Abstract
The oral route is considered to be the main entry site of several transmissible spongiform encephalopathies or prion diseases of animals and man. Following natural and experimental oral exposure to scrapie, sheep first accumulate disease associated prion protein (PrPd) in Peyer’s patch (PP) lymphoid follicles. In this study, recombinant ovine prion protein (rPrP) was inoculated into gut loops of young lambs and the transportation across the intestinal wall studied. In particular, the immunohistochemical phenotypes of cells bearing the inoculated prion protein were investigated. The rPrP was shown to be transported across the villi of the gut, into the lacteals and submucosal lymphatics, mimicking the transport route of PrPd from scrapie brain inoculum observed in a previous intestinal loop experiment. The cells bearing the inoculated rPrP were mainly mononuclear cells, and multicolor immunofluorescence procedures were used to show that the rPrP bearing cells were professional antigen presenting cells expressing Major histocompatibility complex II (MHCII). In addition, the rPrP bearing cells labeled with CD205, CD11b and the macrophage marker CD68, and not with the dendritic cell markers CD11c and CD209. Others have reported that cells expressing CD205 and CD11b in the absence of CD11c have been shown to induce T cell tolerance or regulatory T cells. Based on this association, it was speculated that the rPrP and by extension PrPd and scrapie infective material may exploit the physiological process of macromolecular uptake across the gut, and that this route of entry may have implications for immune surveillance.
Introduction
The transmissible spongiform encephalopathies (TSEs) are caused by prions composed of an infectious conformer of the host-coded prion protein. The conformational differences between infectious and non-infectious disease associated forms of prion protein are not fully described. The natural route of infection for TSEs is widely accepted to be the alimentary tract. The initial investigations of the pathogenesis of scrapie in sheep recognized that the early presence of infectivity in gut tissues was evidence for an oral route of infection.Citation1 The accumulation of the disease-associated forms of prion protein (PrPd) as revealed by immunohistochemistry in gut-associated lymphoid tissues in many natural and experimental TSEs,Citation2-Citation7 suggest that Peyer’s patches (PPs) are primary sites of amplification of the scrapie agent. The detailed study of PP lymphoid follicles during the early phase of TSE infections revealed the presence of PrPd on follicular dendritic cellsCitation8 and in tingible body macrophages.Citation9-Citation11 These findings have been cited as indirect evidence that the scrapie agent is transported across the follicle-associated epithelium (FAE), possibly through M cells.Citation12
Multiple pathways exist for the uptake of foreign material from the gut lumen. Macromolecules can be taken up across the absorptive epithelium (AE) of villi,Citation13-Citation16 by dendritic cells (DCs) extending dendrites into the intestinal lumen to sample gut content,Citation17 or by M cells.Citation18 M cells are known to deliver samples of foreign material through active transepithelial vesicular transport to intraepithelial lymphoid cells and to the subjacent organized mucosal lymphoid tissues and this function has been exploited by various pathogens to gain entry to the intestinal mucosa.Citation18,Citation19 The involvement of M cells in the uptake of the infectious scrapie agent is supported by in vitro studies demonstrating that transepithelial transport of scrapie prions occurred in cultured M cells and not in Caco-2 cultures without M cells.Citation20 However, M cell independent pathways have also been suggested.Citation21 The transportation of infection associated PrP in complex with ferritin has been found to occur across Caco-2 epithelial cells in vesicular structures.Citation22 Moreover, Jeffrey et al.Citation23 showed, in an in vivo model, that PrPd positive material was transported across intestinal villi, into villi lacteals and submucosal lymphatics and not through the FAE, dome and into the PP follicles.
There is very little information available about the fate of specific antigens and pathogens immediately after intestinal uptake. Migratory bone-marrow derived DCs are believed to function as sentinels at mucosal surfaces where they encounter both infectious agents and innocuous antigens derived from food components and the normal microflora. The present understanding is that DCs enter the intestinal wall from the blood stream and form a dense layer just beneath the AE in the villi or beneath the FAE of the domed follicle, in close contact with the M cells.Citation24,Citation25 So positioned, DCs acquire antigens that have been transcytosed from the gut lumen and deliver these antigens to lymphoid tissues to initiate appropriate immune responses. Åkesson et al.Citation26 surveyed the distribution of DC markers on cell populations in the intestinal mucosa of sheep. In an immunohistochemical study, DC population in sheep intestine were shown to co-express CD11c, CD205 and MHCII. The triple labeled DC population was often strategically located in the lamina propria underneath the epithelium and in the sub-FAE dome. Åkesson and coworkers speculated that this DC population was involved in sampling antigens that cross the intestinal mucosa of sheep. Experimental studies in mice have shown that DCs in intestinal lymph can acquire infection associated PrP and participate in the dissemination of scrapie infectionCitation27 but the association of PrPd with DCs in the intestinal mucosa of experimental animals or domestic ruminants is less well documented.
A limitation to the characterization of cell populations bearing PrPd has been the conflict between the harsh treatment of tissues used in conventional immunohistochemical techniques for the detection of PrPd and the inability of many cell marker epitopes to withstand the immunohistochemical protocol used for PrPd-detection. In prion diseases, the normal host coded cellular prion protein is converted into an abnormal, detergent insoluble, variably proteinase-resistant isoform, which often co-purifies with infectivity.Citation28 The production of recombinant proteins in microorganisms enables the creation of species-specific prion protein that is better suited to fluorescent labeling than crude brain homogenates from sheep with clinical scrapie.
To study the cell types that acquire prion protein in the intestinal mucosa, we used recombinant E. coli synthesized inclusion bodies of ovine prion protein (rPrP) as a biologically safe surrogate marker for PrPd. By using the in vivo intestinal loop model, we undertook to investigate the route of uptake of rPrP and to assess the relevance of this recombinant protein by comparing the transport of rPrP with the transport of PrPd in similar experiments where intestinal loops were exposed to scrapie brain homogenates from naturally infected sheep.Citation23 The rPrP was labeled with the fluorescent marker Texas red prior to inoculation, and the transportation of the inoculum followed using a fluorescence and/or confocal microscope. Labeling of cells in combination with Texas red labeled rPrP enabled the phenotype of the cells bearing the rPrP to be investigated.
Results
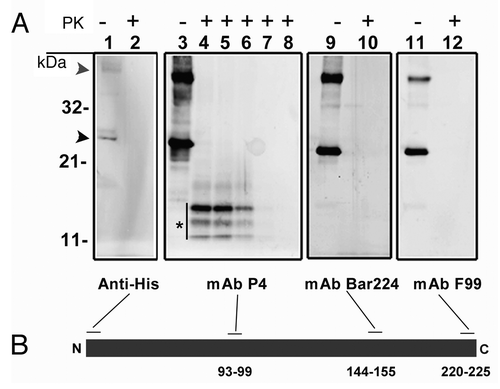
Western blot analysis of rPrP, with antibodies covering the termini of the protein showed that the recombinant protein was full length and intact. Treatment with proteinase K revealed that the aggregates of rPrP used in this study were generally proteinase sensitive. However, as shown in , a triplet of bands detected only with the P4 antibody, which binds to a centrally located epitope in PrP (aa 93–99), sustained proteinase K treatment for about 5 min. These weakly proteinase K resistant protein bands, of which the heaviest was dominant, covered apparent masses of 11 to 14 kDa, suggesting peptide sizes of about 100 to 130 amino acids. Since these bands were neither detected with BAR 224, which binds to aa 144–155, nor with anti-His (N-terminal tag), it can be inferred that proteinase K-digestion has occurred from both ends of the protein, sparing fragments spanning a larger part of the N-terminal and central domain. Further epitope-mapping of these fragments was not pursued.
