified by the reagents, resulting in the loss of signal apparent in the western blot analysis (). The lysine in the unstructured region of the hamster PrP (3F4 epitope) and the tyrosine and/or serine between β-sheet 2 and α-helix B of mouse PrP (AH6 epitope) are not surface exposed in the respective PrPSc isoforms, but are in the respective PrPC isoforms.
The epitopes for the GE8 (IKQHTVTTT) and 6D11 (THNQWNKPSKP) are common to both mouse and hamster PrP.Citation14,Citation16 The covalent modification of the lysine by these reagents is the most likely cause of the signal loss observed in the western blots probed with GE8 ( and ), although covalent modification of one or more of the threonines cannot be ruled out. Again, the covalent modification of one or both of the lysines present is the most likely cause of the signal loss observed in the western blots probed with 6D11 ( and ), although covalent modification of the serine or threonine is possible. These results suggest that the lysine residue near the glycosylation site present in α-helix B (GE8 epitope) and the lysine residues nearest the site of PK cleavage (6D11 epitope) are not surface exposed in the PrPSc isoform, but are in the PrPC isoform in both mouse and hamster PrP. This suggests common structural elements in the mouse-adapted (Me7) and hamster-adapted (263K) forms of scrapie.
The epitopes for the AG4, 7D9, and MAB5424 antibodies have been characterized ().Citation16,Citation17 Although none was empirically determined to have an epitope that reacted with either reagents, they proved to be useful controls (, , and ). These results indicate that the effect of the reagents is a specific chemical modification and not a non-specific destruction of the mouse or hamster PrP.
The intensity of the signals from the western blots is dependent upon the structure of the reagent. tMAB-NHS and Ac-NHS both possess the same activated ester functionality; they differ in terms of charge and steric bulk. Both Me7-infected mouse brain extracts and 263K-infected hamster brain extracts react equivalently with the epitope recognized by the GE8 antibody ( and ). In 263K-infected hamster brain extract, the epitope recognized by the antibodies 3F4 and 6D11 is much more reactive to Ac-NHS than to tMAB-NHS. A similar situation is observed in Me7-infected mouse brain extracts when the western blots are probed with the antibodies AH6 and 6D11. This indicates that the PrPSc conformation, the GE8 epitope has limited availability to either reagent. In contrast, the 3F4, AH6, and 6D11 epitopes are more susceptible to reaction with the Ac-NHS than tMAB-NHS. It is not clear if this is due to the steric bulk or charge differences between these molecules. It does indicate that exploiting steric bulk and charge may provide additional structural information about PrPSc.
The difference in the reduction of the WB signal for the Ac-NHS as opposed to that of tMAB-NHS may be due to changes in the secondary structure of the protein as a result of covalent modification. Reaction with Ac-NHS would covalently convert a polar group (amine or hydroxyl) into a non-polar species. The tMAB-NHS reagent would convert those polar groups to a positively charged species, although the charge would be a further five carbons distant. Such changes could disrupt the secondary structure of PrPSc and, thereby, expose formerly buried amino acid side chains.
The Dy and 263K strains differ in their susceptibility to PK and denaturation by guanidine hydrochloride solutions.Citation3,Citation29-Citation31 PK digestion of Dy yields a PrP 27–30 that has at least 10 fewer N-terminal amino acids than does the PrP 27–30 from the 263K strain.Citation30 Dy is much more readily disaggregated by guanidine hydrochloride, than is the 263K strain.Citation3,Citation31 This suggests that of the two strains, the Dy strain should be more susceptible to changes in the secondary structure induced by the covalent modification of amino acid side chains. When these strains are reacted with Ac-NHS and tMAB-NHS, it is the Dy strain that reacts less with these reagents than does the 263K strain (). This is especially apparent when the reaction mixture is probed with the mAb 6D11 (), whose epitope is nearest the N-terminus of PrP 27–30 (). In contrast, the signal intensities are comparable when the reaction mixtures are probed with the GE8 antibody, whose epitope is contained in α-helix B and far from the N-terminus (). These results suggest that the changes in secondary structure are minimal.