Figure 1. (A) western blot analysis of recombinant ovine PrP (rPrP), derived from bacterial inclusion bodies, with regard to proteinase sensitivity. A panel of four mAbs was used to map the N- and C-terminal ends (Anti-His and F99 respectively), the central (P4) and globular (BAR 224) domains. As shown in lanes 1, 3, 9 and 11, which were not treated with proteinase K, rPrP was detected by all mAbs with prominent dimeric (gray arrowhead) and monomeric (black arrowhead) forms. In lanes 2, 4, 10 and 12, samples were treated with proteinase K (5 µg/ml) at 22°C for 30 sec, which resulted in complete loss of signal from all mAbs except P4 which revealed a distinct triplet of bands (*), in the range of 11–14 kDa. In lanes 5, 6, 7 and 8 Proteinase K digestion continued for 1, 2, 5 and 10 min respectively. A rapid decline of signal was evident with no signal left after 10 min of Proteinase K treatment. (B) Schematic representation of rPrP with binding-sites for mAbs. Numbers refer to mAb epitopes in ovine PrP.
Light microscopic investigation of hematoxylin and eosin-stained formalin fixed intestinal tissues showed no differences between the rPrP inoculated loops and the controls (not shown). Irrespective of the inoculum, mild oedematous changes in the lamina propria were occasionally observed, as was a mild dilation of the lacteals and submucosal lymphatics in some of the loops. Both AE and FAE showed normal morphology. There were sparse to moderate amounts of polymorphonuclear and mononuclear leukocytes observed in the gut tissues, but there were no differences in the presence of these cells between the intestinal loops within the same animal. Variation between animals in the number of cells in the intestinal mucosa was therefore interpreted as nonspecific and not a result of the inoculated material (not shown).
By immunohistochemistry, the PrP-antibodies L42, F99, BAR224, and R145 were used to detect the inoculated rPrP. Labeling was not observed in any intestinal compartment in any of the intestinal control loops inoculated with NaCl or phosphate buffered saline (PBS) (not shown), hence the immunohistochemical labeling detected in the loops injected with rPrP was interpreted as labeling of the injected material. Serial sections labeled with the different PrP-antibodies showed the same pattern of labeling, although there was some variation in intensity.
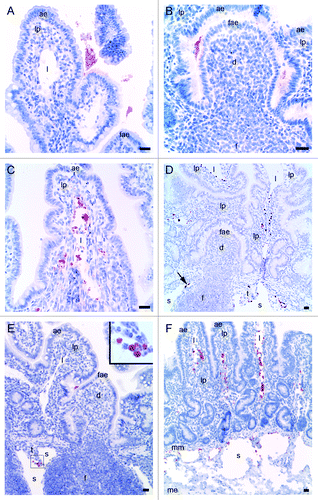
Eleven animals were given Texas red labeled rPrP in one or more intestinal loops for 10, 30 or 60 min (for a total number of six, seven and six loops respectively) (). With immunoperoxidase histochemical techniques, using the antibodies against rPrP, labeling was demonstrated in various amounts in the different loops. Four loops were completely negative in all intestinal compartments (F102 60 min, F261 30 and 60 min, F262 10 min). In the rest of the loops, irrespective of the time points, the presence of rPrP-labeling was observed in a sparse to large amount in the lumen, both covering the AE of villi and the FAE of the dome overlying the PP follicles (). Although rPrP-labeling was found on luminal material, covering the surface of FAE, no immunolabelling was detected in FAE, domes or PP follicles at any of the time points studied. In the intestinal tissues of the loops inoculated with rPrP for ten minutes, two animals showed a moderate to large amount of rPrP-labeling in villi lacteals and interfollicular T-cell area, in addition to a sparse amount in lamina propria and submucosal lymphatics. The other four animals in this ten minute group showed only labeling in the lumen. Of the seven loops exposed to rPrP for 30 min, six animals showed sparse to moderate labeling in lamina propria (). Three of these six loops also showed a moderate to large amount of labeling in the lacteals (), interfollicular T-cell area and submucosal lymphatics (). Of the loops exposed for 60 min, one of six loops showed rPrP-labeling in interfollicular T-cell area and submucosal lymphatics (). In addition, rPrP was present in villi lacteals and lymphatics in intestinal tissue without PP follicles ().
Table 1. Immunohistochemical labeling for rPrP in intestinal compartments at different time points
Figure 2. Light microscopical studies of rPrP immunohistochemical labeling (red staining) in intestinal loops of lambs 10, 30 and 60 min after inoculation. (A) rPrP labeling was detected in the lumen in close association with the apex of villous AE (07/F103 10 min BAR224), (B) and FAE of the domed PP follicle (07/F262 30 min F99). (C) rPrP labeling was observed in the lamina propria of the villi and was frequently present in the lacteals of the villi (09/F607 30 min BAR224). (D) A lower magnification shows rPrP labeled material in the villi lacteals, lamina propria, T-cell area and the submucosal lymphatics (arrow) between the PP follicles (09/607 30 min BAR 224). (E) rPrP labeling in the lumen in close contact with the FAE, and in cells in the submucosal lymphatic area (insert, magnification rPrP-positive cells) (07/F102 60 min BAR 224). (F) rPrP-labeling was also observed in intestinal tissues without PPs (07/F607 30 min BAR 224). Note that rPrP labeling was never observed in the FAE, dome or PP follicles (B, D and E). Scale bars, 50 µm, (ae) AE, (fae) FAE, (lp) lamina propria, (l) lacteal, (d) dome, (f) follicle, (t) T-cell area, (s) submucosal lymphatics, (mm) muscularis mucosa, (me) muscularis externa.
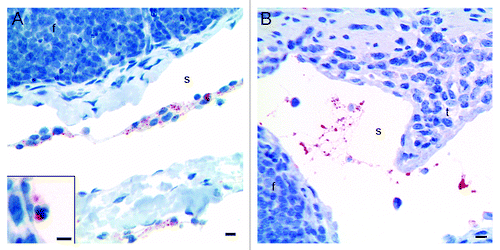
The rPrP-labeling was present in association with cells () in the villi lacteals, interfollicular T cell areas, as well as in submucosal lymphatics, but rPrP-labeling was also observed on aggregates of non-cellular material (). The nuclei of the cells bearing rPrP were often obscured by the abundant rPrP-labeling, but when observable, most nuclei were round to oval (). Occasionally polymorphonuclear cells showed rPrP-labeling (, inset).
Figure 3. Immunohistochemical labeling with the PrP-antibody BAR224 showed that the inoculated rPrP was present both in cells (A) and cell-free (B) in the submucosal lymphatics. (A) The rPrP was mainly in mononuclear cells (07/F103 30 min BAR224) but also in a few polymorphonuclear cells (A, inset. 09/F607 30 min BAR224). (B) The rPrP was present in cell-free material in submucosal lymphatics (07/F289 30 min BAR224). Scale bars, 10 µm.
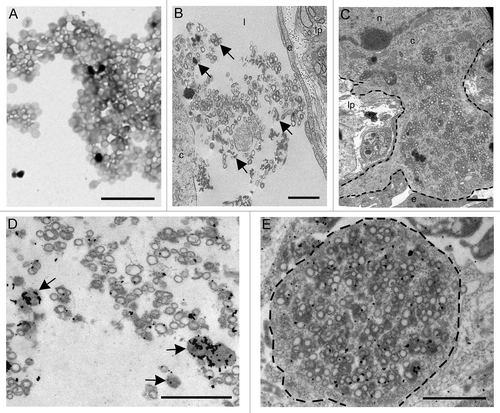
Both unlabelled and Texas red conjugated rPrP was studied by electron microscopy prior to intestinal inoculation. The rPrP preparation contained small and amorphous electron dense aggregates in addition to circular and ovoidal membrane profiles of unconfirmed origins (). After intestinal inoculation, amorphous electron dense aggregates and circular membrane profiles similar to the structures originally identified in the rPrP preparations was found as cell free aggregates within the lacteals () and within the cytoplasm of mononuclear cells in the lamina propria (). Immunogold labeling showed that rPrP was located in lacteals and inside mononuclear cells in association with amorphous electron dense aggregates and circular membrane profiles (). These findings showed that the mucosa could absorb both molecular rPrP and larger macromolecular structures within the inoculum.
Figure 4. (A) Electron microscopy on the preparation containing rPrP demonstrated circular membrane profiles of unconfirmed origins in addition to electron dense amorphous material. (B and C) After inoculation into intestinal loops, electron dense amorphous material and circular structures were observed non-cell-associated in the lacteals (arrows) (B) and in the cytoplasm of a mononuclear cell (delineated) (C). Immunogold labeling showed rPrP labeling of inconspicuous electron dense amorphous aggregates (arrows) and on membranes of circular profiles in a lacteal (D) and in the cytoplasm of a mononuclear cell (delineated) (E). Scale bars, 1 µm. (l) lacteal, (lp) lamina propria, (e) endothelial cell, (c) cytoplasm, (n) nucleus.