This approach of combining chemical modification with antibody-based detection is a convenient means of analyzing PrPSc. It can be used to identify accessible amino acids, based on antibody recognition. The approach is much simpler than a mass spectrometry-based approach and has been successfully employed to identify a chemical modification that would have been very difficult to detect using mass spectrometry.Citation32 This indicated that this approach is not limited to the NHS activated ester chemistry. It can be used to detect PrPSc in the presence of PrPC without the need to use PK. This approach can be used to distinguish among strains. It provides an additional use for the large number of available anti-PrP antibodies.
Materials and Methods
Chemicals
The 3F4 antibody was obtained from Covance (Princeton, NJ). The MAB5424 antibody was obtained from Millipore Corporation (Billerica, MA). The antibodies AG4, AH6 and GE8 were obtained from the TSE Resource Centre (now in the Roslin Institute, Scotland, UK). The antibodies 6D11 and 7D9 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A secondary antibody, AP-Goat Anti-Mouse IgG (H+L), was obtained from Invitrogen (Carlsbad, CA). The other two secondary antibodies, Anti-Mouse IgG (Fab specific)–AP and Anti-Mouse IgG (Fc specific)–AP (both antibodies produced in goat), were purchased from Sigma-Aldrich (St. Louis, MO). PNGaseF was obtained from New England Biolabs (Ipswich, MA). Proteinase K was acquired from Invitrogen (Carlsbad, CA). The MOPS and MES running buffers and LDS loading buffers were obtained from Invitrogen (Carlsbad, CA). PVDF Membranes (0.2mm pore size) were obtained from Invitrogen (Carlsbad, CA). Novex NuPAGE 4–12% Bis-Tris Gels, either 1.5mm x 15 well or 1.5mm x 10 well were obtained from Invitrogen (Carlsbad, CA). Precise 8–16% protein gels 1mm x 10 wells were obtained from Thermo Scientific (Rockford, IL). StartingBlock T20 (PBS) blocking buffer was obtained from Thermo Scientific (Rockford, IL). HPLC grade water was purchased from Burdick and Jackson (Muskegon, MI). Acetonitrile, HPLC grade, was from Fisher Scientific (Fairlawn, NJ). Trypsin (porcine, sequencing grade, modified) was purchased from Promega (Madison, WI). All other reagents were from Sigma-Aldrich (St. Louis, MO).
Synthesis of the Ac-NHS and tMAB-NHS reagents Citation 33
One gram of dicyclohexylcarbodiimide (4.8 mM) was dissolved in 20 mL of ethyl acetate and allowed to stir at room temperature. 250 mL of acetic acid (4.3 mM) and 0.5 g of N-hydroxysuccinimide (4.3 mM) were added to the stirred solution. The resulting solution was allowed to stir overnight. The white precipitate from the reaction mixture was removed by filtration. The ethyl acetate was removed in vacuo to yield a yellow oil. The oil was dissolved in a small volume of 10% ethyl acetate in hexanes and chromatographed on silica gel to yield a fraction containing the desired product. The solvent was removed in vacuo to yield a white solid. The synthesis of the N-hydroxysuccinimide ester of 4-trimethylamoniumbutyric acid [(3-carboxypropyl)trimethylammonium chloride] (tMAB) has been described previously.Citation34 The structures of these synthetic compounds were verified by NMR. Each reagent was dissolved in DMSO to make a 200 mM stock solution.
Production of recombinant PrP
Recombinant PrP was obtained from plasmids expressing the protein sequence. The plasmids (pET-11a; Merck KGaA; Darmstadt, Germany) expressing the hamster and mouse sequences were a gift from Prof. Dr. Carsten Korth. The plasmids were cloned into BL21 cells (EMD Chemicals, Inc.; Gibbstown, NJ). The molecular weight of each protein was verified by mass spectrometry. All of the recombinant proteins contained an N-terminal methionine.Citation35 The recombinant proteins were purified by standard procedures.