By using the Leica DM RXA light microscope equipped with fluorescence filters, rPrP was shown to be bound to Texas red. Light and fluorescence microscopy were used to evaluate co-localization and by switching between the rPrP-labeling visualized in the normal light microscope and the Texas red fluorescence visualized by using fluorescence excitation light 594, the immunohistochemical rPrP labeling and the Texas red fluorescence signal were shown to be co-localized (). To demonstrate that the transportation of rPrP was independent of the Texas red labeling, immunohistochemical detection of rPrP with PrP-antibodies in sections from loops given rPrP without Texas red showed that unlabelled rPrP was localized in the same compartments as Texas red labeled rPrP. In tissues from loops inoculated with rPrP without Texas red labeling, the rPrP was negative in all fluorescence excitation lights ().
Figure 5. Immunohistochemical labeling for rPrP and Texas red fluorescence were co-localized. (A) rPrP labeling (red color) was observed in lamina propria and lacteals of a villus (09/F607 30 min BAR224). (B) The same section as shown in (A) viewed with a fluorescence microscope demonstrated that Texas red fluorescence co-localized with the immunohistochemical rPrP-labeling (arrows in both panels) (Texas red immunofluorescence filter 594). (C) immunoperoxidase labeling for rPrP without Texas red labeling in lamina propria of a villus (09/F610 30 min R145). (D) The same section as in (C) demonstrating rPrP without Texas red labeling is negative when viewed with fluorescence filter 594. Scale bar, 10 µm. (ae) AE, (lp) lamina propria, (l) lacteal, the basal lamina between AE and lamina propria is delineated.
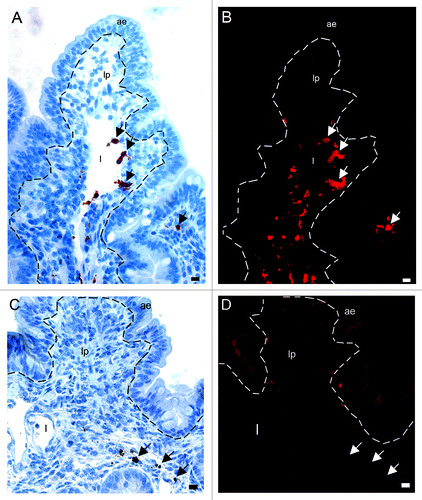
As with the immunoperoxidase and electron microscopy studies, Texas red fluorescence was observed both cell-bound and free in the villi, lamina propria, lacteals, interfollicular T-cell areas and submucosal lymphatics. In addition, sparse puncta of fluorescence were observed in the AE. No rPrP-labeling was found in the FAE, domes or PP follicles.
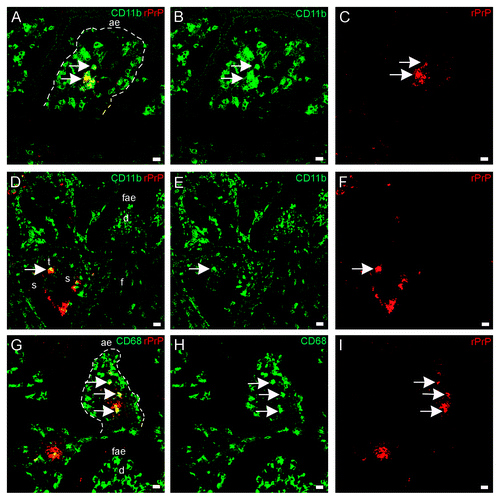
To be able to distinguish the cell phenotypes bearing rPrP, we used tissue sections from loops inoculated with the fluorescent Texas red labeled rPrP for 30 min (F103 and F607), and co-labeled tissue sections with antibodies against various cell markers (Major histocompatibility complex II (MHCII), CD205, CD11c, CD11b, CD209, CD68, CD21, CD3, see ). There were many CD11c+/CD205+ cells, which were mainly present in lamina propria and dome, and these cells did not show a fluorescence signal for rPrP (). Many CD11c + cells were observed in lamina propria and in dome, and these cells were not associated with rPrP (). In addition, the DC marker DC-sign/CD209 was found to label cells in lamina propria and interfollicular T-cell areas, but was not found to co-localize with rPrP (). CD205+/rPrP+ cells were observed in the villi lacteals (), interfollicular T-cell areas, and submucosal lymphatics (). The antigen presenting cell marker MHCII labeled cells that had acquired rPrP and were observed in villi lacteals () and submucosal lymphatics (not shown). CD11b+/rPrP+ cells were present in lacteals (), interfollicular T-cell areas (), and submucosal lymphatics. Double labeled CD68+/rPrP+ cells were observed in lamina propria and villi lacteals (). CD3, the pan T-cell marker, labeled cells in AE, lamina propria, FAE, dome, interfollicular T-cell area, follicle capsule and cells in submucosal lymphatics. Even though rPrP bearing cells and CD3+ T cells could be observed in the same areas, and often in close contact, the rPrP+ cells did not show labeling for CD3 (not shown). The pan-B-cell and follicular dendritic cell marker CD21 labeled cells diffusely in the follicles and neck-region of the PPs, which were areas negative for rPrP (not shown).
Table 2. Primary antibodies used to characterize cells in the present study
Figure 6. In situ co-localization studies of mucosal cell subsets and rPrP. (A) Multicolor immunofluorescence labeling for CD11c (blue), CD205 (green) and Texas red-labeled rPrP (red) demonstrated that rPrP was not present in double labeled CD11c+/CD205+ cells (light blue)(07/F103 30 min). Scale bar, 20 µm. A few CD205 single-labeled cells showed rPrP-labeling (yellow) in the T-cell area (arrows). (B) Despite the presence of many CD11c single labeled cells (green) in lamina propria and the dome area, these cells were not associated with rPrP (red) in any of the intestinal compartments studied. rPrP was not observed in the FAE, dome or PP follicle. (blue, hoechst nuclear staining) (09/F607 30 min). Scale bar, 50 µm. (C) Villus. The specific DC marker CD209 (green) did not show co-localization with rPrP(red) (07/F103 30 min). Scale bar, 5 µm. (D-F) Villus. Paired immunofluorescence. CD205+ cells (E, green) were found to contain rPrP (F, red). (D) The merging of panels E and F demonstrated the co-localization (yellow) of rPrP with CD205+ cells (09/F607 30 min). Scale bar, 10 µm. (G-I) T-cell area. Paired immunofluorescence. CD205+ cells (H, green) and rPrP (I, red). (G) The merging of panels H and I demonstrated the co-localization (yellow) of rPrP in a few CD205+ cells in the T-cell area (arrows) and submucosal lymphatics (arrowhead) (07/F103 30 min). Scale bar, 10 µm. (J-L) Villus. Paired immunofluorescence. MHCII+ cells (K, green) and rPrP (L, red). (J) The merging of panels K and L demonstrated rPrP in MHCII+ cells (yellow, arrows) (09/F607 30 min). Scale bar 5 µm. (ae) AE, (lp) lamina propria, (l) lacteal, (d) dome, (f) follicle, (t) T-cell area, (s) submucosal lymphatics, dashed lines indicate the basal lamina between the AE and the lamina propria.