Isolation and purification of recombinant PrP Citation 36
Washed inclusion bodies were isolated according to established procedures.Citation37 The inclusion body pellet was suspended in 1 mL of denaturing buffer (6M guanidine hydrochloride, 100 mM sodium phosphate, 10 mM Tris, pH 8.0) and sonicated for 3 min. After sonication the suspension was centrifuged at 20,000 x g for 5 min to remove any insoluble material. The supernatant was applied to a 1 mL HIS-Select cartridge (Sigma Corporation, Milwaukee, Wisconsin) that had previously been stripped and recharged with copper according to the manufacturer’s instructions. The denatured recombinant protein bound to the cartridge was renatured by the application of a five hour linear gradient (0.04 mL/min) starting with 100% denaturing buffer and ending with 100% refolding buffer (100 mM sodium phosphate, 10 mM Tris, pH 8.0). After the gradient was completed the cartridge was washed for a further hour with refolding buffer at a flow rate of 0.05 mL/min. The refolded protein was eluted with 5 mL of 0.1 M EDTA and immediately dialyzed against 1L of 100 mM ammonium acetate pH 4.5 overnight at room temperature, using a dialysis cassette (7000 MWCO; Thermo Scientific; Rockford, IL). The next day the dialysis buffer was discarded and replaced with 1L of fresh buffer and allowed to dialyze for 2 h. Aliquots were lyophilized and quantitated by BCA protein assay (Thermo Scientific; Rockford, IL). The molecular weight of the proteins predicted by the prnp gene sequences matched that observed by mass spectrometric analysis.
Animal Handling and Obtaining infected brains
LVG Syrian golden hamsters and Swiss CD-1 mice were obtained from commercial sources (Charles River Laboratories; Wilmington, MA). Uninfected hamster and mouse brains were harvested from uninfected animals obtained from commercial sources (Charles River Laboratories; Wilmington, MA).
The 263K ( = Sc237)Citation38,Citation39 and the drowsy (Dy)Citation40 strains of hamster-adapted scrapie were obtained from InPro Biotechnology (South San Francisco, CA) and passaged once through LVG Syrian golden hamsters (Charles River Laboratories, Wilmington, MA). The Me7 strain of mouse-adapted scrapieCitation41 was obtained from InPro Biotechnology (South San Francisco, CA) and passaged once through Swiss CD-1 mice (Charles River Laboratories; Wilmington, MA).
One brain each from an uninfected hamster, a 263K-infected hamster, a Dy-infected hamster, an uninfected mouse, and an Me7-infected mouse was separately homogenized using an Omni GLH general laboratory homogenizer and disposable Omni Tip plastic generator probes in phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4) to make a 10% brain homogenate in a 15 mL plastic tube. An aliquot of each of these five brain homogenates was transferred into a plastic microcentrifuge tube and mixed with a sufficient volume of a 10% (w/v) solution of the detergent β-octylglucopyransoide (BOG) and a sufficient volume of 500mM sodium phosphate solution (pH 7.2) to make a detergent-solubilized brain extract in 2% (w/v) BOG and 20 mM phosphate pH 7.2. Each of the five resulting detergent solutions was centrifuged (20,000 x g, 15 min) at room temperature to clarify the solutions and yield the respective detergent-solubilized brain extracts. These five clarified detergent-solubilized brain extracts were used for the described reactions.
Reaction of the brain extracts with Ac-HNS or tMAB-NHS reagents
The five clarified brain extracts were used in the reactions described below. Three 270 μL aliquots of each of the brain extracts were placed in 15 separate microcentrifuge tubes. Thirty μL of the tMAB-NHS reagent stock was added to one tube from each of the five clarified extracts and mixed. The same amount of the Ac-NHS stock solution was added to another tube from each of the five clarified extracts and mixed. The last of the three aliquots was left as a control. The reaction was allowed to proceed for 15 min at room temperature. After 15 min, 30 μL of a 1 M Tris solution (5 equivalents) was added to each of the 15 tubes and mixed. These solutions were allowed to stand for 15 min at room temperature.
In addition to the previously described reactions, separate samples of the Me7-infected or 263K-infected clarified brain extracts were spiked with mouse or hamster recombinant PrP, respectively. Two aliquots of 270 μL of the Me7-infected mouse or 263K-infected clarified brain extracts were transferred to four microcentrifuge tubes. One microliter of a solution containing either mouse or hamster recombinant PrP (~600 ng) was added to the two Me7-infected mouse or two 263K-infected hamster samples, respectively. Thirty μL of the tMAB-NHS reagent was added to one of the recombinant spiked samples of Me7-infected mouse and 263K-infected hamster clarified brain extracts. The reaction was allowed to proceed for 15 min at room temperature. After 15 min, 30 μL of 1M Tris was added and allowed to react for 15 min.
The solutions derived from the 263K-infected and Dy-infected clarified hamster brain extracts and Me7-infected clarified mouse brain extracts (spiked and unspiked) were denatured by addition of a sufficient amount of 8M guanidine hydrochloride to achieve a final concentration of 6M.Citation42 The resulting solution was allowed to stand for 24 h at room temperature.
Protein was precipitated with ice-cold methanol (85% methanol to 15% protein solution) in microcentrifuge tubes. The tubes were centrifuged (20,000 x g for 20 min) in a chilled rotor (-9°C) with an Eppendorf Model 5417R centrifuge (Hamburg, Germany). The methanol supernatant was discarded and the pellet washed with methanol and centrifuged again under the same conditions. The methanol supernatant was discarded and the pellet was allowed to air dry for 15 min. The pellet was dissolved in 240 μL of 2x LDS buffer (Invitrogen; Carlsbad, CA) and put in boiling water for 10 min. These solutions in LDS were used for western blotting.
Mass spectrometry
Whole protein mass spectrometry (NanoLC/ESI/MS) was performed with an Applied Biosystems (ABI/MDS SCIEX; Toronto, Canada) model QStar Pulsar equipped with a Proxeon Biosystems (Odense, Denmark) nanoelectrospray source. The instrument was equipped with a NanoLC system (Dionex; Sunnyvale, CA) that includes an autosampler, column switching device, loading pump, and nanoflow solvent delivery system. The signals were processed with the Bayesian protein reconstruct feature of the software.
Ten μg of recombinant Syrian hamster PrP (rSHaPrP) was dissolved in 90 μL of reaction buffer (2% w/v β-octylglucopyranoside, 20 mM sodium phosphate pH 7.2, in water). The solution was split into two portions; one portion was reacted with the Ac-NHS reagent and the other with the tMAB-NHS reagent (vide supra). After quenching, the two samples were precipitated with methanol (vide supra). The sample reacted with the Ac-NHS reagent was analyzed by mass spectrometry. The sample reacted with tMAB-NHS or Ac-NHS was analyzed by SDS-PAGE (Precise 8–16% protein gels 1mm x 10 wells) and stained with Coomassie brilliant blue.
Densitometry
Densitometry measurements were made using the AlphaEaseFC version 4.0.0 software (Alpha Innotec Corporation, San Leandro, CA) with black having a value of 255 and white having a value of 0. The statistical analysis was performed with Microsoft Excel.