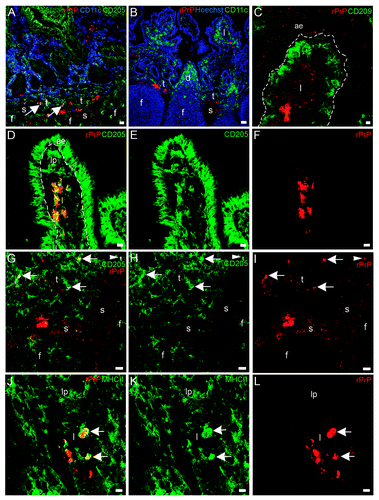
Figure 7. In situ co-localization studies of mucosal cell subsets and rPrP. (A-F) Paired immunofluorescence. CD11b+ cells (B and E, green) were found to contain rPrP (C and F, red). (A) The merging of panels B and C demonstrated the presence of rPrP in CD11b+ cells in the villus (yellow, arrows); and (D) the merging of panels E and F demonstrated co-localization in a T-cell area (arrow). Note that the FAE, dome and follicle are rPrP-negative (A-C 07/F103 30 min and D-F 09/F607 30 min). Scale bars, 10 µm (A-C) and 20 µm (D-F). (G-I) Lamina propria and dome. Paired immunofluorescence. CD68+ cells (H, green) and rPrP (I, red). (G) The merging of panels H and I demonstrates co-localization (yellow, arrows) in lamina propria, note rPrP positive material in the lumen covering FAE, but no rPrP in FAE or in the dome (09/F607 30 min). The basal lamina between the AE and the lamina propria is delineated. Scale bar, 10 µm. (ae) AE (fae) FAE, (d) dome, (f) follicle, (t) T-cell area, (s) submucosal lymphatics.
Studying the control tissues by using the fluorescence and confocal microscope, loops given NaCl or PBS were all negative when scanned with the different lasers and filters (not shown). Intestinal tissues inoculated with Texas red labeled rPrP material showed red fluorescence when scanned with the 543 nm wavelength laser corresponding to the Texas red fluorochrome, and no signal when scanned with the 488 nm wavelength laser.
Discussion
The pattern of uptake of the rPrP aggregates was similar to that observed previously for PrPd from crude brain homogenates of confirmed classical scrapie cases.Citation23 As found with PrPd, rPrP was localized to the villi and submucosa and not found in the dome or lymphoid follicles of the PPs. These findings strongly suggest that the trans-mucosal uptake of rPrP is biologically relevant for the uptake of prions. Bacterial inclusion bodies consist of protein aggregates that, despite their amorphous appearance, can have structural and functional similarities to amyloid.Citation29 These protein aggregates can readily be prepared to a purity of about 90% and allow incorporation of various molecular tags, such as peptide tags embedded in the protein structure or e.g., fluorochromes covalently attached to the protein aggregates. Thus, such relatively homogenous protein aggregates may serve as versatile substitutes for prions, which are normally only available in crude and ill-defined preparations but where the most infectious forms also consists of PrP aggregates.Citation30 Here, we employed bacterially produced ovine prion protein carrying an N-terminal His10 tag, which was further labeled with the fluorescent tag Texas Red to facilitate fluorescent co-localization studies. The present study showed that labeling rPrP with fluorescent Texas red did not alter the distribution of the protein in the gut. The Texas red labeled rPrP was used further in immunofluorescence co-localization studies to phenotype cells involved in the transport of PrP aggregates across the intestinal mucosa.
The rPrP was acquired by antigen-presenting cells (MHCII+ cells) and was not taken up by cells bearing T cell (CD3) or B cell and follicular dendritic cell (CD21) markers. If B cells are excluded from the mucosal transport of prion protein, the remaining professional antigen presenting cells can be broadly divided into DCs and macrophages (mononuclear phagocytes). To distinguish between these two cell populations, various molecular markers were used that had previously been found to be expressed on DCs and macrophages in the intestinal mucosa of sheep.Citation26 The labeled rPrP was found to co-localize with the typical macrophage marker CD68 but not with the classical DC markers; CD11c and CD209. Furthermore, there was a clear localization of the labeled rPrP in cells expressing CD205 and in cells expressing CD11b. Åkesson and coworkersCitation26 identified a major DC population in the sheep intestine as bearing CD11c/CD205/MHCII. However, in the present study, rPrP did not co-localize with CD11c+/CD205+ cells. The phenotype of antigen presenting cells bearing rPrP that emerges from the present study is thus more consistent with that of macrophages or immature DCs rather than of classical DCs. It should be stated that the division between DCs and macrophages is becoming less clear. HumeCitation31 went so far as to consider DCs as macrophages that display a particular state of activation. This assertion was an acknowledgment of the mounting evidence that DCs and macrophages share many phenotypic and functional characteristics, in addition to a common bone marrow progenitor.Citation32 Given the absence of exclusive cell markers, it is therefore important to interpret the function of antigen presenting cells in the intestine on the basis of anatomical location and the activation status of the intestinal immune system, in addition to the expression of accessory molecules.
rPrP was associated with cells expressing the CD68 molecule, a marker of lysosomes useful to identify monocytes and macrophages.Citation33 Macrophages ingest excess PrPd in earlyCitation4 and late stages of disease. Both in murine scrapieCitation10 and sheep scrapie,Citation34 PrPd accumulates in lysosomes. In vitro studies using scrapie exposed macrophages have shown that these cells could restrict the amount of agent initiating the infection.Citation35 In addition, depletion of macrophages enhanced the in vivo neosynthesis of infection associated PrP or reduced the elimination of newly synthesized PrPd molecules,Citation36,Citation37 demonstrating the importance of macrophages in reducing infectivity. Nevertheless, macrophages may passively contribute to the propagation of the infectious agent as they may not clear the infection completely or sufficiently quickly.Citation35,Citation38
The distinguishing feature of the majority of antigen presenting cells bearing rPrP was their expression of CD205 or CD11b. CD205 is an integral membrane protein that belongs to the macrophage mannose receptor family of the C-type lectin endocytic receptors and functions to direct antigen to compartments for antigen processing and presentation.Citation39 CD205 was initially described by KraalCitation40 as a receptor specifically labeling nonlymphoid DCs, in addition to thymic cortex epithelia and epithelia of the apical intestinal villi. However later work has found CD205 to be expressed on a wide range of leukocytesCitation41,Citation42 including monocytes and myeloid blood DCs.Citation43 In cattle, CD205 is expressed at high levels on various subpopulations of DCs, but is also present at lower levels on B cells, T cells and macrophages.Citation44 Gliddon and coworkersCitation45 found that large mononuclear cells in afferent lymph draining from the skin of cattle expressed high levels of CD205 and also expressed high levels of MHC II. Interestingly, antigens presented by CD205+ DCs in the absence of inflammatory stimuli have been found to induce tolerance.Citation46,Citation47 Studies targeting antigen to CD205+ antigen presenting cells in the steady (non-inflammatory) state have reported the deletion of T cells and induction of tolerance. When these antigens were delivered along with a pro-inflammatory stimulus that promoted DC maturation, the response was converted to immunity.Citation48 In the present study, the rPrP was transported across the mucosa by CD205+ antigen presenting cells and likely to be delivered to T-cell rich areas under non-inflammatory conditions.
CD11b was also expressed on cells transporting the rPrP across the mucosa in the present study. CD11b is a glycoprotein, and together with CD11c, is a member of the β2-integrin family, which are surface adhesion molecules exclusively expressed on leukocytes. The β2-integrins are important for recruitment and transendothelial migration of leukocytes to inflamed tissues as well as for T-cell activation during antigen presentation.Citation49,Citation50 Despite abundant expression of β2 integrins on antigen presenting cells, their functional relevance for antigen presentation is not completely understood. Studies have shown that, when activated, murine antigen presenting cells can bind to T-cells via CD11b and inhibit T-cell activation.Citation51 CD11c was not shown to have this effect. Intestinal lamina propria CD11b+ /CD11c- antigen presenting cells of mice induced Foxp3+ regulatory T-cell differentiation while neither lamina propria CD11b+/CD11c+ nor CD11b-/CD11c+ cells were capable of inducing this type of T-cell differentiation.Citation52 As the rPrP in our study was acquired by mainly CD11b+ and not CD11c+ cells, it is open to speculation whether the cells transporting rPrP across the intestinal mucosa and to the T cell area of the PPs could induce regulatory T cell responses to the injected prion protein. This speculation suggests that the involvement of T regulatory cells and tolerance in the uptake and persistence of PrPd in animals orally infected with scrapie requires further investigation.