Western blots
The protein gels (NuPAGE 1.5 mm thick 10 or 15 well) were purchased from commercial vendors and used according to manufacturer’s instructions. The gels, thick blotting paper and PVDF membrane were soaked in transfer buffer (25 mM Tris, 250 mM glycine in 15% methanol) for 10 min prior to using the BIO-RAD Trans-Blot SD semi-dry transfer cell (Hercules, CA) for 50 min at 100 mA. After the transfer was complete, the PVDF membrane was placed into a square Petri dish (10 x 10 cm) and gently agitated in 20 mL of wash buffer (PBS + 0.05 Tween 20) for 5 min. After 5 min the wash buffer was discarded and the membrane was blocked for 15 min in 10 mL of StartingBlock T20 with gentle agitation. After 15 min, one microliter of the primary antibody (3F4, AH6, GE8 or MAB5424; concentration 1–2 mg/mL) was added to 10 mL of blocking buffer and incubated with the membrane for 1 h at room temperature. After 1 h the blocking buffer containing the primary antibody was discarded and 20 mL of wash buffer was added and gently agitated for 5 min. The washing buffer was discarded and three additional wash steps were performed. After the four wash steps were completed, 10 mL of blocking buffer containing 10 μL of the secondary antibody, AP-Goat Anti-Mouse IgG (H+L), was added. The membrane was incubated at room temperature for 45 min. After 45 min the membrane was washed four times at five minute intervals.
After the washing was complete the blot was placed in 10 mL of a Sigma/FAST BCIP/NBT solution (1 tablet per 10mL per manufacturer’s instructions) and incubated at 37°C. After development, the blots were dried and scanned.
| Abbreviations: | ||
| TSE | = | transmissible spongiform encephalopathy |
| NHS | = | N-hydroxysuccinimide |
| tMAB | = | 4-trimethylamoniumbutyric acid |
| PVDF | = | Polyvinylidene fluoride |
| MOPS | = | 3(N-morpholino) propanesulfonic acid |
| MES | = | 2-(N-morpholino) ethanesulfonic acid |
| DMSO | = | dimethylsulfoxide |
| PK | = | proteinase K |
| PrP | = | prion protein |
| PrPSc | = | prion protein scrapie isoform |
| PrPC | = | prion protein normal isoform |
| BOG | = | β-octylglucopyranoside |
| MWCO | = | molecular weight cut-off |
| Tris | = | tris(hydroxymethyl)aminomethane |
| LVG | = | Lakeview Golden |
| tMAB-NHS | = | NHS ester of tMAB |
| Ac-NHS | = | NHS ester of acetic acid |
| kDa | = | kilodalton |
| GPI | = | glycophosphatidylinositol |
| SDS-PAGE | = | sodiumdodecyl sulfate-polyacrylamide gel electrophoresis |
| PNGase F | = | Peptide N-Glycosidase F |
| HIS | = | histidine |
| DTT | = | dithiothreitol |
| CV | = | coefficient of variation |
| Da | = | dalton |
| MRM | = | multiple reaction monitoring |
Acknowledgments
The author wishes to acknowledge the technical assistance of Ms. Kirby L. Keller. The author wishes to acknowledge the assistance of Ms. Melissa L. Erickson in preparing this manuscript. The US. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, age, disability, and where applicable, sex, marital status, familial status, parental status, religion, sexual orientation, genetic information, political beliefs, reprisal, or because all or part of an individual's income is derived from any public assistance program. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA's TARGET Center at (202) 720–2600 (voice and TDD). To file a reportt discrimination, write to USDA, Director, Office of Civil Rights, 1400 Independence Avenue, S.W., Washington, DC 20250–9410, or call (800) 795–3272 (voice) or (202) 720–6382 (TDD). USDA is an equal opportunity provider and employer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363 - 83; http://dx.doi.org/10.1073/pnas.95.23.13363; PMID: 9811807
- Stahl N, Baldwin M, Teplow DB, Hood LE, Beavis R, Chait B, et al. Cataloging post-translational modifications of the scrapie prion protein by mass spectrometry. In: Prusiner SB, Collinge J, Powell J, Anderton B, eds. Prion diseases of humans and animals. New York: Ellis Horwood, 1992:361-79.
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157 - 65; http://dx.doi.org/10.1038/2654; PMID: 9771749
- Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry 1991; 30:7672 - 80; http://dx.doi.org/10.1021/bi00245a003; PMID: 1678278
- Onisko BC, Silva CJ, Requena JR. Methods to differentiate protein conformers Application number 20060110785. 2006.
- Carter JM, Dynin I, Erickson ML, Onisko BC, Requena JR, Silva CJP. 1.8 Conformation-dependent covalent modification of PrP. Prion2009 September 23-25, 2009 Porto Carras, Chalkidiki Greece Book of Abstracts, 2009:54.