The molecular mechanisms of rPrP uptake from the gut were not addressed in the present study. It was found that the rPrP was in direct contact with the luminal border of both the FAE and the villous AE, and since the antigen was not found to be transported across the FAE, it can be assumed that a receptor-mediated mechanism for uptake of PrPd, as described for many other pathogens,Citation53 is not present in the FAE of sheep. M-cells have been implicated in the uptake of prionsCitation20,Citation54 but the present findings would argue against their involvement. It would appear that prion protein is taken up across the villous epithelium and transported in lymphatics in a cell-free fraction or by antigen presenting cells bearing CD205, CD11b and/or CD68. The major route of entry for pathogenic organisms to the immune system has been stated to be through selective uptake by PP M cells and transport to subepithelial APCs. In this study, the rPrP uptake followed the pattern of uptake demonstrated for other macromolecules and not through the major route for pathogenic organisms. Small quantities of intact macromolecules can cross through intestinal enterocytes in animals and humans,Citation13,Citation55-Citation57 in a process linked to the suppression of the systemic immune response to luminal antigens.Citation58,Citation59 The phenotype of cells transporting rPrP across the intestinal mucosa of sheep suggests that PrPd of scrapie may be exploiting this normal physiological process.
Material and Methods
Animals
Eleven scrapie free lambs of the Norwegian white breeds were used in this study, seven females and four males. The age ranged from 7 to 32 d. To reduce the possibility of stress, the animals were not fasted before surgery, and they were sedated by an intramuscular injection of midazolam 1 mg/kg under transportation, prior to the operation. Anaesthesia was induced by intra muscular injection of ketamine 20 mg/kg mixed with midazolam 1 mg/kg. In addition, the animals were given an initial intravenous injection of the analgesic butorphanol tartrate 0.05 mg/kg. The animals were kept under anesthesia with symptomatic intravenous injections comprising a mixture of ketamine (20 mg) to midazolam (1 mg) throughout the experiment. At the end of the experiment (one hour), the animals were euthanised with an intravenous overdose of pentobarbital followed by exsanguination.
The animals were placed on their left side; a venous catheter was placed in the right jugular vein for intra-venous injections of anesthetics and continuous fluid infusion of ringer’s solution throughout the operation. After sterile preparation, a right flank laparotomy was performed. The animal’s reflexes, temperature, respiratory rate and mucous membranes were continuously monitored. Arterial blood was sampled for blood-gas analysis immediately before exsanguination.
The distal ileum with its continuous PP was divided into 15 cm long loops separated by five to ten cm of intestine between each loop. To reduce the amount of tissue damage and irritation to the serosa elastic drain tubes were used to close the intestinal segments. The loops were inoculated at different time points (10, 30 and 60 min) prior to euthanasia ( and ). After every inoculation, the intestine was replaced into the abdomen and the abdominal wall clamped until the next inoculation or until euthanasia and tissue sampling.
Table 3. Multiple intestinal loops in sheep were inoculated with recombinant ovine prion protein (rPrP) and tissue samples were collected after 10, 30 or 60 min
Ileum containing loops were removed immediately after euthanasia. Tissue specimens were fixed for light and fluorescence microscopy. The formalin fixed samples were embedded in paraffin wax before cutting according to standard procedures for light microscopy. The intestinal tissue samples for cryostat sectioning were placed with the mucosal side onto thin slices of liver before freezing to protect the mucosa from freezing artifacts. The frozen tissues were then stored at -70°C until further preparation.
Procedures have been conducted in accordance with the laws and regulations controlling experiments using live animals in Norway, that is the Animal Welfare Act of 20 December 1974 and the Regulation of Animal Experimentation of 15 January, 1996.
Inoculum
To clone and purify the ovine recPrPHis10 (rPrP), cDNA corresponding to the mature ovine PrP (amino acids 25–231) was amplified using forward primer 5′GGCATTCCATATGAAGAAGCGACCAAAACC and reverse primer ´GCGGATCCTATCATGCCCCCCGTTGGTAAT, containing a NdeI and a BamHI restriction endonuclease cleavage site, respectively. The PrP amplification product was inserted into the NdeI and BamHI sites of the expression vector pET-16b (Novagen, Cat. No. 69662–3) and expressed in E.coli BL21(DE3)pLysS (Novagen, Cat. No. 69451–4) grown in 2x YT-medium (tryptone 16 g/l, yeast extract 10 g/l, NaCl 5g/l, pH 7,0) in the presence of ampicillin (50 mg/ml) and chloramphenicol (25 mg/ml). At OD600 = 0.6–0.9, the expression of rPrP was induced by addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 1 mM. After incubation for another 4 h, cells were pelleted by centrifugation at 8000 rpm for 10 min and stored at -20°C. Two cell pellets derived from a total of 250 ml culture were thawed at 50°C and diluted by addition of 12.5 ml NaCl-Tris buffer (150 mM NaCl, 50 mM Tris pH 8.0), supplemented with lysozyme, MgCl2 and DNaseI to final concentrations of 0.3 mg/ml, 5 mM and 7µg/ml, respectively. The suspensions were stirred with a glass rod until the viscosity disappeared. The inclusion bodies were sedimented at 8000 rpm for 20 min at 4°C, and washed twice in 300 ml NaCl-Tris buffer containing 1% Triton X-100 by magnetic stirring for 1 h. The pellets were solubilised during the washing steps by brief sonications with an analogous water bath sonicator (Transsonic 310). The final pellets containing purified inclusion bodies were resuspended in 12.5 ml 10 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA and stored at -20°C.
Western blot analysis of the rPrP with and without proteinase K was done by leaving aliquots of purified inclusion bodies untreated or subjected to proteinase K (Fluka BioChemika, Cat. No. 82456) treatment (5 µg/ml) at 22°C for up to 10 h. Samples were drawn from the Proteinase K treated vial at different time-intervals during incubation (30 sec, 1, 2, 5, 10, 30, 60, 300 and 600 min) and transferred directly to boiling SDS sample buffer to stop the digestion and prepare for electrophoresis. Proteins were resolved on precast 12% Bis-Tris polyacrylamide gels (BioRad, Cat. No. 345–0119) with 3-(N-morpholino) propanesulfonic acid as running buffer, electroblotted onto polyvinylidene difluoride membranes (Hybond-P, GE Healthcare, Cat. No. RPN2020F) and probed with the monoclonal antibodies: Anti-His (Molecular Probes, Cat. No. P-21315), P4 (R-Biopharm AG, Cat. No. R8007), BAR224 (BioNovus Life Science, Cat. No. SP-A03211), and F99 (VMRD Inc., Cat. No F99/97.6.1) over night at 4°C. Secondary antibodies (goat anti mouse) labeled with alkaline phosphatase were used to visualize bands with a variable mode fluorescence imager (Typhoon 9200, GE Healthcare) after incubation with the ALP substrate (ECFTM, Cat. No. RPN5781).
To label the rPrP with Texas red-X, purified inclusion bodies of rPrP were thawed and solubilised by brief sonication as previously described, followed by centrifugation at 8000 rpm for 20 min at 4°C. The resulting pellets were resuspended in 200 mM NaHCO3 buffer pH 8.3 to reach a final protein concentration of about 2 mg/ml. Texas red-X succiniminyl ester was solubilised by addition of 500 µl of dimetyl sulfoxide to 5 mg of Texas red-X, followed by a brief vortex. The Texas red-X solution, 500µl, was added to 8.5 ml of the rPrP solution and incubated at 18°C for 1 h with gentle stirring. Unbound Texas red-X was removed by centrifugation at 8000 rpm for 20 min. The supernatant was removed and the resulting pellet was resuspended by sonication in 8.5 ml PBS and diluted in 34 ml cell culture medium (DMEM, Cambrex, Cat. No. 15–604D), supplemented with 10% fetal calf serum (Euroclone, Cat. No. ECS 0180L) prior intestinal inoculation, which was performed immediately after preparation.
Histochemistry
Routine histological examination was performed in formalin-fixed and wax embedded tissue samples stained by hematoxylin and eosin.
For immunohistochemical detection of the inoculated rPrP an avidin-biotin complex (Vectastain ABC Kit, Vector Laboratories, Cat. No. PK-6200) procedure was performed. Prior to immunolabelling of the paraffin wax sections of selected tissues, 4 μm thick sections were placed on positively charged slides and dried at 59°C. After a standard dewaxing procedure, sections were autoclaved in citrate buffer (0.01M citric acid monohydrate, pH 6.0) at 120°C. Endogenous peroxidase was inhibited by treatment with H2O2 3% in methanol or water for 20 min. To avoid non-specific binding of the biotinylated antibody, a blocking solution containing either normal horse serum (for the monoclonal antibodies) or normal goat serum (for the polyclonal antibodies) was diluted in bovine serum albumin in Tris-buffered saline (BSA/TBS) or in PBS, and further applied to the sections for 20 min at room temperature. After gentle removal of the blocking solution, sections were incubated with the primary monoclonal antibodies F99, BAR224, L42 (kindly provided by M. H. Groschup), R145 (kindly provided by L. Terry) all diluted in BSA/TBS or in PBS. All sections were incubated in a humid chamber at room temperature (20°C) over night. The next day secondary biotinylated antibody (anti mouse or anti rat) was applied to the sections and incubated for 60 min. The sections were incubated with the ABC-horseradish peroxidase complex solution. Peroxidase activity was visualized by incubation with a solution consisting of 4 mg 3-amino-9-ethylcarbazole, 800 μL N,N-dimethylformamide, 14 mL 0.1 M acetate buffer, pH 5.2, and 150 μL 3% H2O2 or by adding 2 ml 3′3 diaminobenzidine tetrachloride to 200 mL PBS with tween (PBST) and 200 μL hydrogen peroxide to the sections for approximately ten minutes. The reaction was stopped by rinsing in PBST. To enhance the diaminobenzidine tetrachloride reaction 0.5% copper sulfate solution was added for 3 min. Contrast staining was performed with Mayers hematoxylin to all sections before applying coverslips. Rinsing between each step was done in H2O or in PBST.
For immunofluorescence labeling 7–14 μm thick cryo sections of intestinal tissue were mounted onto poly-lysine-coated slides and stored at -70°C before use. When ready for labeling, the sections were air-dried at room temperature for one hour, fixed in acetone for ten minutes, and then air-dried for another ten minutes. The sections were rinsed and rehydrated in PBS. 20% BSA/TBS was applied to block non-specific binding. The blocking solution was tapped off and primary antibodies (see ) were diluted in 1% BSA/TBS and applied to the sections for 1 h at room temperature. Alexa fluorescent secondary antibodies against rabbit IgG and/or against various mouse immunoglobulin isotypes corresponding to the primary antibodies were diluted in 1% BSA/TBS and centrifuged at 1400 rpm for 5 min before application of the supernatants to the sections for 1 h. Hoechst (Molecular Probes, Cat. No. H1399) were applied to a selection of sections to visualize cell nuclei. The slides were mounted in polyvinyl alcohol at pH 8. The sections were then examined in a confocal microscope equipped with an LSM 510 laser confocal unit (Carl Zeiss, Jena, Germany) or sections were examined in the Leica DM RXA microscope (Leica Microsytems) equipped with fluorescence filters which suited the excitation and emission spectrum of the specimens. The emitted signal was recorded in separate monochrome digital images, one for each fluorochrome.
Tissue sections labeled by immunohistochemical procedures were examined in a Leica DM 2000 microscope (Leica Microsytems) and digital images recorded with a Leica EC3 digital camera (Leica Microsytems), or examined in the Leica DM RXA microscope equipped with fluorescence filters, and digital images recorded using a Spot RX slider digital camera (Diagnostic instruments, Inc.). To control for non-specific binding all runs included a control section where the primary antibodies were replaced by 1% BSA/TBS. Tissue sections from loops inoculated with NaCl or PBS were used as negative controls. To control for the possibility of Texas red labeling affecting the intestinal uptake of rPrP, loops were inoculated with labeled and unlabelled rPrP and the two were compared. To confirm that the Texas red fluorochrome was co-localized with rPrP during uptake, tissue sections from loops inoculated with Texas red labeled rPrP were subjected to immunohistochemical detection for rPrP and studied by light and fluorescence microscopy by using the Leica DM RXA microscope with and without fluorescence filters. The co-localization of the PrP-labeling visualized in the normal light microscope and the Texas red fluorescence visualized by using fluorescence excitation light 594 was documented. Sections fluorescently labeled with antibodies against cell markers were studied in fluorescent filters that did not correspond with the relevant fluorochrome, to control for and exclude autofluorescence and bleed through.
For EM, samples of distal ileum containing PPs were immediately removed at necropsy and 1 mm wide strips across the PP were fixed in 0.5% glutaraldehyde and 4% paraformaldehyde and post fixed in 1% osmium tetroxide. The strips of PPs were further sectioned to 1 mm cubes and routinely processed to araldite (Taab laboratories, Cat. No. E009/1). Resin embedded sections were cut at 1 µm and stained with toluidine blue or labeled with the PrP antibodies 1A8,Citation60 or 523.7 (J. Langeveld, Lelystad, Netherlands)Citation61,Citation62 as previously described. Resin blocks containing representation of follicles, domes and villi from each loop of each sheep were then sectioned at 60 nm and routinely stained using uranyl acetate and lead citrate. Sections were then immunolabelled for PrPd using the above antibodies by immunogold methods as previously described.Citation61 Positive control material used in immunogold EM included brain from a confirmed scrapie-positive animal containing abundant PrPd. Samples of the prepared inoculum containing rPrP with and without conjugation to Texas red were prepared for electron microscopy by adsorbing the protein in suspension to Formvar-coated 300-mesh copper grids for 5 min. Grids were then negatively stained with 2% uranyl acetate for 5 min, washed and allowed to dry overnight before viewing.
| Abbreviations: | ||
| TSE | = | transmissible spongiform encephalopathy |
| PrPd | = | disease-associated prion protein |
| rPrP | = | recombinant prion protein |
| PP | = | Peyer’s patch |
| FAE | = | follicle associated epithelium |
| AE | = | absorptive epithelium |
| DC | = | dendritic cell |
| MHC II | = | Major histocompatibility complex II |
| ALP | = | Alkaline phosphatase |
| SDS | = | Sodium-dodecyl-Sulphate |
| PBS | = | phosphate buffered saline |
| PBST | = | PBS with tween |
| BSA | = | bovine serum albumin |
| TBS | = | tris-buffered saline |
| EM | = | electron microscopy |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors wish to thank Henning Mørch, Laila Aune, Inger Rudshaug, Lene Hermansen, Gillian McGovern, Callum Donnelly, Susan Skogtvedt, and Berit Christophersen for invaluable technical assistance.