- Lehto MT, Peery HE, Cashman NR. Current and future molecular diagnostics for prion diseases. Expert Rev Mol Diagn 2006; 6:597 - 611; http://dx.doi.org/10.1586/14737159.6.4.597; PMID: 16824033
- Williamson RA, Peretz D, Pinilla C, Ball H, Bastidas RB, Rozenshteyn R, et al. Mapping the prion protein using recombinant antibodies. J Virol 1998; 72:9413 - 8; PMID: 9765500
- Miller BT, Collins TJ, Rogers ME, Kurosky A. Peptide biotinylation with amine-reactive esters: differential side chain reactivity. Peptides 1997; 18:1585 - 95; http://dx.doi.org/10.1016/S0196-9781(97)00225-8; PMID: 9437720
- Anjaneyulu PS, Staros JV. Reactions of N-hydroxysulfosuccinimide active esters. Int J Pept Protein Res 1987; 30:117 - 24; http://dx.doi.org/10.1111/j.1399-3011.1987.tb03319.x; PMID: 3667072
- Onisko B, Fernández EG, Freire ML, Schwarz A, Baier M, Camiña F, et al. Probing PrPSc structure using chemical cross-linking and mass spectrometry: evidence of the proximity of Gly90 amino termini in the PrP 27-30 aggregate. Biochemistry 2005; 44:10100 - 9; http://dx.doi.org/10.1021/bi0501582; PMID: 16042387
- Pimenova T, Nazabal A, Roschitzki B, Seebacher J, Rinner O, Zenobi R. Epitope mapping on bovine prion protein using chemical cross-linking and mass spectrometry. J Mass Spectrom 2008; 43:185 - 95; http://dx.doi.org/10.1002/jms.1280; PMID: 17924399
- Kouassi GK, Irudayaraj J. A nanoparticle-based immobilization assay for prion-kinetics study. J Nanobiotechnology 2006; 4:8; http://dx.doi.org/10.1186/1477-3155-4-8; PMID: 16916458
- Bibby DF, Gill AC, Kirby L, Farquhar CF, Bruce ME, Garson JA. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J Virol Methods 2008; 151:107 - 15; http://dx.doi.org/10.1016/j.jviromet.2008.03.013; PMID: 18433888
- Kascsak RJ, Rubenstein R, Merz PA, Tonna-DeMasi M, Fersko R, Carp RI, et al. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol 1987; 61:3688 - 93; PMID: 2446004
- Laurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature 2009; 457:1128 - 32; http://dx.doi.org/10.1038/nature07761; PMID: 19242475
- Maddison BC, Whitelam GC, Gough KC. Cellular prion protein in ovine milk. Biochem Biophys Res Commun 2007; 353:195 - 9; http://dx.doi.org/10.1016/j.bbrc.2006.12.006; PMID: 17174270
- Novitskaya V, Makarava N, Bellon A, Bocharova OV, Bronstein IB, Williamson RA, et al. Probing the conformation of the prion protein within a single amyloid fibril using a novel immunoconformational assay. J Biol Chem 2006; 281:15536 - 45; http://dx.doi.org/10.1074/jbc.M601349200; PMID: 16569635
- Spinner DS, Kascsak RB, Lafauci G, Meeker HC, Ye X, Flory MJ, et al. CpG oligodeoxynucleotide-enhanced humoral immune response and production of antibodies to prion protein PrPSc in mice immunized with 139A scrapie-associated fibrils. J Leukoc Biol 2007; 81:1374 - 85; http://dx.doi.org/10.1189/jlb.1106665; PMID: 17379700
- Dumpitak C. Untersuchungen zu Struktur und Funktion von Polysacchariden und alterungsassoziierten Proteinmodifikationen bei Prionen. Dissertation. Düsseldorf: Heinrich-Heine-Universität, 2003.
- Prusiner SB. Prion Biology and Diseases. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 2004.