References
- Hadlow WJ, Kennedy RC, Race RE. Natural infection of Suffolk sheep with scrapie virus. J Infect Dis 1982; 146:657 - 64; http://dx.doi.org/10.1093/infdis/146.5.657; PMID: 6813384
- Sigurdson CJ, Williams ES, Miller MW, Spraker TR, O’Rourke KI, Hoover EA. Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). J Gen Virol 1999; 80:2757 - 64; PMID: 10573172
- Andréoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, van Keulen LJ, et al. Early accumulation of PrP(Sc) in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J Gen Virol 2000; 81:3115 - 26; PMID: 11086143
- van Keulen LJ, Vromans ME, van Zijderveld FG. Early and late pathogenesis of natural scrapie infection in sheep. APMIS 2002; 110:23 - 32; http://dx.doi.org/10.1034/j.1600-0463.2002.100104.x; PMID: 12064252
- Ryder SJ, Dexter GE, Heasman L, Warner R, Moore SJ. Accumulation and dissemination of prion protein in experimental sheep scrapie in the natural host. BMC Vet Res 2009; 5:9; http://dx.doi.org/10.1186/1746-6148-5-9; PMID: 19243608
- Beekes M, McBride PA. Early accumulation of pathological PrP in the enteric nervous system and gut-associated lymphoid tissue of hamsters orally infected with scrapie. Neurosci Lett 2000; 278:181 - 4; http://dx.doi.org/10.1016/S0304-3940(99)00934-9; PMID: 10653023
- Kimberlin RH, Walker CA. Pathogenesis of experimental scrapie. Ciba Found Symp 1988; 135:37 - 62; PMID: 3137002
- McBride PA, Eikelenboom P, Kraal G, Fraser H, Bruce ME. PrP protein is associated with follicular dendritic cells of spleens and lymph nodes in uninfected and scrapie-infected mice. J Pathol 1992; 168:413 - 8; http://dx.doi.org/10.1002/path.1711680412; PMID: 1362440
- van Keulen LJ, Schreuder BE, Meloen RH, Mooij-Harkes G, Vromans ME, Langeveld JP. Immunohistochemical detection of prion protein in lymphoid tissues of sheep with natural scrapie. J Clin Microbiol 1996; 34:1228 - 31; PMID: 8727908
- Jeffrey M, McGovern G, Goodsir CM, Brown KL, Bruce ME. Sites of prion protein accumulation in scrapie-infected mouse spleen revealed by immuno-electron microscopy. J Pathol 2000; 191:323 - 32; http://dx.doi.org/10.1002/1096-9896(200007)191:3<323::AID-PATH629>3.0.CO;2-Z; PMID: 10878556
- Jeffrey M, McGovern G, Martin S, Goodsir CM, Brown KL. Cellular and sub-cellular localisation of PrP in the lymphoreticular system of mice and sheep. Arch Virol Suppl 2000; 23 - 38; PMID: 11214927
- Press CM, Heggebø R, Espenes A. Involvement of gut-associated lymphoid tissue of ruminants in the spread of transmissible spongiform encephalopathies. Adv Drug Deliv Rev 2004; 56:885 - 99; http://dx.doi.org/10.1016/j.addr.2003.09.008; PMID: 15063596
- Gardner ML. Gastrointestinal absorption of intact proteins. Annu Rev Nutr 1988; 8:329 - 50; http://dx.doi.org/10.1146/annurev.nu.08.070188.001553; PMID: 3060169
- Hazzard RA, Hodges GM, Scott JD, McGuinness CB, Carr KE. Early intestinal microparticle uptake in the rat. J Anat 1996; 189:265 - 71; PMID: 8886948
- Doyle-McCullough M, Smyth SH, Moyes SM, Carr KE. Factors influencing intestinal microparticle uptake in vivo. Int J Pharm 2007; 335:79 - 89; http://dx.doi.org/10.1016/j.ijpharm.2006.10.043; PMID: 17197140
- Hodges GM, Carr EA, Hazzard RA, Carr KE. Uptake and translocation of microparticles in small intestine. Morphology and quantification of particle distribution. Dig Dis Sci 1995; 40:967 - 75; http://dx.doi.org/10.1007/BF02064184; PMID: 7729286
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2001; 2:361 - 7; http://dx.doi.org/10.1038/86373; PMID: 11276208
- Owen RL. Sequential uptake of horseradish peroxidase by lymphoid follicle epithelium of Peyer’s patches in the normal unobstructed mouse intestine: an ultrastructural study. Gastroenterology 1977; 72:440 - 51; PMID: 832793
- Neutra MR. Current concepts in mucosal immunity. V Role of M cells in transepithelial transport of antigens and pathogens to the mucosal immune system. Am J Physiol 1998; 274:G785 - 91; PMID: 9612256
- Heppner FL, Christ AD, Klein MA, Prinz M, Fried M, Kraehenbuhl JP, et al. Transepithelial prion transport by M cells. Nat Med 2001; 7:976 - 7; http://dx.doi.org/10.1038/nm0901-976; PMID: 11533681
- Ghosh S. Mechanism of intestinal entry of infectious prion protein in the pathogenesis of variant Creutzfeldt-Jakob disease. Adv Drug Deliv Rev 2004; 56:915 - 20; http://dx.doi.org/10.1016/j.addr.2003.10.035; PMID: 15063598
- Mishra RS, Basu S, Gu Y, Luo X, Zou WQ, Mishra R, et al. Protease-resistant human prion protein and ferritin are cotransported across Caco-2 epithelial cells: implications for species barrier in prion uptake from the intestine. J Neurosci 2004; 24:11280 - 90; http://dx.doi.org/10.1523/JNEUROSCI.2864-04.2004; PMID: 15601934
- Jeffrey M, González L, Espenes A, Press CM, Martin S, Chaplin M, et al. Transportation of prion protein across the intestinal mucosa of scrapie-susceptible and scrapie-resistant sheep. J Pathol 2006; 209:4 - 14; http://dx.doi.org/10.1002/path.1962; PMID: 16575799
- Maric I, Holt PG, Perdue MH, Bienenstock J. Class II MHC antigen (Ia)-bearing dendritic cells in the epithelium of the rat intestine. J Immunol 1996; 156:1408 - 14; PMID: 8568241
- Kelsall BL, Strober W. Distinct populations of dendritic cells are present in the subepithelial dome and T cell regions of the murine Peyer’s patch. J Exp Med 1996; 183:237 - 47; http://dx.doi.org/10.1084/jem.183.1.237; PMID: 8551227
- Akesson CP, McL Press C, Espenes A, Aleksandersen M. Phenotypic characterisation of intestinal dendritic cells in sheep. Dev Comp Immunol 2008; 32:837 - 49; http://dx.doi.org/10.1016/j.dci.2007.12.004; PMID: 18222542
- Huang FP, Farquhar CF, Mabbott NA, Bruce ME, MacPherson GG. Migrating intestinal dendritic cells transport PrP(Sc) from the gut. J Gen Virol 2002; 83:267 - 71; PMID: 11752724
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136 - 44; http://dx.doi.org/10.1126/science.6801762; PMID: 6801762
- Wang L. Towards revealing the structure of bacterial inclusion bodies. Prion 2009; 3:139 - 45; http://dx.doi.org/10.4161/pri.3.3.9922; PMID: 19806034
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature 2005; 437:257 - 61; http://dx.doi.org/10.1038/nature03989; PMID: 16148934
- Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol 2008; 181:5829 - 35; PMID: 18941170
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science 2010; 327:656 - 61; http://dx.doi.org/10.1126/science.1178331; PMID: 20133564
- Ackermann MR, DeBey BM, Stabel TJ, Gold JH, Register KB, Meehan JT. Distribution of anti-CD68 (EBM11) immunoreactivity in formalin-fixed, paraffin-embedded bovine tissues. Vet Pathol 1994; 31:340 - 8; http://dx.doi.org/10.1177/030098589403100307; PMID: 8053129
- McGovern G, Jeffrey M. Scrapie-specific pathology of sheep lymphoid tissues. PLoS One 2007; 2:e1304; http://dx.doi.org/10.1371/journal.pone.0001304; PMID: 18074028
- Carp RI, Callahan SM. Effect of mouse peritoneal macrophages on scrapie infectivity during extended in vitro incubation. Intervirology 1982; 17:201 - 7; http://dx.doi.org/10.1159/000149289; PMID: 6813286
- Beringue V, Demoy M, Lasmézas CI, Gouritin B, Weingarten C, Deslys JP, et al. Role of spleen macrophages in the clearance of scrapie agent early in pathogenesis. J Pathol 2000; 190:495 - 502; http://dx.doi.org/10.1002/(SICI)1096-9896(200003)190:4<495::AID-PATH535>3.0.CO;2-T; PMID: 10700001