- Plummer TH Jr., Elder JH, Alexander S, Phelan AW, Tarentino AL. Demonstration of peptide:N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J Biol Chem 1984; 259:10700 - 4; PMID: 6206060
- Maley F, Trimble RB, Tarentino AL, Plummer TH Jr.. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem 1989; 180:195 - 204; http://dx.doi.org/10.1016/0003-2697(89)90115-2; PMID: 2510544
- Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, et al. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 2002; 41:12868 - 75; http://dx.doi.org/10.1021/bi025958g; PMID: 12379130
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature 2005; 437:257 - 61; http://dx.doi.org/10.1038/nature03989; PMID: 16148934
- Zou WQ, Langeveld J, Xiao X, Chen S, McGeer PL, Yuan J, et al. PrP conformational transitions alter species preference of a PrP-specific antibody. J Biol Chem 2010; 285:13874 - 84; http://dx.doi.org/10.1074/jbc.M109.088831; PMID: 20194495
- Kascsak R. Reiterating the epitope specificity of prion-specific mAb 3F4. J Biol Chem 2010; 285:le5, author reply le6.
- Bolton DC, Seligman SJ, Bablanian G, Windsor D, Scala LJ, Kim KS, et al. Molecular location of a species-specific epitope on the hamster scrapie agent protein. J Virol 1991; 65:3667 - 75; PMID: 1710287
- Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 1992; 66:2096 - 101; PMID: 1347795
- Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol 1994; 68:7859 - 68; PMID: 7966576
- Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, et al. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci 2001; 10:854 - 63; http://dx.doi.org/10.1110/ps.39201; PMID: 11274476
- Gong B, Ramos A, Vázquez-Fernández E, Silva CJ, Alonso J, Liu Z, et al. Probing structural differences between PrP(C) and PrP(Sc) by surface nitration and acetylation: evidence of conformational change in the C-terminus. Biochemistry 2011; 50:4963 - 72; http://dx.doi.org/10.1021/bi102073j; PMID: 21526750
- Neises B, Steglich W. Simple Method for the Esterification of Carboxylic Acids. Angew Chem Int Ed Engl 1978; 17:522 - 4; http://dx.doi.org/10.1002/anie.197805221
- Che FY, Fricker LD. Quantitative peptidomics of mouse pituitary: comparison of different stable isotopic tags. J Mass Spectrom 2005; 40:238 - 49; http://dx.doi.org/10.1002/jms.743; PMID: 15706629
- Flinta C, Persson B, Jörnvall H, von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem 1986; 154:193 - 6; http://dx.doi.org/10.1111/j.1432-1033.1986.tb09378.x; PMID: 3080313
- Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods 2007; 4:645 - 50; http://dx.doi.org/10.1038/nmeth1066; PMID: 17643109
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1989.
- Kimberlin RH, Walker C. Characteristics of a short incubation model of scrapie in the golden hamster. J Gen Virol 1977; 34:295 - 304; http://dx.doi.org/10.1099/0022-1317-34-2-295; PMID: 402439
- Marsh RF, Kimberlin RH. Comparison of scrapie and transmissible mink encephalopathy in hamsters. II. Clinical signs, pathology, and pathogenesis. J Infect Dis 1975; 131:104 - 10; http://dx.doi.org/10.1093/infdis/131.2.104; PMID: 1167884
- Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol 1992; 73:329 - 34; http://dx.doi.org/10.1099/0022-1317-73-2-329; PMID: 1531675
- Zlotnik I, Rennie JC. Further observations on the experimental transmission of scrapie from sheep and goats to laboratory mice. J Comp Pathol 1963; 73:150 - 62; PMID: 14003830
- Prusiner SB, Groth D, Serban A, Stahl N, Gabizon R. Attempts to restore scrapie prion infectivity after exposure to protein denaturants. Proc Natl Acad Sci U S A 1993; 90:2793 - 7; http://dx.doi.org/10.1073/pnas.90.7.2793; PMID: 8464892