- Maignien T, Shakweh M, Calvo P, Marcé D, Salès N, Fattal E, et al. Role of gut macrophages in mice orally contaminated with scrapie or BSE. Int J Pharm 2005; 298:293 - 304; http://dx.doi.org/10.1016/j.ijpharm.2005.02.042; PMID: 15964722
- Sassa Y, Inoshima Y, Ishiguro N. Bovine macrophage degradation of scrapie and BSE PrPSc. Vet Immunol Immunopathol 2010; 133:33 - 9; http://dx.doi.org/10.1016/j.vetimm.2009.06.018; PMID: 19647878
- Jiang W, Swiggard WJ, Heufler C, Peng M, Mirza A, Steinman RM, et al. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature 1995; 375:151 - 5; http://dx.doi.org/10.1038/375151a0; PMID: 7753172
- Kraal G, Breel M, Janse M, Bruin G. Langerhans’ cells, veiled cells, and interdigitating cells in the mouse recognized by a monoclonal antibody. J Exp Med 1986; 163:981 - 97; http://dx.doi.org/10.1084/jem.163.4.981; PMID: 3950549
- Guo M, Gong S, Maric S, Misulovin Z, Pack M, Mahnke K, et al. A monoclonal antibody to the DEC-205 endocytosis receptor on human dendritic cells. Hum Immunol 2000; 61:729 - 38; http://dx.doi.org/10.1016/S0198-8859(00)00144-0; PMID: 10980384
- Witmer-Pack MD, Swiggard WJ, Mirza A, Inaba K, Steinman RM. Tissue distribution of the DEC-205 protein that is detected by the monoclonal antibody NLDC-145. II. Expression in situ in lymphoid and nonlymphoid tissues. Cell Immunol 1995; 163:157 - 62; http://dx.doi.org/10.1006/cimm.1995.1110; PMID: 7758126
- Kato M, McDonald KJ, Khan S, Ross IL, Vuckovic S, Chen K, et al. Expression of human DEC-205 (CD205) multilectin receptor on leukocytes. Int Immunol 2006; 18:857 - 69; http://dx.doi.org/10.1093/intimm/dxl022; PMID: 16581822
- Gliddon DR, Hope JC, Brooke GP, Howard CJ. DEC-205 expression on migrating dendritic cells in afferent lymph. Immunology 2004; 111:262 - 72; http://dx.doi.org/10.1111/j.0019-2805.2004.01820.x; PMID: 15009426
- Carreno-Gómez B, Woodley JF, Florence AT. Studies on the uptake of tomato lectin nanoparticles in everted gut sacs. Int J Pharm 1999; 183:7 - 11; http://dx.doi.org/10.1016/S0378-5173(99)00050-2; PMID: 10361144
- Hawiger D, Inaba K, Dorsett Y, Guo M, Mahnke K, Rivera M, et al. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J Exp Med 2001; 194:769 - 79; http://dx.doi.org/10.1084/jem.194.6.769; PMID: 11560993
- Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, Steinman RM. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J Exp Med 2002; 196:1627 - 38; http://dx.doi.org/10.1084/jem.20021598; PMID: 12486105
- Steinman RM, Hawiger D, Liu K, Bonifaz L, Bonnyay D, Mahnke K, et al. Dendritic cell function in vivo during the steady state: a role in peripheral tolerance. Ann N Y Acad Sci 2003; 987:15 - 25; http://dx.doi.org/10.1111/j.1749-6632.2003.tb06029.x; PMID: 12727620
- Hogg N, Takacs L, Palmer DG, Selvendran Y, Allen C. The p150,95 molecule is a marker of human mononuclear phagocytes: comparison with expression of class II molecules. Eur J Immunol 1986; 16:240 - 8; http://dx.doi.org/10.1002/eji.1830160306; PMID: 3456894
- Anderson DC, Miller LJ, Schmalstieg FC, Rothlein R, Springer TA. Contributions of the Mac-1 glycoprotein family to adherence-dependent granulocyte functions: structure-function assessments employing subunit-specific monoclonal antibodies. J Immunol 1986; 137:15 - 27; PMID: 3519773
- Varga G, Balkow S, Wild MK, Stadtbaeumer A, Krummen M, Rothoeft T, et al. Active MAC-1 (CD11b/CD18) on DCs inhibits full T-cell activation. Blood 2007; 109:661 - 9; http://dx.doi.org/10.1182/blood-2005-12-023044; PMID: 17003381
- Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol 2007; 8:1086 - 94; http://dx.doi.org/10.1038/ni1511; PMID: 17873879
- Kyd JM, Cripps AW. Functional differences between M cells and enterocytes in sampling luminal antigens. Vaccine 2008; 26:6221 - 4; http://dx.doi.org/10.1016/j.vaccine.2008.09.061; PMID: 18852006
- Prinz M, Huber G, Macpherson AJ, Heppner FL, Glatzel M, Eugster HP, et al. Oral prion infection requires normal numbers of Peyer’s patches but not of enteric lymphocytes. Am J Pathol 2003; 162:1103 - 11; http://dx.doi.org/10.1016/S0002-9440(10)63907-7; PMID: 12651603
- Walker WA, Cornell R, Davenport LM, Isselbacher KJ. Macromolecular absorption. Mechanism of horseradish peroxidase uptake and transport in adult and neonatal rat intestine. J Cell Biol 1972; 54:195 - 205; http://dx.doi.org/10.1083/jcb.54.2.195; PMID: 5040858
- Limpanussorn J, Simon L, Dayan AD. Transepithelial transport of large particles in rat: a new model for the quantitative study of particle uptake. J Pharm Pharmacol 1998; 50:753 - 60; http://dx.doi.org/10.1111/j.2042-7158.1998.tb07136.x; PMID: 9720624
- Warshaw AL, Walker WA, Cornell R, Isselbacher KJ. Small intestinal permeability to macromolecules. Transmission of horseradish peroxidase into mesenteric lymph and portal blood. Lab Invest 1971; 25:675 - 84; PMID: 5129766
- Swarbrick ET, Stokes CR, Soothill JF. Absorption of antigens after oral immunisation and the simultaneous induction of specific systemic tolerance. Gut 1979; 20:121 - 5; http://dx.doi.org/10.1136/gut.20.2.121; PMID: 428823
- Stokes CR, Swarbrick ET, Soothill JF. Genetic differences in immune exclusion and partial tolerance to ingested antigens. Clin Exp Immunol 1983; 52:678 - 84; PMID: 6872322
- Landsverk T, Halleraker M, Aleksandersen M, McClure S, Hein W, Nicander L. The intestinal habitat for organized lymphoid tissues in ruminants; comparative aspects of structure, function and development. Vet Immunol Immunopathol 1991; 28:1 - 16; http://dx.doi.org/10.1016/0165-2427(91)90038-E; PMID: 1905075
- McGovern G, Brown KL, Bruce ME, Jeffrey M. Murine scrapie infection causes an abnormal germinal centre reaction in the spleen. J Comp Pathol 2004; 130:181 - 94; http://dx.doi.org/10.1016/j.jcpa.2003.11.001; PMID: 15003476
- Jeffrey M, McGovern G, Goodsir CM, Síso S, González L. Strain-associated variations in abnormal PrP trafficking of sheep scrapie. Brain Pathol 2009; 19:1 - 11; http://dx.doi.org/10.1111/j.1750-3639.2008.00150.x; PMID: 18400047
- Puri NK, Brandon MR. Sheep MHC class II molecules. II. Identification and characterization of four distinct subsets of sheep MHC class II molecules. Immunology 1987; 62:575 - 80; PMID: 3480873
- Howard CJ, Morrison WI, Bensaid A, Davis W, Eskra L, Gerdes J, et al. Summary of workshop findings for leukocyte antigens of cattle. Vet Immunol Immunopathol 1991; 27:21 - 7; http://dx.doi.org/10.1016/0165-2427(91)90072-K; PMID: 1902342
- Gupta VK, McConnell I, Hopkins J. Reactivity of the CD11/CD18 workshop monoclonal antibodies in the sheep. Vet Immunol Immunopathol 1993; 39:93 - 102; http://dx.doi.org/10.1016/0165-2427(93)90168-4; PMID: 8310662
- Bielefeldt-Ohmann H, Sabara M, Lawman MJ, Griebel P, Babiuk LA. A monoclonal antibody detects macrophage maturation antigen which appears independently of class II antigen expression. Reactivity of monoclonal EBM11 with bovine macrophages. J Immunol 1988; 140:2201 - 9; PMID: 3280679
- Geijtenbeek TB, Engering A, Van Kooyk Y. DC-SIGN, a C-type lectin on dendritic cells that unveils many aspects of dendritic cell biology. J Leukoc Biol 2002; 71:921 - 31; PMID: 12050176
- Jones M, Cordell JL, Beyers AD, Tse AG, Mason DY. Detection of T and B cells in many animal species using cross-reactive anti-peptide antibodies. J Immunol 1993; 150:5429 - 35; PMID: 8515069
- Hein WR, Dudler L, Mackay CR. Surface expression of differentiation antigens on lymphocytes in the ileal and jejunal Peyer’s patches of lambs. Immunology 1989; 68:365 - 70; PMID: 2592011